A New Biomarker Profiling Strategy for Gut Microbiome Research: Valid Association of Metabolites to Metabolism of Microbiota Detected by Non-Targeted Metabolomics in Human Urine
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Preparation
2.3. Metabolites Profiling by Liquid-Chromatography Mass Spectrometry (LC-MS)
2.4. Data Processing
2.5. Metabolite Annotation
3. Results
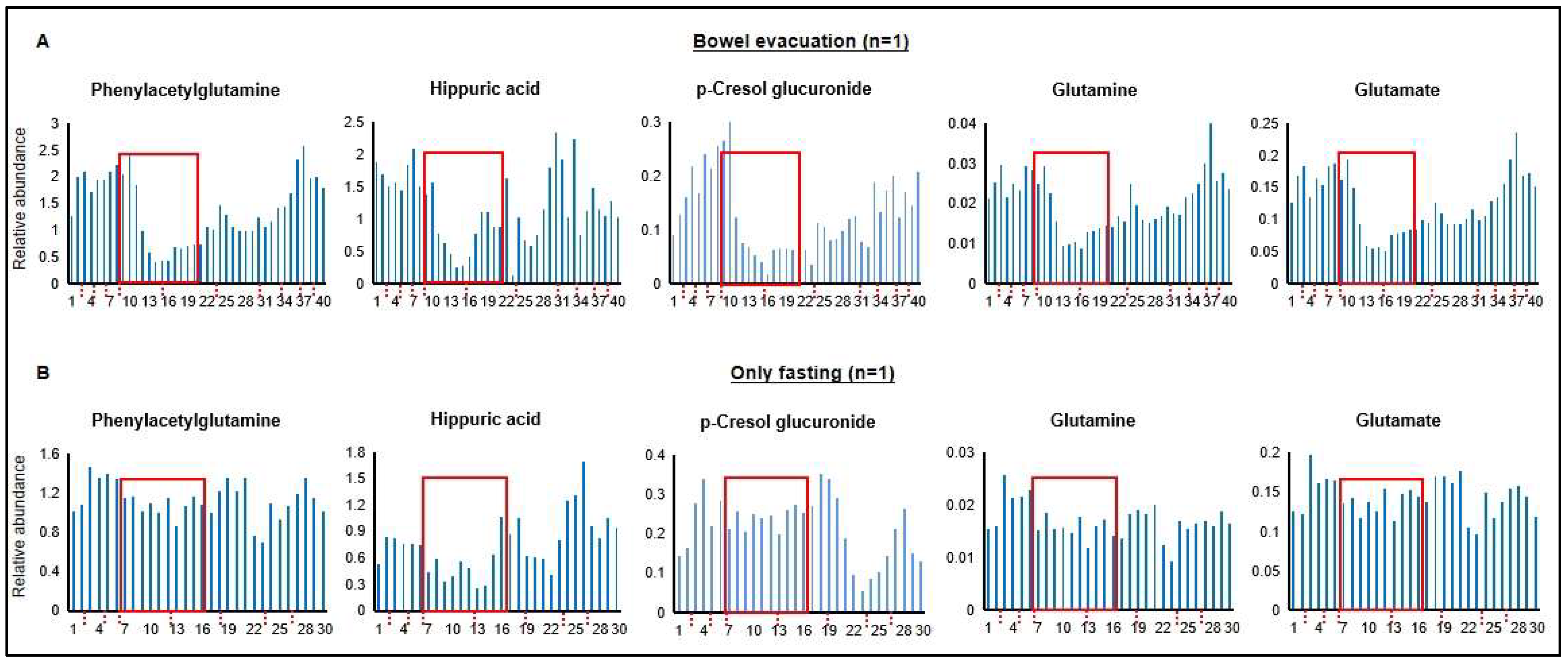
3.1. Distinct Metabolites Are Decreased Subsequent to Laxative-Induced Bowel Evacuation
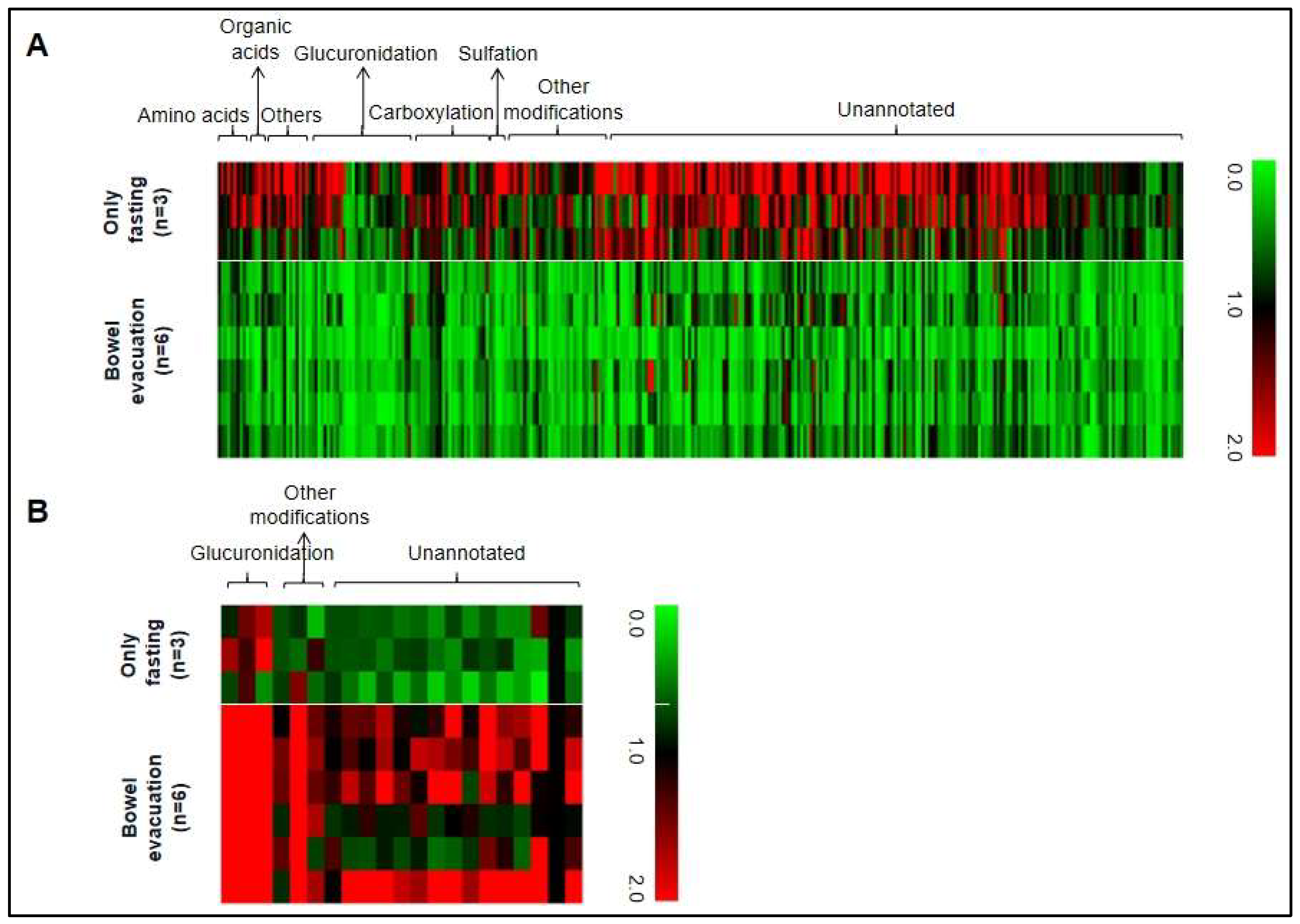
3.2. Confirmation of the Findings of Luminal (Fecal) Microbiota-Associated Metabolites in Human Urine
3.3. A Considerable Number of Metabolites in Human Urine Are Associated to the Luminal (Fecal) Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Selber-Hnatiw, S.; Sultana, T.; Tse, W.; Abdollahi, N.; Abdullah, S.; Al Rahbani, J.; Alazar, D.; Alrumhein, N.J.; Aprikian, S.; Arshad, R.; et al. Metabolic networks of the human gut microbiota. Microbiology 2020, 166, 96–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X.; et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017, 23, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Whon, T.W.; Shin, N.R.; Kim, J.Y.; Roh, S.W. Omics in gut microbiome analysis. J. Microbiol. 2021, 59, 292–297. [Google Scholar] [CrossRef]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef]
- Han, S.; Van Treuren, W.; Fischer, C.R.; Merrill, B.D.; DeFelice, B.C.; Sanchez, J.M.; Higginbottom, S.K.; Guthrie, L.; Fall, L.A.; Dodd, D.; et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 2021, 595, 415–420. [Google Scholar] [CrossRef]
- Wu, H.; Tremaroli, V.; Schmidt, C.; Lundqvist, A.; Olsson, L.M.; Kramer, M.; Gummesson, A.; Perkins, R.; Bergstrom, G.; Backhed, F. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab. 2020, 32, 379–390.e3. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zeng, B.; Liu, M.; Chen, J.; Pan, J.; Han, Y.; Liu, Y.; Cheng, K.; Zhou, C.; Wang, H.; et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci. Adv. 2019, 5, eaau8317. [Google Scholar] [CrossRef] [PubMed]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e22. [Google Scholar] [CrossRef] [PubMed]
- Pruss, K.M.; Chen, H.; Liu, Y.; Van Treuren, W.; Higginbottom, S.K.; Jarman, J.B.; Fischer, C.R.; Mak, J.; Wong, B.; Cowan, T.M.; et al. Host-microbe co-metabolism via MCAD generates circulating metabolites including hippuric acid. Nat. Commun. 2023, 14, 512. [Google Scholar] [CrossRef]
