Potential of Cheese-Associated Lactic Acid Bacteria to Metabolize Citrate and Produce Organic Acids and Acetoin
Abstract
:1. Introduction
2. Materials and Methods
2.1. LAB Strains
2.2. Growth Evaluation of the LAB Strains
2.3. Citrate Utilization by the LAB Strains
2.4. LAB Strains Proteolytic Activity
2.5. Inoculum Preparation and Fermentation
2.5.1. Determination of pH Values
2.5.2. Analysis of Organic Acids, Acetoin, and Diacetyl
2.5.3. Principal Component Analysis
3. Results
3.1. LAB Growth, Citrate Fermentation, and Production of Extracellular Protease
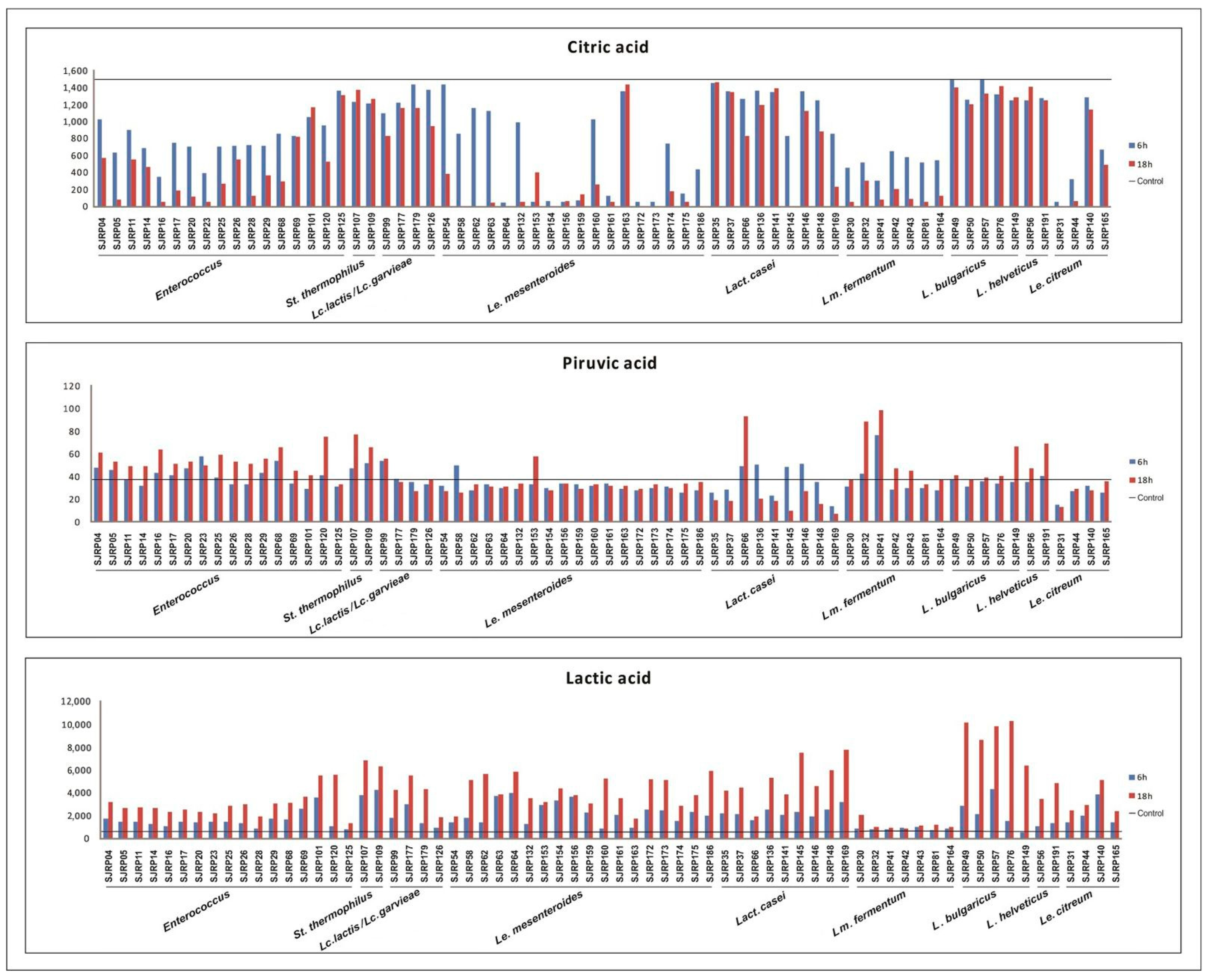
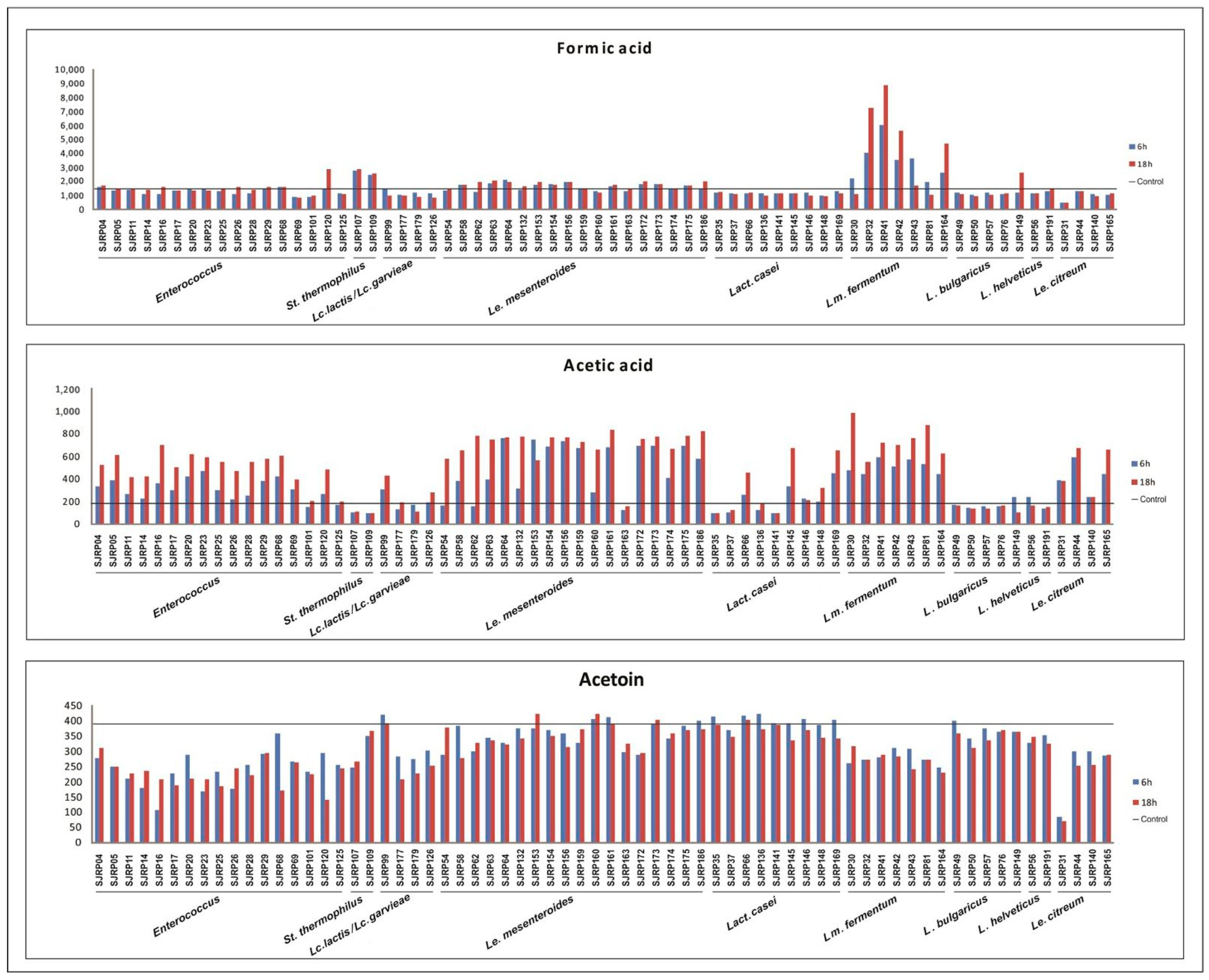
3.2. Reduction in pH and Production of Organic Compounds
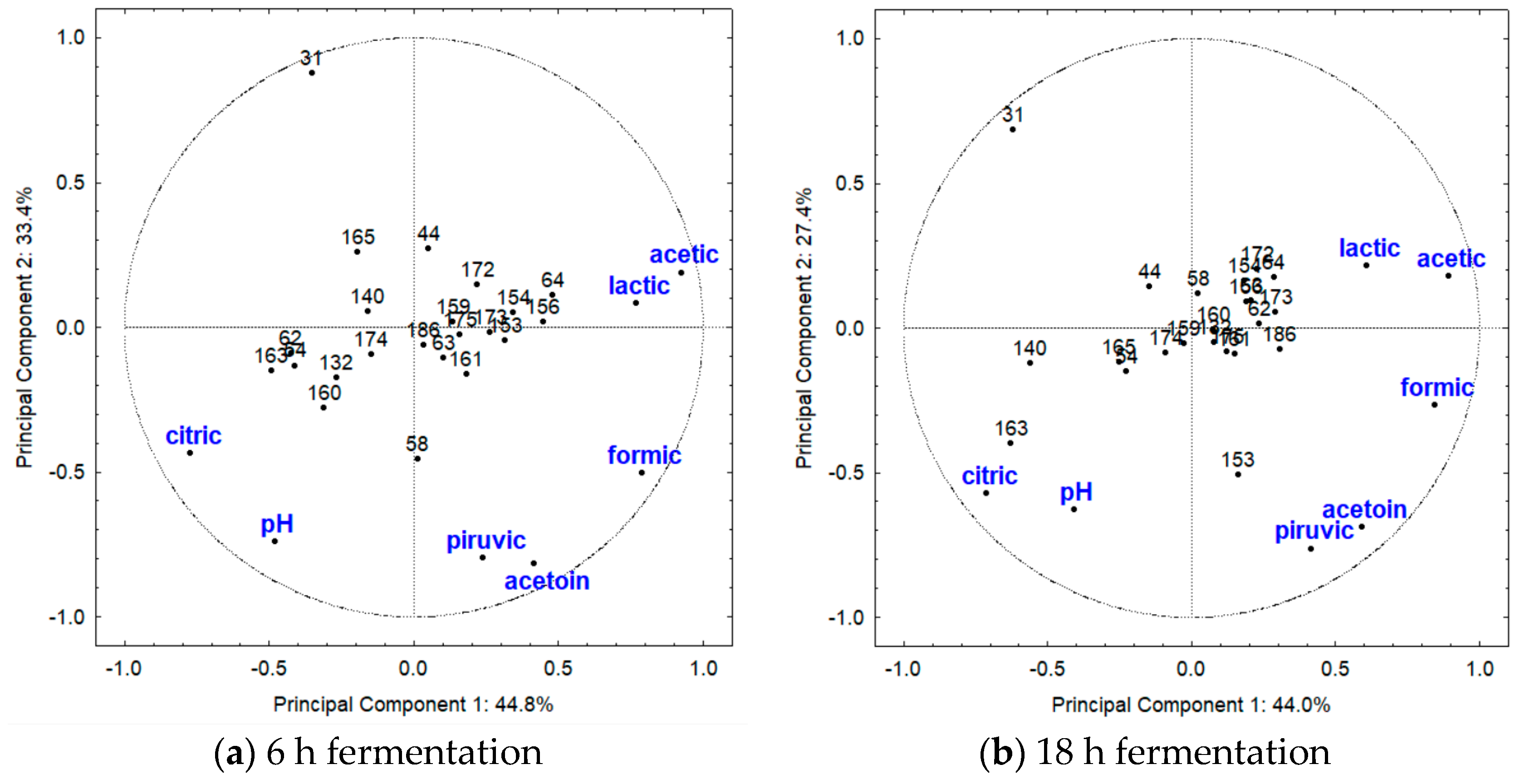
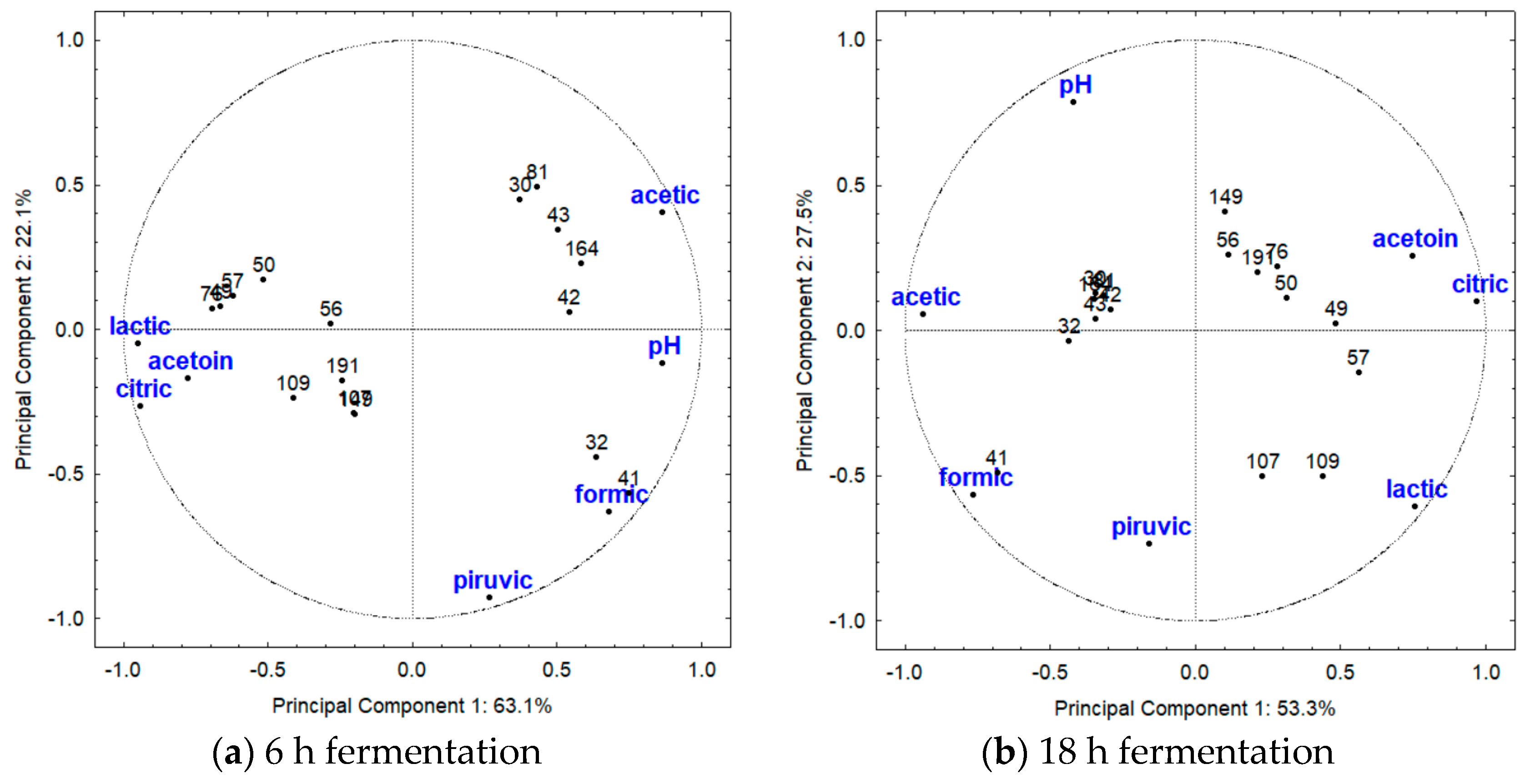
3.3. Principal Component Analysis (PCA)
4. Discussion
4.1. LAB Growth, Citrate Fermentation, and Production of Extracellular Protease
4.2. Reduction in pH and Production of Organic Compounds and Acetoin
4.3. Principal Component Analysis (PCA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; da Silva, R.C.; Ibrahim, S.A. Lactic acid bacteria: Food safety and human health applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Plavec, T.V.; Berlec, A. Safety aspects of genetically modified lactic acid bacteria. Microorganisms 2020, 8, 297. [Google Scholar] [CrossRef] [PubMed]
- Suissa, R.; Oved, R.; Jankelowitz, G.; Turjeman, S.; Koren, O.; Kolodkin-Gal, I. Molecular genetics for probiotic engineering: Dissecting lactic acid bacteria. Trends Microbiol. 2022, 30, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Pang, H.; Zhang, H.; Cai, Y. Biodiversity of lactic acid bacteria. In Lactic Acid Bacteria; Zhang, H., Cai, Y., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 103–203. ISBN 978-94-017-8841-0. [Google Scholar]
- Buron-Moles, G.; Chailyan, A.; Dolejs, I.; Forster, J.; Mikš, M.H. Uncovering carbohydrate metabolism through a genotype-phenotype association study of 56 lactic acid bacteria genomes. Appl. Microbiol. Biotechnol. 2019, 103, 3135–3152. [Google Scholar] [CrossRef] [PubMed]
- Punia Bangar, S.; Suri, S.; Trif, M.; Ozogul, F. Organic acids production from lactic acid bacteria: A preservation approach. Food Biosci. 2022, 46, 101615. [Google Scholar] [CrossRef]
