Identification of Potential Leishmania N-Myristoyltransferase Inhibitors from Withania somnifera (L.) Dunal: A Molecular Docking and Molecular Dynamics Investigation
Abstract
1. Introduction
1.1. Overview of the Antileishmanial Properties of Withania somnifera
1.2. Immunomodulatory Effects of W. somnifera in Leishmaniasis Infections
1.3. Molecular Docking Studies in the Development of Antileishmanial Drugs
2. Experimental
2.1. Phytochemical Review and Data Collection
2.2. Molecular-Docking Simulation
2.3. Molecular-Dynamics Simulation
3. Results
4. Discussion
Molecular-Dynamics Simulation Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orabi, M.A.; Zidan, S.A.; Sakagami, H.; Murakami, Y.; Ali, A.A.; Alyami, H.S.; Alshabi, A.M.; Matsunami, K. Antileishmanial and lung adenocarcinoma cell toxicity of Withania somnifera (Linn.) dunal root and fruit extracts. Nat. Prod. Res. 2022, 36, 4231–4237. [Google Scholar] [CrossRef]
- Brannigan, J.A.; Wilkinson, A.J. Drug discovery in leishmaniasis using protein lipidation as a target. Biophys. Rev. 2021, 13, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Riezk, A.; Raynes, J.G.; Yardley, V.; Murdan, S.; Croft, S.L. Activity of chitosan and its derivatives against Leishmania major and Leishmania mexicana in vitro. Antimicrob. Agents Chemother. 2020, 64, e01772-19. [Google Scholar] [CrossRef] [PubMed]
- Zidan, S.A.; Abdelhamid, R.A.; Alian, A.; Fouad, M.A.; Matsunami, K.; Orabi, M.A. Diterpenes and sterols from the Red Sea soft coral Sarcophyton trocheliophorum and their cytotoxicity and anti-leishmanial activities. J. Asian Nat. Prod. Res. 2022, 24, 794–802. [Google Scholar] [CrossRef]
- Corpas-Lopez, V.; Moniz, S.; Thomas, M.; Wall, R.J.; Torrie, L.S.; Zander-Dinse, D.; Tinti, M.; Brand, S.; Stojanovski, L.; Manthri, S. Pharmacological validation of N-myristoyltransferase as a drug target in Leishmania donovani. ACS Infect. Dis. 2018, 5, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.S.; Srinivas, K. Modern drug discovery process: An in silico approach. J. Bioinform. Seq. Anal. 2011, 2, 89–94. [Google Scholar]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Alamzeb, M.; Ali, S.; Mamoon-Ur-Rashid; Khan, B.; Ihsanullah; Adnan; Omer, M.; Ullah, A.; Ali, J.; Setzer, W.N. Antileishmanial Potential of Berberine Alkaloids from Berberis glaucocarpa Roots: Molecular Docking Suggests Relevant Leishmania Protein Targets. Nat. Prod. Commun. 2021, 16, 1934578X211031148. [Google Scholar] [CrossRef]
- Istanbullu, H.; Bayraktar, G. Toward New Antileishmanial Compounds: Molecular Targets for Leishmaniasis Treatment. In Leishmaniasis—General Aspects of a Stigmatized Disease; de Azevedo Calderon, L., Ed.; IntechOpen: London, UK, 2021; p. 256. [Google Scholar]
- Hassan, A.A.; Khalid, H.E.; Abdalla, A.H.; Mukhtar, M.M.; Osman, W.J.; Efferth, T. Antileishmanial Activities of Medicinal Herbs and Phytochemicals In Vitro and In Vivo: An Update for the Years 2015 to 2021. Molecules 2022, 27, 7579. [Google Scholar] [CrossRef]
- Wright, M.H.; Paape, D.; Storck, E.M.; Serwa, R.A.; Smith, D.F.; Tate, E.W. Global analysis of protein N-myristoylation and exploration of N-myristoyltransferase as a drug target in the neglected human pathogen Leishmania donovani. Chem. Biol. 2015, 22, 342–354. [Google Scholar] [CrossRef] [PubMed]
- McKean, P.G.; Delahay, R.; Pimenta, P.F.; Smith, D.F. Characterisation of a second protein encoded by the differentially regulated LmcDNA16 gene family of Leishmania major. Mol. Biochem. Parasitol. 1997, 85, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Towler, D.; Eubanks, S.; Towery, D.; Adams, S.; Glaser, L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J. Biol. Chem. 1987, 262, 1030–1036. [Google Scholar] [CrossRef] [PubMed]