- Kikuchi, K.; Saigusa, D.; Kanemitsu, Y.; Matsumoto, Y.; Thanai, P.; Suzuki, N.; Mise, K.; Yamaguchi, H.; Nakamura, T.; Asaji, K.; et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat. Commun. 2019, 10, 1835. [Google Scholar] [CrossRef]
- Dekkers, K.F.; Sayols-Baixeras, S.; Baldanzi, G.; Nowak, C.; Hammar, U.; Nguyen, D.; Varotsis, G.; Brunkwall, L.; Nielsen, N.; Eklund, A.C.; et al. An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat. Commun. 2022, 13, 5370. [Google Scholar] [CrossRef]
- Zierer, J.; Jackson, M.A.; Kastenmuller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Hu, J.; Ding, J.; Li, X.; Li, J.; Zheng, T.; Xie, L.; Li, C.; Tang, Y.; Guo, K.; Huang, J.; et al. Distinct signatures of gut microbiota and metabolites in different types of diabetes: A population-based cross-sectional study. EClinicalMedicine 2023, 62, 102132. [Google Scholar] [CrossRef]
- Hryhorczuk, L.M.; Novak, E.A.; Gershon, S. Gut flora and urinary phenylacetic acid. Science 1984, 226, 996. [Google Scholar] [CrossRef][Green Version]
- Goodwin, B.L.; Ruthven, C.R.; Sandler, M. Gut flora and the origin of some urinary aromatic phenolic compounds. Biochem. Pharmacol. 1994, 47, 2294–2297. [Google Scholar] [CrossRef]
- Li, R.J.; Jie, Z.Y.; Feng, Q.; Fang, R.L.; Li, F.; Gao, Y.; Xia, H.H.; Zhong, H.Z.; Tong, B.; Madsen, L.; et al. Network of Interactions between Gut Microbiome, Host Biomarkers, and Urine Metabolome in Carotid Atherosclerosis. Front. Cell Infect. Microbiol. 2021, 11, 708088. [Google Scholar] [CrossRef] [PubMed]
- Ballet, C.; Correia, M.S.P.; Conway, L.P.; Locher, T.L.; Lehmann, L.C.; Garg, N.; Vujasinovic, M.; Deindl, S.; Lohr, J.M.; Globisch, D. New enzymatic and mass spectrometric methodology for the selective investigation of gut microbiota-derived metabolites. Chem. Sci. 2018, 9, 6233–6239. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Li, X.H.; Chen, W.N. An untargeted fecal and urine metabolomics analysis of the interplay between the gut microbiome, diet and human metabolism in Indian and Chinese adults. Sci. Rep. 2019, 9, 9191. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhao, X.; Lu, X.; Lin, X.; Xu, G. A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis. Front. Mol. Biosci. 2015, 2, 4. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, Z.; Chen, A.; Lu, X.; Zhao, C.; Hu, C.; Zhou, L.; Liu, X.; Wang, X.; Hou, X.; et al. Comprehensive Strategy to Construct In-House Database for Accurate and Batch Identification of Small Molecular Metabolites. Anal. Chem. 2018, 90, 7635–7643. [Google Scholar] [CrossRef]