- Pretorius, N.; Engelbrecht, L.; Du Toit, M. Influence of sugars and ph on the citrate metabolism of different lactic acid bacteria strains in a synthetic wine matrix. J. Appl. Microbiol. 2019, 127, 1490–1500. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Özcan, E.; Selvi, S.S.; Nikerel, E.; Teusink, B.; Toksoy Öner, E.; Çakır, T. A Genome-scale metabolic network of the aroma bacterium Leuconostoc mesenteroides subsp. cremoris. Appl. Microbiol. Biotechnol. 2019, 103, 3153–3165. [Google Scholar] [CrossRef] [PubMed]
- Smid, E.J.; Kleerebezem, M. Production of aroma compounds in lactic fermentations. Annu. Rev. Food Sci. Technol. 2014, 5, 313–326. [Google Scholar] [CrossRef]
- Sharma, H.; Ozogul, F.; Bartkiene, E.; Rocha, J.M. Impact of lactic acid bacteria and their metabolites on the techno-functional properties and health benefits of fermented dairy products. Crit. Rev. Food Sci. Nutr. 2021, 63, 4819–4841. [Google Scholar] [CrossRef]
- Giacon, T.G.; de Gois e Cunha, G.C.; Eliodório, K.P.; Oliveira, R.P.d.S.; Basso, T.O. Homo- and heterofermentative lactobacilli are distinctly affected by furanic compounds. Biotechnol. Lett. 2022, 44, 1431–1445. [Google Scholar] [CrossRef] [PubMed]
- Bansal, V.; Veena, N. Understanding the role of ph in cheese manufacturing: General aspects of cheese quality and safety. J. Food Sci. Technol. 2022, 1–11. [Google Scholar] [CrossRef]
- Borges, D.O.; Matsuo, M.M.; Bogsan, C.S.B.; da Silva, T.F.; Casarotti, S.N.; Penna, A.L.B. Leuconostoc mesenteroides subsp. mesenteroides SJRP55 reduces listeria monocytogenes growth and impacts on fatty acids profile and conjugated linoleic acid content in fermented cream. LWT 2019, 107, 264–271. [Google Scholar] [CrossRef]
- Possas, A.; Bonilla-Luque, O.M.; Valero, A. From cheese-making to consumption: Exploring the microbial safety of cheeses through predictive microbiology models. Foods 2021, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xing, S.; He, L.; Li, C.; Wang, X.; Zeng, X.; Dai, Y. Characterization, high-density fermentation, and the production of a directed vat set starter of lactobacilli used in the food industry: A Review. Foods 2022, 11, 3063. [Google Scholar] [CrossRef] [PubMed]
- Battelli, G.; Scano, P.; Albano, C.; Cagliani, L.R.; Brasca, M.; Consonni, R. Modifications of the volatile and nonvolatile metabolome of goat cheese due to adjunct of non-starter lactic acid bacteria. LWT—Food Sci. Technol. 2019, 116, 108576. [Google Scholar] [CrossRef]