- Frearson, J.A.; Brand, S.; McElroy, S.P.; Cleghorn, L.A.; Smid, O.; Stojanovski, L.; Price, H.P.; Guther, M.L.S.; Torrie, L.S.; Robinson, D.A. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature 2010, 464, 728–732. [Google Scholar] [CrossRef]
- Price, H.P.; Menon, M.R.; Panethymitaki, C.; Goulding, D.; McKean, P.G.; Smith, D.F. Myristoyl-CoA: Protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J. Biol. Chem. 2003, 278, 7206–7214. [Google Scholar] [CrossRef] [PubMed]
- Glover, C.J.; Hartman, K.D.; Felsted, R.L. Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J. Biol. Chem. 1997, 272, 28680–28689. [Google Scholar] [CrossRef] [PubMed]
- Herrera, L.J.; Brand, S.; Santos, A.; Nohara, L.L.; Harrison, J.; Norcross, N.R.; Thompson, S.; Smith, V.; Lema, C.; Varela-Ramirez, A. Validation of N-myristoyltransferase as potential chemotherapeutic target in mammal-dwelling stages of Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2016, 10, e0004540. [Google Scholar] [CrossRef]
- Hutton, J.A.; Goncalves, V.; Brannigan, J.A.; Paape, D.; Wright, M.H.; Waugh, T.M.; Roberts, S.M.; Bell, A.S.; Wilkinson, A.J.; Smith, D.F. Structure-based design of potent and selective Leishmania N-myristoyltransferase inhibitors. J. Med. Chem. 2014, 57, 8664–8670. [Google Scholar] [CrossRef]
- Åsberg, P.; Hammer, K.; Olsson, J.; Henriksson, M. Novel Compounds and Their Use in Therapy. U.S. Patent No WO2013009259, 17 January 2013. [Google Scholar]
- Rackham, M.D.; Yu, Z.; Brannigan, J.A.; Heal, W.P.; Paape, D.; Barker, K.V.; Wilkinson, A.J.; Smith, D.F.; Leatherbarrow, R.J.; Tate, E.W. Discovery of high affinity inhibitors of Leishmania donovani N-myristoyltransferase. MedChemComm 2015, 6, 1761–1766. [Google Scholar] [CrossRef]
- Bell, A.S.; Yu, Z.; Hutton, J.A.; Wright, M.H.; Brannigan, J.A.; Paape, D.; Roberts, S.M.; Sutherell, C.L.; Ritzefeld, M.; Wilkinson, A.J. Novel thienopyrimidine inhibitors of Leishmania N-myristoyltransferase with on-target activity in intracellular amastigotes. J. Med. Chem. 2020, 63, 7740–7765. [Google Scholar] [CrossRef]
- Olaleye, T.O.; Brannigan, J.A.; Roberts, S.M.; Leatherbarrow, R.J.; Wilkinson, A.J.; Tate, E.W. Peptidomimetic inhibitors of N-myristoyltransferase from human malaria and leishmaniasis parasites. Org. Biomol. Chem. 2014, 12, 8132–8137. [Google Scholar] [CrossRef]
- Paul, S.; Chakraborty, S.; Anand, U.; Dey, S.; Nandy, S.; Ghorai, M.; Saha, S.C.; Patil, M.T.; Kandimalla, R.; Proćków, J. Withania somnifera (L.) Dunal (Ashwagandha): A comprehensive review on ethnopharmacology, pharmacotherapeutics, biomedicinal and toxicological aspects. Biomed. Pharmacother. 2021, 143, 112175. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.C.; Wadhwa, R. Science of Ashwagandha: Preventive and Therapeutic Potentials; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Sharma, U.; Velpandian, T.; Sharma, P.; Singh, S. Evaluation of anti-leishmanial activity of selected Indian plants known to have antimicrobial properties. Parasitol. Res. 2009, 105, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- El-On, J.; Ozer, L.; Gopas, J.; Sneir, R.; Enav, H.; Luft, N.; Davidov, G.; Golan-Goldhirsh, A. Antileishmanial activity in Israeli plants. Ann. Trop. Med. Parasitol. 2009, 103, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.; Banerjee, B.; Das, B.; Ganguly, A.; Sen, T.; Pramanik, S.; Mukhopadhyay, S.; Majumder, H. Apoptosis is induced in leishmanial cells by a novel protein kinase inhibitor withaferin A and is facilitated by apoptotic topoisomerase I–DNA complex. Cell Death Differ. 2007, 14, 358–367. [Google Scholar] [CrossRef]