- Zheng, S.; Zhang, X.; Li, Z.; Hoene, M.; Fritsche, L.; Zheng, F.; Li, Q.; Fritsche, A.; Peter, A.; Lehmann, R.; et al. Systematic, Modifying Group-Assisted Strategy Expanding Coverage of Metabolite Annotation in Liquid Chromatography-Mass Spectrometry-Based Nontargeted Metabolomics Studies. Anal. Chem. 2021, 93, 10916–10924. [Google Scholar] [CrossRef]
- Janik, R.; Thomason, L.A.M.; Stanisz, A.M.; Forsythe, P.; Bienenstock, J.; Stanisz, G.J. Magnetic resonance spectroscopy reveals oral Lactobacillus promotion of increases in brain GABA, N-acetyl aspartate and glutamate. Neuroimage 2016, 125, 988–995. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef]
- Ticinesi, A.; Guerra, A.; Nouvenne, A.; Meschi, T.; Maggi, S. Disentangling the Complexity of Nutrition, Frailty and Gut Microbial Pathways during Aging: A Focus on Hippuric Acid. Nutrients 2023, 15, 1138. [Google Scholar] [CrossRef]
- Penczynski, K.J.; Krupp, D.; Bring, A.; Bolzenius, K.; Remer, T.; Buyken, A.E. Relative validation of 24-h urinary hippuric acid excretion as a biomarker for dietary flavonoid intake from fruit and vegetables in healthy adolescents. Eur. J. Nutr. 2017, 56, 757–766. [Google Scholar] [CrossRef]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 2018, 94, fiy125. [Google Scholar] [CrossRef] [PubMed]
- Pellock, S.J.; Redinbo, M.R. Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J. Biol. Chem. 2017, 292, 8569–8576. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Zhao, D.; Peng, C.; Tan, C.; Wang, Q.; Gong, J. Effects of theabrownin on serum metabolites and gut microbiome in rats with a high-sugar diet. Food Funct. 2019, 10, 7063–7080. [Google Scholar] [CrossRef] [PubMed]
- Mosele, J.I.; Martin-Pelaez, S.; Macia, A.; Farras, M.; Valls, R.M.; Catalan, U.; Motilva, M.J. Faecal microbial metabolism of olive oil phenolic compounds: In vitro and in vivo approaches. Mol. Nutr. Food Res. 2014, 58, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sui, L.; Zhao, H.; Zhang, W.; Gao, L.; Hu, W.; Song, M.; Liu, X.; Kong, F.; Gong, Y.; et al. Differences in the Establishment of Gut Microbiota and Metabolome Characteristics between Balb/c and C57BL/6J Mice after Proton Irradiation. Front. Microbiol. 2022, 13, 874702. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, N.; Nigam, S.K. Drug transporters OAT1 and OAT3 have specific effects on multiple organs and gut microbiome as revealed by contextualized metabolic network reconstructions. Sci. Rep. 2022, 12, 18308. [Google Scholar] [CrossRef]
- McCallum, G.; Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 2023; ahead of print. PMID: 37740073. [Google Scholar] [CrossRef]