- Suzuki-Iwashima, A.; Matsuura, H.; Iwasawa, A.; Shiota, M. Metabolomics analyses of the combined effects of lactic acid bacteria and penicillium camemberti on the generation of volatile compounds in model mold-surface-ripened cheeses. J. Biosci. Bioeng. 2020, 129, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.F.; Casella, T.; Gomes, E.S.; Nogueira, M.C.L.; De Dea Lindner, J.; Penna, A.L.B. Diversity of lactic acid bacteria isolated from brazilian water buffalo mozzarella cheese. J. Food Sci. 2015, 80, M411–M417. [Google Scholar] [CrossRef]
- Kempler, G.M.; McKay, L.L. Improved medium for detection of citrate-fermenting Streptococcus lactis subsp. diacetylactis. Appl. Environ. Microbiol. 1980, 39, 926–927. [Google Scholar] [CrossRef]
- Pailin, T.; Kang, D.H.; Schmidt, K.; Fung, D.Y.C. Detection of extracellular bound proteinase in eps-producing lactic acid bacteria cultures on skim milk agar. Lett. Appl. Microbiol. 2001, 33, 45–49. [Google Scholar] [CrossRef]
- Donkor, O.N.; Nilmini, S.L.I.; Stolic, P.; Vasiljevic, T.; Shah, N.P. Survival and activity of selected probiotic organisms in set-type yoghurt during cold storage. Int. Dairy J. 2007, 17, 657–665. [Google Scholar] [CrossRef]
- Zeppa, G.; Conterno, L.; Gerbi, V. Determination of organic acids, sugars, diacetyl, and acetoin in cheese by high-performance liquid chromatography. J. Agric. Food Chem. 2001, 49, 2722–2726. [Google Scholar] [CrossRef] [PubMed]
- De Paula, A.T.; Jeronymo-Ceneviva, A.B.; Silva, L.F.; Todorov, S.D.; Franco, B.D.G.d.M.; Choiset, Y.; Haertlé, T.; Chobert, J.-M.; Dousset, X.; Penna, A.L.B. Leuconostoc mesenteroides SJRP55: A bacteriocinogenic strain isolated from brazilian water buffalo mozzarella cheese. Probiotics Antimicrob. Proteins 2014, 6, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Amiri, S.; Aghamirzaei, M.; Mostashari, P.; Sarbazi, M.; Tizchang, S.; Madahi, H. The impact of biotechnology on dairy industry. In Microbial Biotechnology in Food and Health; Elsevier: Cambridge, MA, USA, 2021; Volume 2, pp. 53–79. ISBN 9780128198131. [Google Scholar]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Qiao, Y.; Zhang, Y.; Leng, C.; Chen, H.; Sun, J.; Fan, X.; Li, A.; Feng, Z. Metabolic profiles of carbohydrates in Streptococcus thermophilus during pH-controlled batch fermentation. Front. Microbiol. 2020, 11, 1131. [Google Scholar] [CrossRef] [PubMed]