- Pramanick, S.; Roy, A.; Ghosh, S.; Majumder, H.K.; Mukhopadhyay, S. Withanolide Z, a new chlorinated withanolide from Withania somnifera. Planta Med. 2008, 74, 1745–1748. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Dayakar, A.; Veronica, J.; Sundar, S.; Maurya, R. An in vitro study of apoptotic like death in Leishmania donovani promastigotes by withanolides. Parasitol. Int. 2013, 62, 253–261. [Google Scholar] [CrossRef]
- Kaur, S.; Chauhan, K.; Sachdeva, H. Protection against experimental visceral leishmaniasis by immunostimulation with herbal drugs derived from Withania somnifera and Asparagus racemosus. J. Med. Microbiol. 2014, 63, 1328–1338. [Google Scholar] [CrossRef]
- Tripathi, C.D.P.; Kushawaha, P.K.; Sangwan, R.S.; Mandal, C.; Misra-Bhattacharya, S.; Dube, A. Withania somnifera chemotype NMITLI 101R significantly increases the efficacy of antileishmanial drugs by generating strong IFN-γ and IL-12 mediated immune responses in Leishmania donovani infected hamsters. Phytomedicine 2017, 24, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, C.; Gupta, R.; Kushawaha, P.; Mandal, C.; Misra Bhattacharya, S.; Dube, A. Efficacy of Withania somnifera chemotypes NMITLI–101R, 118R and Withaferin A against experimental visceral leishmaniasis. Parasite Immunol. 2014, 36, 253–265. [Google Scholar] [CrossRef]
- Sachdeva, H.; Sehgal, R.; Kaur, S. Studies on the protective and immunomodulatory efficacy of Withania somnifera along with cisplatin against experimental visceral leishmaniasis. Parasitol. Res. 2013, 112, 2269–2280. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Veronica, J.; Sundar, S.; Maurya, R. Alcoholic fractions F5 and F6 from Withania somnifera leaves show a potent Antileishmanial and Immunomodulatory activities to control experimental Visceral Leishmaniasis. Front. Med. 2017, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Katiyar, S.P.; Jeyakanthan, J.; Dubey, V.K.; Sundar, D. Blocking Protein kinase C signaling pathway: Mechanistic insights into the anti-leishmanial activity of prospective herbal drugs from Withania somnifera. BMC Genom. 2012, 13, S20. [Google Scholar] [CrossRef] [PubMed]
- Phadke, S.; Pathak, D.; Somani, D. Design and In silico Studies of 2, 5-Disubstituted 1, 2, 4-Triazole and 1, 3, 4-Thiadiazole Derivatives as Pteridine Reductase 1 Inhibitors. J. Pharm. Res. Int. 2021, 33, 166–178. [Google Scholar] [CrossRef]
- El-Hawary, S.S.; Mohammed, R.; Taher, M.A.; AbouZid, S.F.; Mansour, M.A.; Almahmoud, S.A.; Huwaimel, B.; Amin, E. Characterization of Promising Cytotoxic Metabolites from Tabebuia guayacan Hemsl.: Computational Prediction and In Vitro Testing. Plants 2022, 11, 888. [Google Scholar] [CrossRef] [PubMed]
- Arif, R.; Ahmad, S.; Mustafa, G.; Mahrosh, H.S.; Ali, M.; Tahir ul Qamar, M.; Dar, H.R. Molecular Docking and Simulation Studies of Antidiabetic Agents Devised from Hypoglycemic Polypeptide-P of Momordica charantia. Biomed. Res. Int. 2021, 2021, 5561129. [Google Scholar] [CrossRef]
- Mahnashi, M.H.; Alyami, B.A.; Alqahtani, Y.S.; Jan, M.S.; Rashid, U.; Sadiq, A.; Alqarni, A.O. Phytochemical profiling of bioactive compounds, anti-inflammatory and analgesic potentials of Habenaria digitata Lindl.: Molecular docking based synergistic effect of the identified compounds. J. Ethnopharmacol. 2021, 273, 113976. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, A.; Gupta, U. Molecular Docking studies on the Anti-fungal activity of Allium sativum (Garlic) against Mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1.0.0. Ann. Antivir. Antiretrovir. 2021, 5, 28–32. [Google Scholar]