- Vaga, S.; Lee, S.; Ji, B.; Andreasson, A.; Talley, N.J.; Agreus, L.; Bidkhori, G.; Kovatcheva-Datchary, P.; Park, J.; Lee, D.; et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci. Rep. 2020, 10, 14977. [Google Scholar] [CrossRef]
- Moldave, K.; Meister, A. Synthesis of phenylacetylglutamine by human tissue. J. Biol. Chem. 1957, 229, 463–476. [Google Scholar] [CrossRef]
- Yang, D.; Brunengraber, H. Glutamate, a window on liver intermediary metabolism. J. Nutr. 2000, 130, 991S–994S. [Google Scholar] [CrossRef]
- Romano, K.A.; Nemet, I.; Prasad Saha, P.; Haghikia, A.; Li, X.S.; Mohan, M.L.; Lovano, B.; Castel, L.; Witkowski, M.; Buffa, J.A.; et al. Gut Microbiota-Generated Phenylacetylglutamine and Heart Failure. Circ. Heart Fail. 2023, 16, e009972. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Su, J.; Liu, R.; Zhao, S.; Li, W.; Xu, X.; Li, D.; Shi, J.; Gu, B.; Zhang, J.; et al. Sexual dimorphism in glucose metabolism is shaped by androgen-driven gut microbiome. Nat. Commun. 2021, 12, 7080. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Chen, Y.Y.; Wu, X.Z.; Bai, P.R.; An, N.; Liu, X.L.; Zhu, Q.F.; Feng, Y.Q. Uncovering the Carboxylated Metabolome in Gut Microbiota-Host Co-metabolism: A Chemical Derivatization-Molecular Networking Approach. Anal. Chem. 2023, 95, 11550–11557. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.S.P.; Jain, A.; Alotaibi, W.; Young Tie Yang, P.; Rodriguez-Mateos, A.; Globisch, D. Comparative dietary sulfated metabolome analysis reveals unknown metabolic interactions of the gut microbiome and the human host. Free Radic. Biol. Med. 2020, 160, 745–754. [Google Scholar] [CrossRef]
- Valeri, F.; Endres, K. How biological sex of the host shapes its gut microbiota. Front. Neuroendocrinol. 2021, 61, 100912. [Google Scholar] [CrossRef]

| No. | Metabolites | Annotation Base | Category | Selected Microbiota-Related References |
|---|---|---|---|---|
| 1 | Phenylalanine | a, b, c | Amino acid | [8] |
| 2 | Glutamine | a, b | Amino acid | [12,27] |
| 3 | Glutamate | a, b | Amino acid | [12,27] |
| 4 | Methionine | a, b | Amino acid | [8] |
| 5 | Tryptophan | a, b | Amino acid | [8] |
| 6 | N-Acetyltryptophan | a, b | Amino acid | [8] |
| 7 | 5-Hydroxytryptophan | a, b | Amino acid | [8] |
| 8 | N-Acetyltyrosine | a, b | Amino acid | [8] |
| 9 | N-(3-Indolylacetyl)-l-alanine | a, b | Amino acid | [8] |
| 10 | N-cyclohexyltaurine | a, b | Amino acid | |
| 11 | Phenylacetylglutamine | a, b, c | Amino acid | [13,28] |
| 12 | Hippuric acid | a, b, c | Organic acid | [29,30] |
| 13 | Hydroxyhippuric acid | a, b | Organic acid | [8] |