- Slattery, L.; O’Callaghan, J.; Fitzgerald, G.F.; Beresford, T.; Ross, R.P. Invited review: Lactobacillus helveticus—A thermophilic dairy starter related to gut bacteria. J. Dairy Sci. 2010, 93, 4435–4454. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, M.; Martino, G.P.; Blancato, V.S.; Espariz, M.; Hartke, A.; Sauvageot, N.; Benachour, A.; Alarcón, S.H.; Magni, C. Diversity of volatile organic compound production from leucine and citrate in Enterococcus faecium. Appl. Microbiol. Biotechnol. 2020, 104, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Repizo, G.D.; Blancato, V.S.; Mortera, P.; Lolkema, J.S.; Magni, C. Biochemical and genetic characterization of the Enterococcus faecalis oxaloacetate decarboxylase complex. Appl. Environ. Microbiol. 2013, 79, 2882–2890. [Google Scholar] [CrossRef] [PubMed]
- Mataragas, M. Investigation of genomic characteristics and carbohydrates’ metabolic activity of Lactococcus lactis subsp. lactis during ripening of a Swiss-type cheese. Food Microbiol. 2020, 87, 103392. [Google Scholar] [CrossRef]
- Zuljan, F.A.; Mortera, P.; Alarcón, S.H.; Blancato, V.S.; Espariz, M.; Magni, C. Lactic acid bacteria decarboxylation reactions in cheese. Int. Dairy J. 2016, 62, 53–62. [Google Scholar] [CrossRef]
- Axelson, L. Lactic acid bacteria: Classification and physiology. In Lactic Acid Bacteria; Salminen, S., von Wright, A., Eds.; CRC Press: New York, NY, USA, 2004; pp. 14–84. ISBN 9780429146442. [Google Scholar]
- Penna, A.L.B.; de Paula, A.T.; Casarotti, S.N.; Silva, L.F.; Diamantino, V.R.; Todorov, S.D. Overview of the functional lactic acid bacteria in fermented milk products. In Beneficial Microbes in Fermented and Functional Foods; Rai, V.R., Bai, J.A., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 113–148. ISBN 9780429189746. [Google Scholar]
- Khattab, A.R.; Guirguis, H.A.; Tawfik, S.M.; Farag, M.A. Cheese ripening: A review on modern technologies towards flavor enhancement, process acceleration and improved quality assessment. Trends Food Sci. Technol. 2019, 88, 343–360. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, L.; Wen, R.; Chen, Q.; Kong, B. Role of lactic acid bacteria in flavor development in traditional chinese fermented foods: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 2741–2755. [Google Scholar] [CrossRef] [PubMed]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of bioactive peptides by species: From gene to application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [PubMed]