- Allam, A.E.; Abouelela, M.E.; Assaf, H.K.; Sayed, A.M.; Nafady, A.M.; El-Shanawany, M.A.; Takano, F.; Ohta, T. Phytochemical and in silico studies for potential constituents from Centaurium spicatum as candidates against the SARS-CoV-2 main protease and RNA-dependent RNA polymerase. Nat. Prod. Res. 2021, 36, 5724–5731. [Google Scholar] [CrossRef]
- Miller, B.R., III; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Majumdar, D.N. Withania somnifera. II. Alkaloidal constituents and their chemical characterization. Indian J. Pharm. 1955, 17, 158–161. [Google Scholar]
- Tetali, S.D.; Acharya, S.; Ankari, A.B.; Nanakram, V.; Raghavendra, A.S. Metabolomics of Withania somnifera (L.) Dunal: Advances and applications. J. Ethnopharmacol. 2021, 267, 113469. [Google Scholar] [CrossRef]
- Remya, C.; Dileep, K.; Variayr, E.; Sadasivan, C. An in silico guided identification of nAChR agonists from Withania somnifera. Front. Life Sci. 2016, 9, 201–213. [Google Scholar] [CrossRef]
- Kannan, N.; Kulandaivelu, G. Novel method to isolate Withaferin A from Withania somnifera roots and its bioactivity. Allelopath. J. 2007, 20, 213. [Google Scholar]
- Khanna, K.; Schwarting, A.; Bobbitt, J. The occurrence of isopelletierine in Withania somnifera. J. Pharm. Sci. 1962, 51, 1194. [Google Scholar] [CrossRef] [PubMed]
- Sivanandhan, G.; Mariashibu, T.S.; Arun, M.; Rajesh, M.; Kasthurirengan, S.; Selvaraj, N.; Ganapathi, A. The effect of polyamines on the efficiency of multiplication and rooting of Withania somnifera (L.) Dunal and content of some withanolides in obtained plants. Acta Physiol. Plant. 2011, 33, 2279–2288. [Google Scholar] [CrossRef]
- Schröter, H.-B.; Neumann, D.; Katritzky, A.; Swinbourne, F. Withasomnine. A pyrazole alkaloid from Withania somnifera Dunl. Tetrahedron 1966, 22, 2895–2897. [Google Scholar] [CrossRef]
- Kandil, F.; El Sayed, N.; Abou-Douh, A.; Ishak, M.; Mabry, T.J. Flavonol glycosides and phenolics from Withania somnifera. Phytochemistry 1994, 37, 1215–1216. [Google Scholar] [CrossRef]
- Bolleddula, J.; Fitch, W.; Vareed, S.K.; Nair, M.G. Identification of metabolites in Withania sominfera fruits by liquid chromatography and high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 1277–1290. [Google Scholar] [CrossRef]
- Lee, S.R.; Lee, B.S.; Yu, J.S.; Kang, H.; Yoo, M.J.; Yi, S.A.; Han, J.-W.; Kim, S.; Kim, J.K.; Kim, J.-C. Identification of anti-adipogenic withanolides from the roots of Indian ginseng (Withania somnifera). J. Ginseng Res. 2022, 46, 357–366. [Google Scholar] [CrossRef]
- Rajalakshmy, M.; Geetha, G. Isolation and identification of withasomnine, withanolides and butein from industrial herbal marc of Withania somnifera (L.) Dunal. Indian J. Nat. Prod. Resour. 2016, 7, 116–124. [Google Scholar] [CrossRef]
- Senthil, K.; Thirugnanasambantham, P.; Oh, T.J.; Kim, S.H.; Choi, H.K. Free radical scavenging activity and comparative metabolic profiling of in vitro cultured and field grown Withania somnifera roots. PLoS ONE 2015, 10, e0123360. [Google Scholar] [CrossRef]
- Poojari, P.; Kiran, K.R.; Swathy, P.S.; Muthusamy, A. Withania somnifera (L.) Dunal: An Overview of Bioactive Molecules, Medicinal Properties and Enhancement of Bioactive Molecules Through Breeding Strategies. In In Vitro Plant Breeding Towards Novel Agronomic Traits; Springer: Singapore, 2019; pp. 1–25. [Google Scholar] [CrossRef]
- Rautela, I.; Sharma, M.D.; Sharma, N.; Kishor, K.; Singh, K.; Sharma, N. Comparative GC-MS analysis of leaf and root extract of medicinal plant Withania somnifera. World J. Pharm. Res. 2018, 7, 956–972. [Google Scholar]
- Abou-Douh, A.M. New withanolides and other constituents from the fruit of Withania somnifera. Arch. Pharm. 2002, 335, 267–276. [Google Scholar] [CrossRef]