| 14 | Hydroxyphenyl lactic acid | a, b | Organic acid | [8] |
| 15 | 5-Hydroxyindole-3-acetic acid | a, b | Organic acid | [8] |
| 16 | Aminobutyric acid | a, b, c | Organic acid | [8] |
| 17 | Dimethyluric acid | a, b, c | Organic acid | [8] |
| 18 | Aminooctanoic acid | a, b | Organic acid | [8] |
| 19 | p-Cresol glucuronide | a, b | Organic acid | [16,31,32] |
| 20 | Dimethylxanthine | a, b | Nucleoside | [33] |
| 21 | Orotidine | a, b | Nucleoside | [8] |
| 22 | 8-Hydroxy-2-deoxyguanosine | a, b | Nucleoside | |
| 23 | Decanoylcarnitine | a, b, c | Others | [8] |
| 24 | Tyrosol | a, b | Others | [34] |
| 25 | Hydroxybenzyl alcohol | a, b | Others | |
| 26 | 2-Methyl-1,2,3,4-tetrahydro-6,7-isoquinolinediol | a, b | Others | |
| 27 | 4-Hydroxyquinoline | a, b | Others | [8] |
| 28 | Hydroxybenzaldehyde | a, b | Others | [35] |
| 29 | Dihydroxyacetone | a, b | Others | [36] |
| 30 | Acetamidophenyl glucuronide | a, b | Others | [32] |
| 31 | 3-Methyloxindole | a, b | Others | [8] |
| 32 | Phenylacetamide | a, b | Others | [8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Zhou, L.; Hoene, M.; Peter, A.; Birkenfeld, A.L.; Weigert, C.; Liu, X.; Zhao, X.; Xu, G.; Lehmann, R. A New Biomarker Profiling Strategy for Gut Microbiome Research: Valid Association of Metabolites to Metabolism of Microbiota Detected by Non-Targeted Metabolomics in Human Urine. Metabolites 2023, 13, 1061. https://doi.org/10.3390/metabo13101061
Zheng S, Zhou L, Hoene M, Peter A, Birkenfeld AL, Weigert C, Liu X, Zhao X, Xu G, Lehmann R. A New Biomarker Profiling Strategy for Gut Microbiome Research: Valid Association of Metabolites to Metabolism of Microbiota Detected by Non-Targeted Metabolomics in Human Urine. Metabolites. 2023; 13(10):1061. https://doi.org/10.3390/metabo13101061
Chicago/Turabian StyleZheng, Sijia, Lina Zhou, Miriam Hoene, Andreas Peter, Andreas L. Birkenfeld, Cora Weigert, Xinyu Liu, Xinjie Zhao, Guowang Xu, and Rainer Lehmann. 2023. "A New Biomarker Profiling Strategy for Gut Microbiome Research: Valid Association of Metabolites to Metabolism of Microbiota Detected by Non-Targeted Metabolomics in Human Urine" Metabolites 13, no. 10: 1061. https://doi.org/10.3390/metabo13101061
APA StyleZheng, S., Zhou, L., Hoene, M., Peter, A., Birkenfeld, A. L., Weigert, C., Liu, X., Zhao, X., Xu, G., & Lehmann, R. (2023). A New Biomarker Profiling Strategy for Gut Microbiome Research: Valid Association of Metabolites to Metabolism of Microbiota Detected by Non-Targeted Metabolomics in Human Urine. Metabolites, 13(10), 1061. https://doi.org/10.3390/metabo13101061