- García-Cano, I.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Wang, K.; Kosmerl, E.; Jiménez-Flores, R. Lactic acid bacteria isolated from dairy products as potential producers of lipolytic, proteolytic and antibacterial proteins. Appl. Microbiol. Biotechnol. 2019, 103, 5243–5257. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Q.; Zheng, X.; Ge, Z.; Lin, K.; Zhang, D.; Chen, Y.; Wang, B.; Shi, X. Investigation of the lactic acid bacteria in kazak cheese and their contributions to cheese fermentation. Front. Microbiol. 2020, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Sarhir, S.T.; Belkhou, R.; Bouseta, A.; Hayaloglu, A.A. Evaluation of techno-functional and biochemical characteristics of selected lactic acid bacteria (Lactococcus lactis and Leuconostoc mesenteroides) used for the production of moroccan fermented milk, Lben. SSRN Electron. J. 2022, 140, 105592. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Nisic, A.; Hyden, P.; Cabal, A.; Sucher, J.; Stöger, A.; Allerberger, F.; Martinović, A. Genetic diversity of Leuconostoc mesenteroides isolates from traditional Montenegrin brine cheese. Microorganisms 2021, 9, 1612. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.Y.; Seo, Y.J.; Chang, H.C. Characterization of the psychrotrophic lactic acid bacterium Leuconostoc gelidum subsp. aenigmaticum LS4 isolated from kimchi based on comparative analyses of its genomic and phenotypic properties. Foods 2021, 10, 1899. [Google Scholar] [CrossRef]
- Hoehnel, A.; Bez, J.; Sahin, A.W.; Coffey, A.; Arendt, E.K.; Zannini, E. Leuconostoc citreum TR116 as a microbial cell factory to functionalise high-protein faba bean ingredients for bakery applications. Foods 2020, 9, 1706. [Google Scholar] [CrossRef]
- Müller, D.C.; Mischler, S.; Schönlechner, R.; Miescher Schwenninger, S. Multiple techno-functional characteristics of Leuconostoc and their potential in sourdough fermentations. Microorganisms 2021, 9, 1633. [Google Scholar] [CrossRef]
- El Hatmi, H.; Jrad, Z.; Oussaief, O.; Nasri, W.; Sbissi, I.; Khorchani, T.; Canabady-Rochelle, L.L.S. Fermentation of Dromedary camel (Camelus dromedarius) milk by Enterococcus faecium, Streptococcus macedonicus as a potential alternative of fermented cow milk. LWT 2018, 90, 373–380. [Google Scholar] [CrossRef]
- Özcan, T.; Ozdemir, T.; Avci, H.R. Survival of Lactobacillus casei and functional characteristics of reduced sugar red beetroot yoghurt with natural sugar substitutes. Int. J. Dairy Technol. 2021, 74, 148–160. [Google Scholar] [CrossRef]
- Ferreira, A.M.; Mendes-Faia, A. The role of yeasts and lactic acid bacteria on the metabolism of organic acids during winemaking. Foods 2020, 9, 1231. [Google Scholar] [CrossRef] [PubMed]