- Marslin, G.; Selvakesavan, R.K.; Franklin, G.; Sarmento, B.; Dias, A.C. Antimicrobial activity of cream incorporated with silver nanoparticles biosynthesized from Withania somnifera. Int. J. Nanomed. 2015, 10, 5955. [Google Scholar]
- Lockley, W.; Roberts, D.; Rees, H.; Goodwin, T. 24-Methylcholesta-5, 24 (25)-dien-3β-ol: A new sterol from Withania somnifera. Tetrahedron Lett. 1974, 15, 3773–3776. [Google Scholar] [CrossRef]
- Takshak, S.; Agrawal, S. Alterations in metabolite profile and free radical scavenging activities of Withania somnifera leaf and root extracts under supplemental ultraviolet-B radiation. Acta Physiol. Plant. 2015, 37, 260. [Google Scholar] [CrossRef]
- Dar, N.J.; Hamid, A.; Ahmad, M. Pharmacologic overview of Withania somnifera, the Indian Ginseng. Cell Mol. Life Sci. 2015, 72, 4445–4460. [Google Scholar] [CrossRef] [PubMed]
- Nile, S.H.; Liang, Y.; Wang, Z.; Zheng, J.; Sun, C.; Nile, A.; Patel, G.; Kai, G. Chemical composition, cytotoxic and pro-inflammatory enzyme inhibitory properties of Withania somnifera (L.) Dunal root extracts. S. Afr. J. Bot. 2021, 151, 46–53. [Google Scholar] [CrossRef]
- Girme, A.; Saste, G.; Pawar, S.; Balasubramaniam, A.K.; Musande, K.; Darji, B.; Satti, N.K.; Verma, M.K.; Anand, R.; Singh, R. Investigating 11 withanosides and withanolides by UHPLC–PDA and mass fragmentation studies from Ashwagandha (Withania somnifera). ACS Omega 2020, 5, 27933–27943. [Google Scholar] [CrossRef]
- Chen, L.-X.; He, H.; Qiu, F. Natural withanolides: An overview. Nat. Prod. Rep. 2011, 28, 705–740. [Google Scholar] [CrossRef]
- Dar, N.J.; Bhat, J.A.; Satti, N.K.; Sharma, P.R.; Hamid, A.; Ahmad, M. Withanone, an active constituent from Withania somnifera, affords protection against NMDA-induced excitotoxicity in neuron-like cells. Mol. Neurobiol. 2017, 54, 5061–5073. [Google Scholar] [CrossRef] [PubMed]
- Nittala, S.S.; Lavie, D. Chemistry and genetics of withanolides in Withania somnifera hybrids. Phytochemistry 1981, 20, 2741–2748. [Google Scholar] [CrossRef]
- Bessalle, R.; Lavie, D. Withanolide C, a chlorinated withanolide from Withania somnifera. Phytochemistry 1992, 31, 3648–3651. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Hussain, S.; Yousuf, S.; Dar, A. Chlorinated and diepoxy withanolides from Withania somnifera and their cytotoxic effects against human lung cancer cell line. Phytochemistry 2010, 71, 2205–2209. [Google Scholar] [CrossRef]
- Nittala, S.S.; Velde, V.V.; Frolow, F.; Lavie, D. Chlorinated withanolides from Withania somnifera and Acnistus breviflorus. Phytochemistry 1981, 20, 2547–2552. [Google Scholar] [CrossRef]
- Tong, X.; Zhang, H.; Timmermann, B.N. Chlorinated withanolides from Withania somnifera. Phytochem. Lett. 2011, 4, 411–414. [Google Scholar] [CrossRef]
- Trivedi, M.K.; Panda, P.; Sethi, K.K.; Jana, S. Metabolite profiling in Withania somnifera roots hydroalcoholic extract using LC/MS, GC/MS and NMR spectroscopy. Chem. Biodivers. 2017, 14, e1600280. [Google Scholar] [CrossRef] [PubMed]
- Misra, L.; Lal, P.; Sangwan, R.S.; Sangwan, N.S.; Uniyal, G.C.; Tuli, R. Unusually sulfated and oxygenated steroids from Withania somnifera. Phytochemistry 2005, 66, 2702–2707. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-M.; Marron, M.T.; Seddon, E.; McLaughlin, S.P.; Ray, D.T.; Whitesell, L.; Gunatilaka, A.L. 2, 3-Dihydrowithaferin A-3β-O-sulfate, a new potential prodrug of withaferin A from aeroponically grown Withania somnifera. Bioorg. Med. Chem. 2009, 17, 2210–2214. [Google Scholar] [CrossRef]
- Subbaraju, G.V.; Vanisree, M.; Rao, C.V.; Sivaramakrishna, C.; Sridhar, P.; Jayaprakasam, B.; Nair, M.G. Ashwagandhanolide, a bioactive dimeric thiowithanolide isolated from the roots of Withania somnifera. J. Nat. Prod. 2006, 69, 1790–1792. [Google Scholar] [CrossRef]
- Jayaprakasam, B.; Strasburg, G.A.; Nair, M.G. Potent lipid peroxidation inhibitors from Withania somnifera fruits. Tetrahedron 2004, 60, 3109–3121. [Google Scholar] [CrossRef]
- Ali, M.; Shuaib, M.; Ansari, S.H. Withanolides from the stem bark of Withania somnifera. Phytochemistry 1997, 44, 1163–1168. [Google Scholar] [CrossRef]
- Khan, M.I.; Maqsood, M.; Saeed, R.A.; Alam, A.; Sahar, A.; Kieliszek, M.; Miecznikowski, A.; Muzammil, H.S.; Aadil, R.M. Phytochemistry, Food Application, and Therapeutic Potential of the Medicinal Plant (Withania coagulans): A Review. Molecules 2021, 26, 6881. [Google Scholar] [CrossRef] [PubMed]
- Vitali, G.; Conte, L.; Nicoletti, M. Withanolide composition and in vitro culture of Italian Withania somnifera. Planta Med. 1996, 62, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Glotter, E.; Kirson, I.; Abraham, A.; Lavie, D. Constituents of Withania somnifera Dun—XIII: The withanolides of chemotype III. Tetrahedron 1973, 29, 1353–1364. [Google Scholar] [CrossRef]
- Xu, Y.-M.; Gao, S.; Bunting, D.P.; Gunatilaka, A.L. Unusual withanolides from aeroponically grown Withania somnifera. Phytochemistry 2011, 72, 518–522. [Google Scholar] [CrossRef]
- Abraham, A.; Kirson, I.; Lavie, D.; Glotte, E. The withanolides of Withania somnifera chemotypes I and II. Phytochemistry 1975, 14, 189–194. [Google Scholar] [CrossRef]
- Gottlied, H.E.; Kirson, I. 13C NMR spectroscopy of the withanolides and other highly oxygenated C28 steroids. Org. Magn. Reson. 1981, 16, 20–25. [Google Scholar] [CrossRef]
- Jayaprakasam, B.; Nair, M.G. Cyclooxygenase-2 enzyme inhibitory withanolides from Withania somnifera leaves. Tetrahedron 2003, 59, 841–849. [Google Scholar] [CrossRef]
- Zhao, J.; Nakamura, N.; Hattori, M.; Kuboyama, T.; Tohda, C.; Komatsu, K. Withanolide derivatives from the roots of Withania somnifera and their neurite outgrowth activities. Chem. Pharm. Bull. 2002, 50, 760–765. [Google Scholar] [CrossRef]
- Joshi, P.; Misra, L.; Siddique, A.A.; Srivastava, M.; Kumar, S.; Darokar, M.P. Epoxide group relationship with cytotoxicity in withanolide derivatives from Withania somnifera. Steroids 2014, 79, 19–27. [Google Scholar] [CrossRef] [PubMed]
- El-Olemy, M.; Kadry, H. Withanolides of Withania somnifera L. Dunal growing wild in Egypt. Bull. Pharm. Sci. 1984, 7, 430–443. [Google Scholar] [CrossRef]
- Anjaneyulyu, A.; Rao, D.S. A new withanolide from the leaves of Withania somnifera. Indian J. Chem. Sec. B 1997, 36, 161–165. [Google Scholar]
- Arshad Jamal, S.; Iqbal Choudhary, M. Two new withanolides from Withania somnifera. Heterocycles 1992, 34, 689–698. [Google Scholar]
- Kundu, A.B.; Mukherjee, A.; Dey, A.K. A new withanolide from the seeds of Withania somnifera Dunal. Indian J. Chem. Sect. B 1976, 14, 434–435. [Google Scholar]
- Kumar, A.; Ali, M.; Mir, S. A new withanolide from the roots of Withania somnifera. Indian J. Chem. Sect. B 2004, 43, 2001–2003. [Google Scholar] [CrossRef]
- Neogi, P.; Kawai, M.; Butsugan, Y.; Mori, Y.; Suzuki, M. Withacoagin, a new withanolide from Withania coagulans roots. Bull. Chem. Soc. Jpn. 1988, 61, 4479–4481. [Google Scholar] [CrossRef]
- Kirson, I.; Cohen, A.; Abraham, A. Withanolides Q and R, two new 23-hydroxy-steroidal lactones. J. Chem. Soc. Perkin Trans. 1 1975, 21, 2136–2138. [Google Scholar] [CrossRef]
- Arshad Jamal, S.; Qureshi, S.; Noor Ali, S.; Choudhary, M.I. Bioactivities and structural studies of withanolides from Withania somnifera. Chem. Heterocycl. Compd. 1995, 31, 1047–1059. [Google Scholar] [CrossRef]
- Jamal, S.A.; Choudhary, M.I.; Asif, E. Two withanolides from Withania somnifera. Phytochemistry 1991, 30, 3824–3826. [Google Scholar]