- Maślak, E.; Złoch, M.; Arendowski, A.; Sugajski, M.; Janczura, I.; Rudnicka, J.; Walczak-Skierska, J.; Buszewska-Forajta, M.; Rafińska, K.; Pomastowski, P.; et al. Isolation and identification of Lactococcus lactis and Weissella cibaria strains from fermented beetroot and an investigation of their properties as potential starter cultures and probiotics. Foods 2022, 11, 2257. [Google Scholar] [CrossRef] [PubMed]
- Fusieger, A.; Martins, M.C.F.; de Freitas, R.; Nero, L.A.; de Carvalho, A.F. Technological properties of Lactococcus lactis subsp. lactis bv. diacetylactis obtained from dairy and non-dairy niches. Braz. J. Microbiol. 2020, 51, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Van Mastrigt, O.; Mager, E.E.; Jamin, C.; Abee, T.; Smid, E.J. Citrate, low pH and amino acid limitation induce citrate utilization in Lactococcus lactis biovar diacetylactis. Microb. Biotechnol. 2018, 11, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, P.; Mahony, J.; Bottacini, F.; Lugli, G.A.; Ventura, M.; Van Sinderen, D. The Lactococcus lactis pan-plasmidome. Front. Microbiol. 2019, 10, 707. [Google Scholar] [CrossRef]
- Silva, L.F.; Sunakozawa, T.N.; Amaral, D.M.F.; Casella, T.; Nogueira, M.C.L.; De Dea Lindner, J.; Bottari, B.; Gatti, M.; Penna, A.L.B. Safety and technological application of autochthonous Streptococcus thermophilus cultures in the buffalo Mozzarella cheese. Food Microbiol. 2020, 87, 103383. [Google Scholar] [CrossRef]
- Markakiou, S.; Gaspar, P.; Johansen, E.; Zeidan, A.A.; Neves, A.R. Harnessing the metabolic potential of Streptococcus thermophilus for new biotechnological applications. Curr. Opin. Biotechnol. 2020, 61, 142–152. [Google Scholar] [CrossRef]
- Xue, Z.P.; Cu, X.; Xu, K.; Peng, J.H.; Liu, H.R.; Zhao, R.T.; Wang, Z.; Wang, T.; Xu, Z.S. The effect of glutathione biosynthesis of Streptococcus thermophilus ST-1 on cocultured Lactobacillus delbrueckii ssp. bulgaricus ATCC11842. J. Dairy Sci. 2022, 106, 884–896. [Google Scholar] [CrossRef]
- Deshwal, G.K.; Tiwari, S.; Kumar, A.; Raman, R.K.; Kadyan, S. Review on factors affecting and control of post-acidification in yoghurt and related products. Trends Food Sci. Technol. 2021, 109, 499–512. [Google Scholar] [CrossRef]
- Hashemi, S.M.B.; Gholamhosseinpour, A.; Abedi, E. Biopreservative potential of Lactobacillus strains in yoghurt dessert. J. Food Meas. Charact. 2021, 15, 1634–1643. [Google Scholar] [CrossRef]