- Choudhary, M.I.; Yousuf, S.; Nawaz, S.A.; Ahmed, S. Cholinesterase inhibiting withanolides from Withania somnifera. Chem. Pharm. Bull. 2004, 52, 1358–1361. [Google Scholar] [CrossRef]
- Chaurasiya, N.D.; Uniyal, G.C.; Lal, P.; Misra, L.; Sangwan, N.S.; Tuli, R.; Sangwan, R.S. Analysis of withanolides in root and leaf of Withania somnifera by HPLC with photodiode array and evaporative light scattering detection. Phytochem. Anal. 2008, 19, 148–154. [Google Scholar] [CrossRef]
- Iguchi, T.; Kuroda, M.; Ishihara, M.; Sakagami, H.; Mimaki, Y. Steroidal constituents isolated from the seeds of Withania somnifera. Nat. Prod. Res. 2021, 35, 2205–2210. [Google Scholar] [CrossRef]
- Misra, L.; Mishra, P.; Pandey, A.; Sangwan, R.S.; Sangwan, N.S.; Tuli, R. Withanolides from Withania somnifera roots. Phytochemistry 2008, 69, 1000–1004. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Abbas, S.; Jamal, S.A. Withania somnifera—A source of exotic withanolides. Heterocycles 1996, 2, 555–563. [Google Scholar]
- Kirson, I.; Glotter, E.; Abraham, A.; Lavie, D. Constituents of Withania somnifera dun—XI: The structure of three new withanolides. Tetrahedron 1970, 26, 2209–2219. [Google Scholar] [CrossRef]
- Kirson, I.; Abraham, A.; Lavie, D. Chemical Analysis of Hybrids of Withania somnifera L. (Dun.). 1. Chemotypes III (Israel) by Indian I (Delhi). Isr. J. Chem. 1977, 16, 20–24. [Google Scholar] [CrossRef]
- Velde, V.V.; Lavie, D. New withanolides of biogenetic interest from Withania somnifera. Phytochemistry 1981, 20, 1359–1364. [Google Scholar] [CrossRef]
- Kirson, I.; Glotter, E.; Lavie, D.; Abraham, A. Constitutents of Withania somnifera Dun. Part XII. The withanolides of an indian chemotype. J. Chem. Soc. C 1971, 2032–2044. [Google Scholar] [CrossRef]
- Nile, S.H.; Nile, A.; Gansukh, E.; Baskar, V.; Kai, G. Subcritical water extraction of withanosides and withanolides from ashwagandha (Withania somnifera L.) and their biological activities. Food Chem. Toxicol. 2019, 132, 110659. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, F.W.; Kirson, I.; Lavie, D.; Abraham, A. New withanolides from a cross of a South African chemotype by chemotype II (Israel) in Withania somnifera. Phytochemistry 1980, 19, 1503–1507. [Google Scholar] [CrossRef]
- Duyu, T.; Khanal, P.; Dey, Y.N.; Jha, S.K. Network pharmacology of Withania somnifera against stress associated neurodegenerative diseases. Adv. Trad. Med. 2021, 21, 565–578. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Singh, P.; Sharma, S.; Singh, T.P.; Ethayathulla, A.; Kaur, P. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J. Biomol. Struct. Dyn. 2021, 39, 5668–5681. [Google Scholar] [CrossRef]
- Moujir, L.M.; Llanos, G.G.; Araujo, L.; Amesty, A.; Bazzocchi, I.L.; Jiménez, I.A. Withanolide-type steroids from Withania aristata as potential anti-leukemic agents. Molecules 2020, 25, 5744. [Google Scholar] [CrossRef]
- Khan, M.A.; Khan, H.; Ali, T. Withanolides isolated from Withania somnifera with α-glucosidase inhibition. Med. Chem. Res. 2014, 23, 2386–2390. [Google Scholar] [CrossRef]
- Bessalle, R.; Lavie, D.; Frolow, F. Withanolide Y, a withanolide from a hybrid of Withania somnifera. Phytochemistry 1987, 26, 1797–1800. [Google Scholar] [CrossRef]
- Nittala, S.S.; Frolow, F.; Lavie, D. Novel occurrence of 14β-hydroxy-group on a withanolide skeleton; X-ray crystal and molecular structure of 14β-hydroxywithanone. J. Chem. Soc. Chem. Commun. 1981, 4, 178–179. [Google Scholar] [CrossRef]
- Zhang, H.; Timmermann, B.N. Withanolide structural revisions by 13C NMR spectroscopic analysis inclusive of the γ-gauche effect. J. Nat. Prod. 2016, 79, 732–742. [Google Scholar] [CrossRef]