- Ayad, E.H.E.; Nashat, S.; El-Sadek, N.; Metwaly, H.; El-Soda, M. Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. 2004, 21, 715–725. [Google Scholar] [CrossRef]
- Zhou, T.; Huo, R.; Kwok, L.Y.; Li, C.; Ma, Y.; Mi, Z.; Chen, Y. Effects of applying Lactobacillus helveticus H9 as adjunct starter culture in yogurt fermentation and storage. J. Dairy Sci. 2019, 102, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Nikoloudaki, O.; Gobbetti, M.; Di Cango, R. Lactic acid bacteria: Lactobacillus helveticus. In Encyclopedia of Dairy Sciences; Elsevier: Cambridge, MA, USA, 2022; pp. 198–205. [Google Scholar]
- Sıçramaz, H.; Güven, O.T.; Can, A.; Ayar, A.; Gül, Y. Impact of different starter cultures and Lactobacillus helveticus on volatile components, chemical and sensory properties of pasta filata cheese. Curr. Res. Food Sci. 2022, 5, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, J.M. The food matrix: Implications in processing, nutrition and health. Crit. Rev. Food Sci. Nutr. 2019, 59, 3612–3629. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Rizzotti, L.; Felis, G.E.; Torriani, S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiol. 2014, 42, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Muelas, R.; Martí de Olives, A.; Romero, G.; Díaz, J.R.; Sayas-Barberá, M.E.; Sendra, E. Evaluation of individual lactic acid bacteria for the fermentation of goat milk: Quality parameters. LWT—Food Sci. Technol. 2018, 98, 506–514. [Google Scholar] [CrossRef]
- Ghosh, K.; Ray, M.; Adak, A.; Halder, S.K.; Das, A.; Jana, A.; Parua (Mondal), S.; Vágvölgyi, C.; Das Mohapatra, P.K.; Pati, B.R.; et al. Role of probiotic Lactobacillus fermentum KKL1 in the preparation of a rice based fermented beverage. Bioresour. Technol. 2015, 188, 161–168. [Google Scholar] [CrossRef]
- Aarnikunnas, J.; Von Weymarn, N.; Rönnholm, K.; Leisola, M.; Palva, A. Metabolic engineering of Lactobacillus fermentum for production of mannitol and pure L-lactic acid or pyruvate. Biotechnol. Bioeng. 2003, 82, 653–663. [Google Scholar] [CrossRef]
- Casarotti, S.N. ; Carneiro, B.M.; Todorov, S.D.; Nero, L.A.; Rahal, P.; Penna, A.L.B. In vitro assessment of safety and probiotic potential characteristics of Lactobacillus strains isolated from water buffalo Mozzarella cheese. Ann. Microbiol. 2017, 67, 289–301. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]

| Characteristic | Species | Strains | Broth/Agar * | Milk | ||
|---|---|---|---|---|---|---|
| Medium/O2 | T °C/48 h | O2 | T °C/18 h | |||
| Mesophilic | Le. mesenteroides | SJRP54, SJRP58, SJRP62, SJRP63, SJRP64, SJRP132, SJRP153, SJRP154, SJRP156, SJRP159, SJRP160, SJRP161, SJRP163, SJRP172, SJRP173, SJRP174, SJRP175, SJRP186 | MRS/AE | 30 | AE | 30 |
| Le. citreum | SJRP31, SJRP44, SJRP140, SJRP165 | MRS/AE | 30 | AE | 30 | |
| Enterococcus spp. | SJRP04, SJRP11, SJRP16, SJRP23, SJRP69, SJRP101, SJRP120, SJRP125 | MRS/AE | 37 | AE | 37 | |
| E. durans | SJRP05, SJRP14, SJRP17, SJRP25, SJRP26, SJRP29, SJRP68 | MRS/AE | 37 | AE | 37 | |
| E. faecium | SJRP20, SJRP28 | MRS/AE | 37 | AE | 37 | |
| Lact. casei | SJRP35, SJRP37, SJRP66, SJRP136, SJRP141, SJRP145, SJRP146, SJRP148, SJRP169 | MRS/AN | 37 | AE | 37 | |
| Lc. lactis and Lc. garvieae | SJRP99, SJRP177, SJRP179, SJRP126 | M17/AE | 37 | AE | 37 | |
| Thermophilic | S. thermophilus | SJRP107, SJRP109 | M17/AE | 42 | AE | 42 |
| L. bulgaricus | SJRP49, SJRP50, SJRP57, SJRP76, SJRP149 | MRS/AN | 42 | AE | 42 | |
| L. helveticus | SJRP56, SJRP191 | MRS/AN | 42 | AE | 42 | |
| Lm. fermentum | SJRP30, SJRP32, SJRP41, SJRP42, SJRP43, SJRP81, SJRP164 | MRS/AN | 42 | AE | 42 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, L.F.; Sunakozawa, T.N.; Monteiro, D.A.; Casella, T.; Conti, A.C.; Todorov, S.D.; Barretto Penna, A.L. Potential of Cheese-Associated Lactic Acid Bacteria to Metabolize Citrate and Produce Organic Acids and Acetoin. Metabolites 2023, 13, 1134. https://doi.org/10.3390/metabo13111134
Silva LF, Sunakozawa TN, Monteiro DA, Casella T, Conti AC, Todorov SD, Barretto Penna AL. Potential of Cheese-Associated Lactic Acid Bacteria to Metabolize Citrate and Produce Organic Acids and Acetoin. Metabolites. 2023; 13(11):1134. https://doi.org/10.3390/metabo13111134
Chicago/Turabian StyleSilva, Luana Faria, Tássila Nakata Sunakozawa, Diego Alves Monteiro, Tiago Casella, Ana Carolina Conti, Svetoslav Dimitrov Todorov, and Ana Lúcia Barretto Penna. 2023. "Potential of Cheese-Associated Lactic Acid Bacteria to Metabolize Citrate and Produce Organic Acids and Acetoin" Metabolites 13, no. 11: 1134. https://doi.org/10.3390/metabo13111134
APA StyleSilva, L. F., Sunakozawa, T. N., Monteiro, D. A., Casella, T., Conti, A. C., Todorov, S. D., & Barretto Penna, A. L. (2023). Potential of Cheese-Associated Lactic Acid Bacteria to Metabolize Citrate and Produce Organic Acids and Acetoin. Metabolites, 13(11), 1134. https://doi.org/10.3390/metabo13111134