- Kim, S.; Yu, J.S.; Lee, J.Y.; Choi, S.U.; Lee, J.; Kim, K.H. Cytotoxic withanolides from the roots of Indian ginseng (Withania somnifera). J. Nat. Prod. 2019, 82, 765–773. [Google Scholar] [CrossRef]
- Maurya, R. Withanolides: A prospective drug for infectious and tropical diseases. In Science of Ashwagandha: Preventive and Therapeutic Potentials; Springer: Berlin/Heidelberg, Germany, 2017; pp. 105–120. [Google Scholar]
- Kuroyanagi, M.; Shibata, K.; Umehara, K. Cell differentiation inducing steroids from Withania somnifera L. (Dun.). Chem. Pharm. Bull. 1999, 47, 1646–1649. [Google Scholar] [CrossRef]
- Glotter, E.; Waitman, R.; Lavie, D. Constituents of Withania somnifera Dun. Part VIII. A new steroidal lactone, 27-deoxy-14-hydroxy-withaferin A. J. Chem. Soc. C 1966, 1765–1767. [Google Scholar] [CrossRef]
- Jayaprakasam, B.; Zhang, Y.; Seeram, N.P.; Nair, M.G. Growth inhibition of human tumor cell lines by withanolides from Withania somnifera leaves. Life Sci. 2003, 74, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Lal, P.; Misra, L.; Sangwan, R.S.; Tuli, R. New withanolides from fresh berries of Withania somnifera. Z. Naturforsch. B 2006, 61, 1143–1147. [Google Scholar] [CrossRef]
- Abbas, S.; Jamal, S.A.; Choudhary, M.I. New withanolides from Withania sp. J. Nat. Prod. 1993, 56, 1000–1006. [Google Scholar]
- Matsuda, H.; Murakami, T.; Kishi, A.; Yoshikawa, M. Structures of withanosides I, II, III, IV, V, VI, and VII, new withanolide glycosides, from the roots of Indian Withania somnifera DUNAL. and inhibitory activity for tachyphylaxis to clonidine in isolated guinea-pig ileum. Bioorg. Med. Chem. 2001, 9, 1499–1507. [Google Scholar] [CrossRef]
- Ghosal, S.; Kaur, R.; Srivastava, R. Sitoindosides IX and X, new glycowithanolides from Withania somnifera. Ind. J. Nat. Prod. 1988, 4, 12–13. [Google Scholar]
- Bhattacharya, S.K.; Goel, R.K.; Kaur, R.; Ghosal, S. Anti-stress activity of sitoindosides VII and VIII, new acylsterylglucosides from Withania somnifera. Phytother. Res. 1987, 1, 32–37. [Google Scholar] [CrossRef]
- Abuzaid, A.A.; Abdoon, A.M.; Aldahan, M.A.; Alzahrani, A.G.; Alhakeem, R.F.; Asiri, A.M.; Alzahrani, M.H.; Memish, Z.A. Cutaneous leishmaniasis in Saudi Arabia: A comprehensive overview. Vector-Borne Zoonotic Dis. 2017, 17, 673–684. [Google Scholar] [CrossRef]
- Tate, E.W.; Bell, A.S.; Rackham, M.D.; Wright, M.H. N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology 2014, 141, 37–49. [Google Scholar] [CrossRef]
- Brannigan, J.A.; Roberts, S.M.; Bell, A.S.; Hutton, J.A.; Hodgkinson, M.R.; Tate, E.W.; Leatherbarrow, R.J.; Smith, D.F.; Wilkinson, A.J. Diverse modes of binding in structures of Leishmania major N-myristoyltransferase with selective inhibitors. IUCrJ 2014, 1, 250–260. [Google Scholar] [CrossRef]
- Brannigan, J.A.; Smith, B.A.; Yu, Z.; Brzozowski, A.M.; Hodgkinson, M.R.; Maroof, A.; Price, H.P.; Meier, F.; Leatherbarrow, R.J.; Tate, E.W. N-myristoyltransferase from Leishmania donovani: Structural and functional characterisation of a potential drug target for visceral leishmaniasis. J. Mol. Biol. 2010, 396, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Abouelela, M.E.; Assaf, H.K.; Abdelhamid, R.A.; Elkhyat, E.S.; Sayed, A.M.; Oszako, T.; Belbahri, L.; El Zowalaty, A.E.; Abdelkader, M.S.A. Identification of potential SARS-CoV-2 main protease and spike protein inhibitors from the genus aloe: An in silico study for drug development. Molecules 2021, 26, 1767. [Google Scholar] [CrossRef] [PubMed]
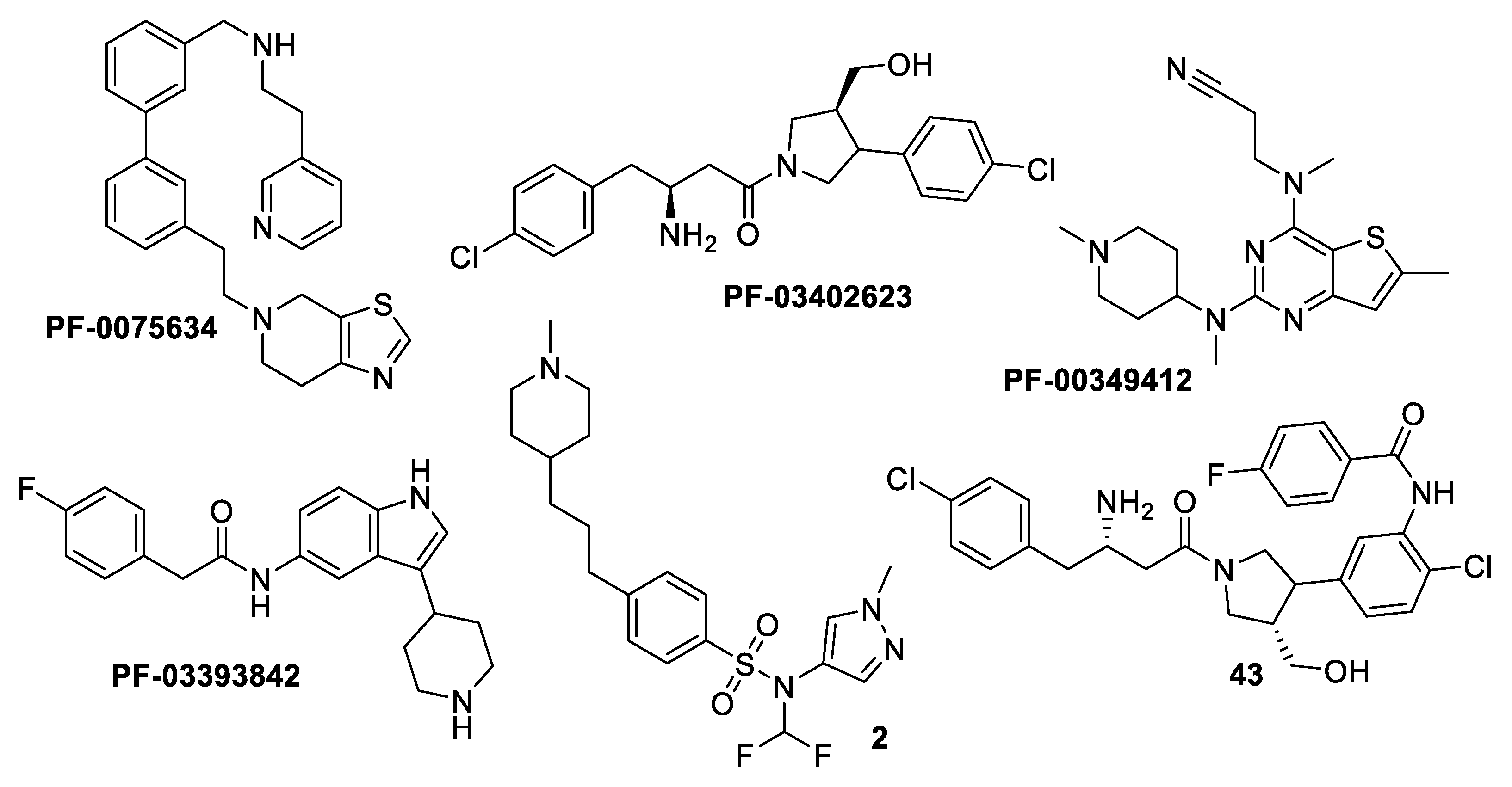
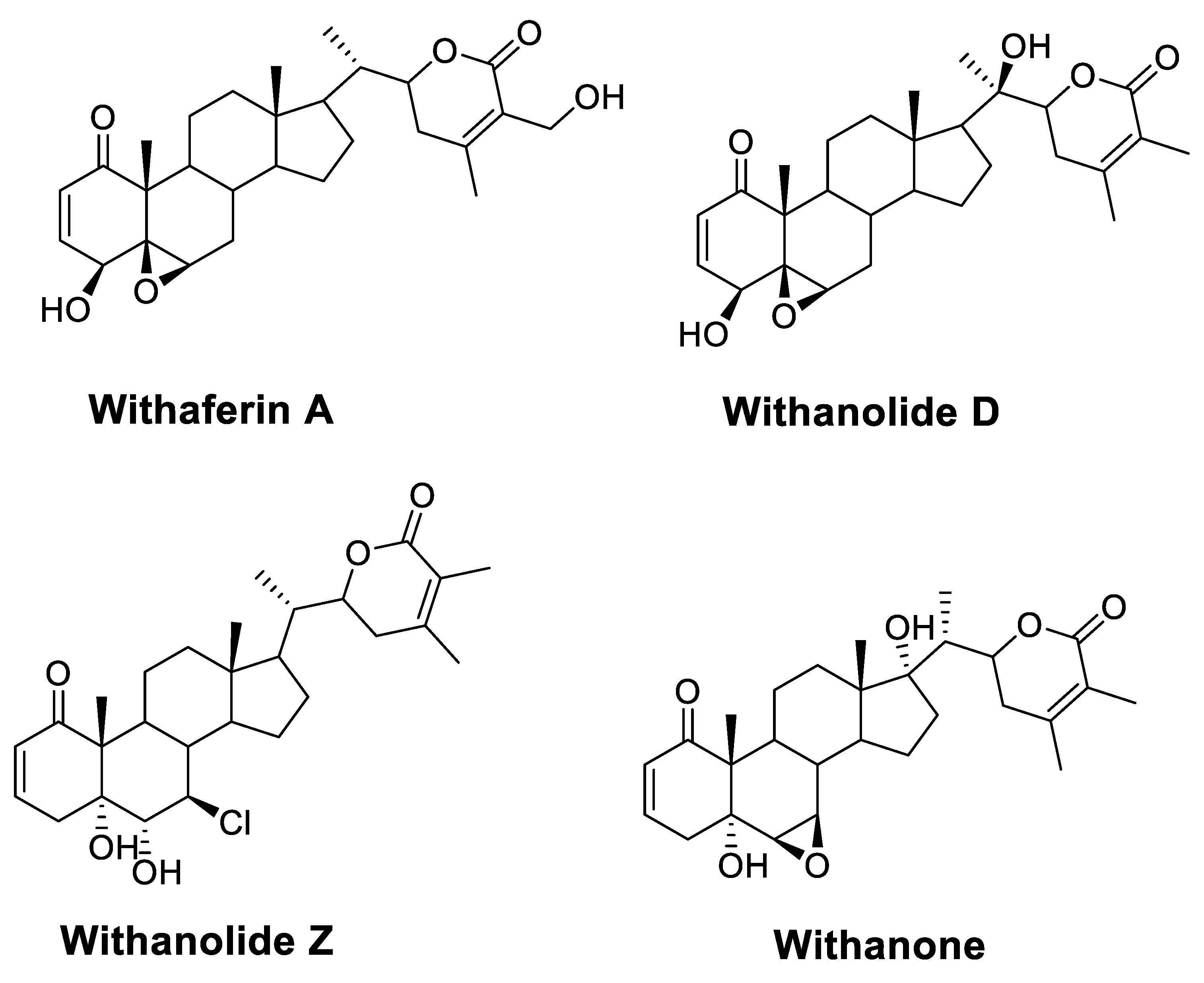
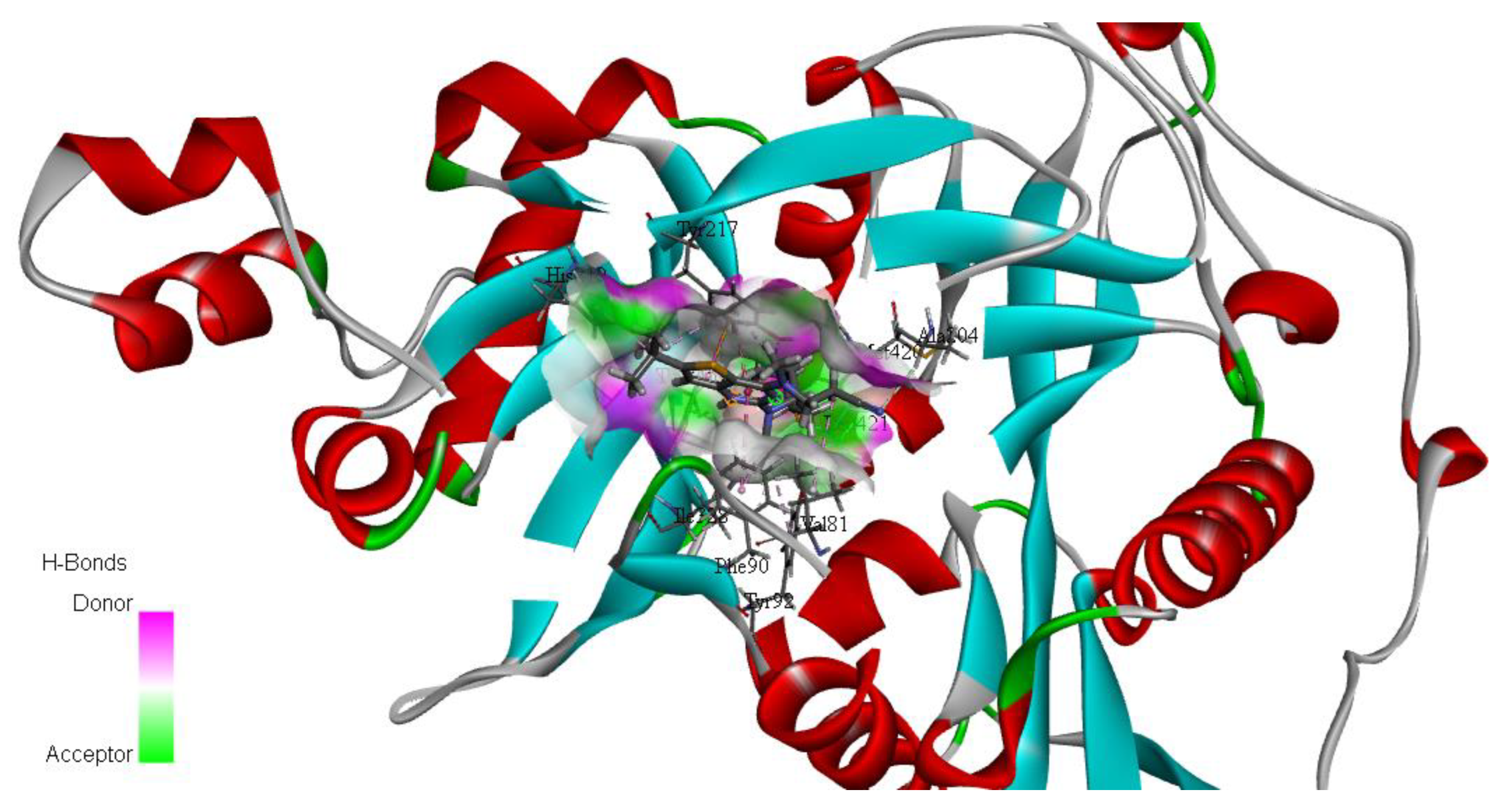


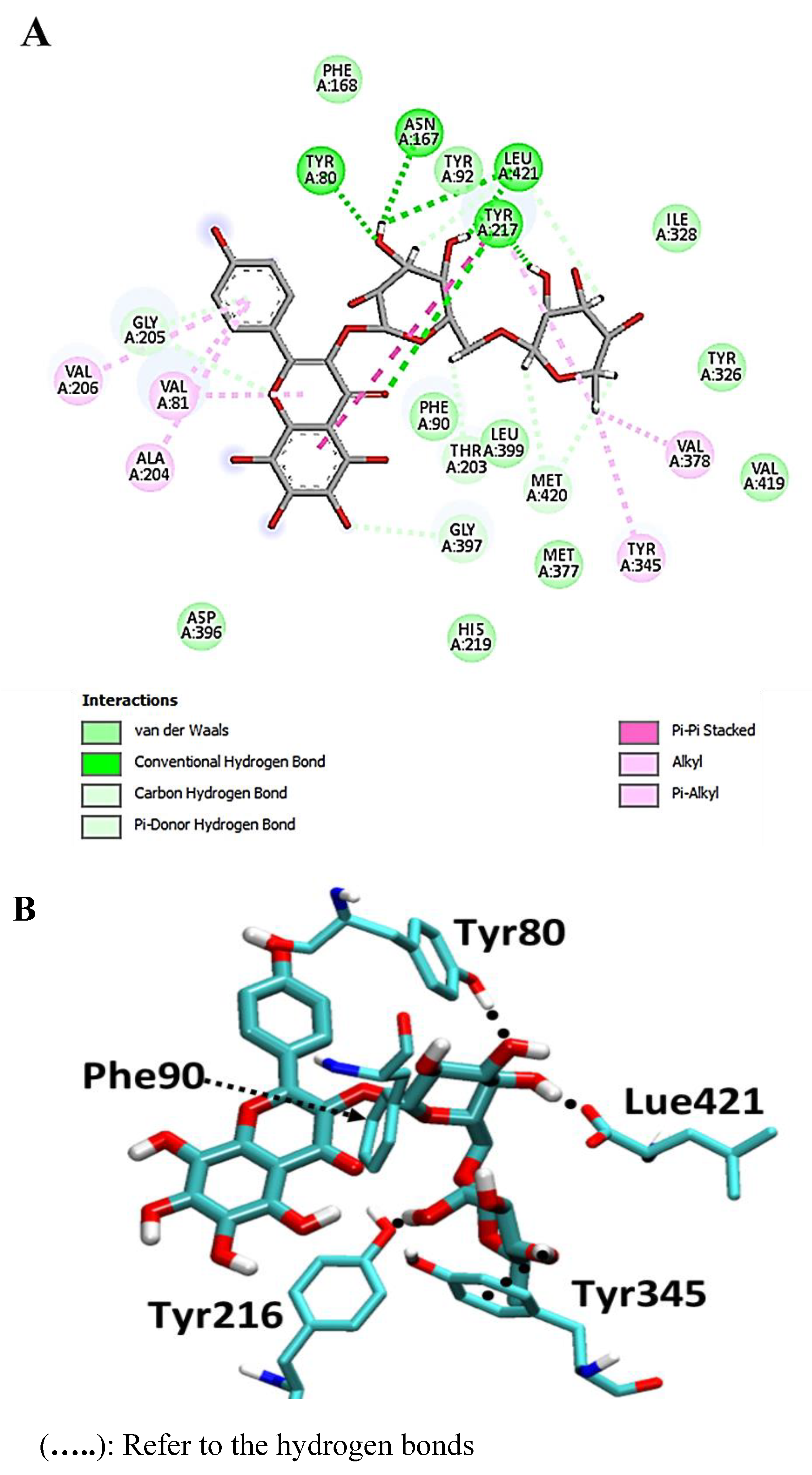
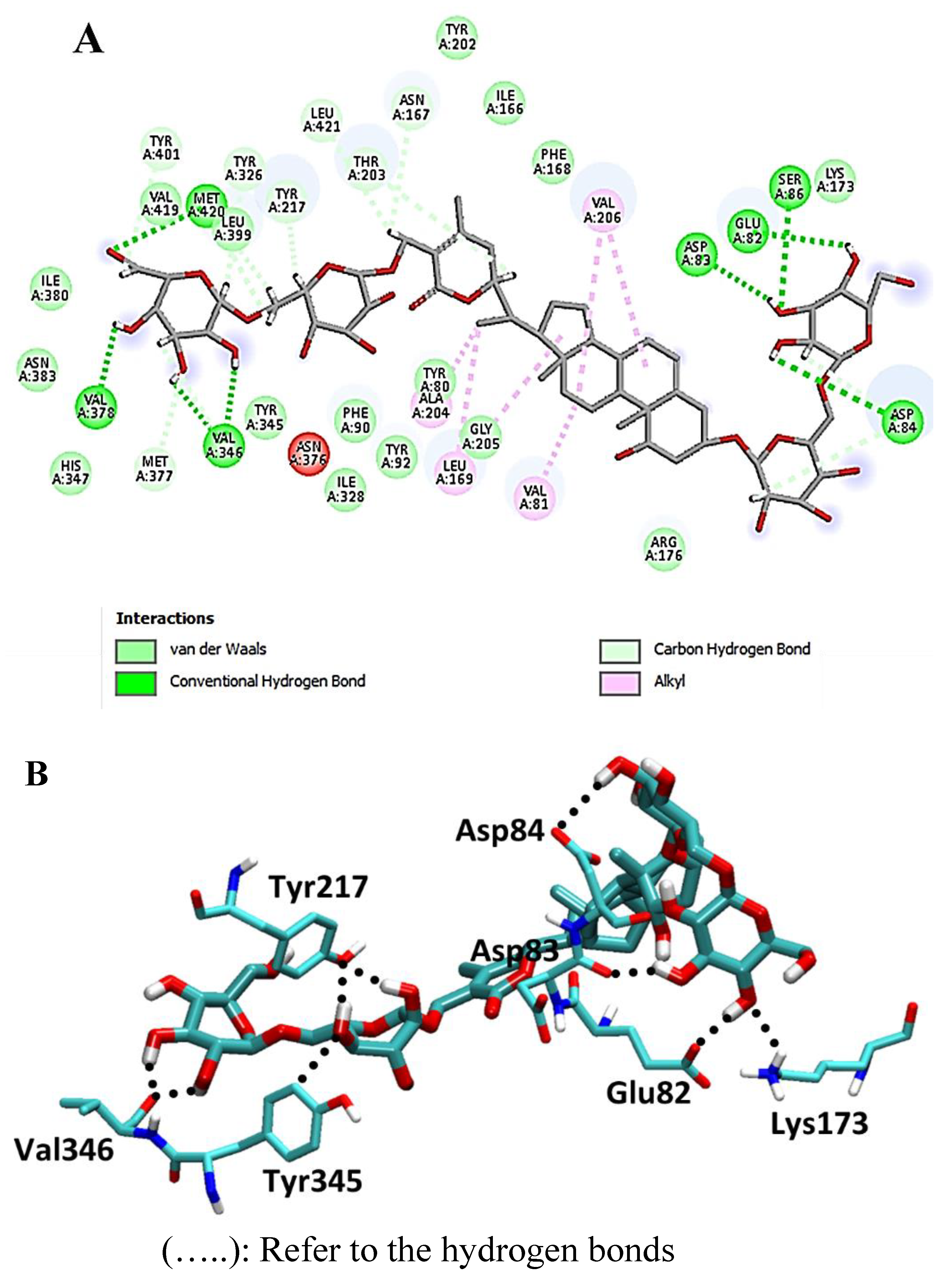
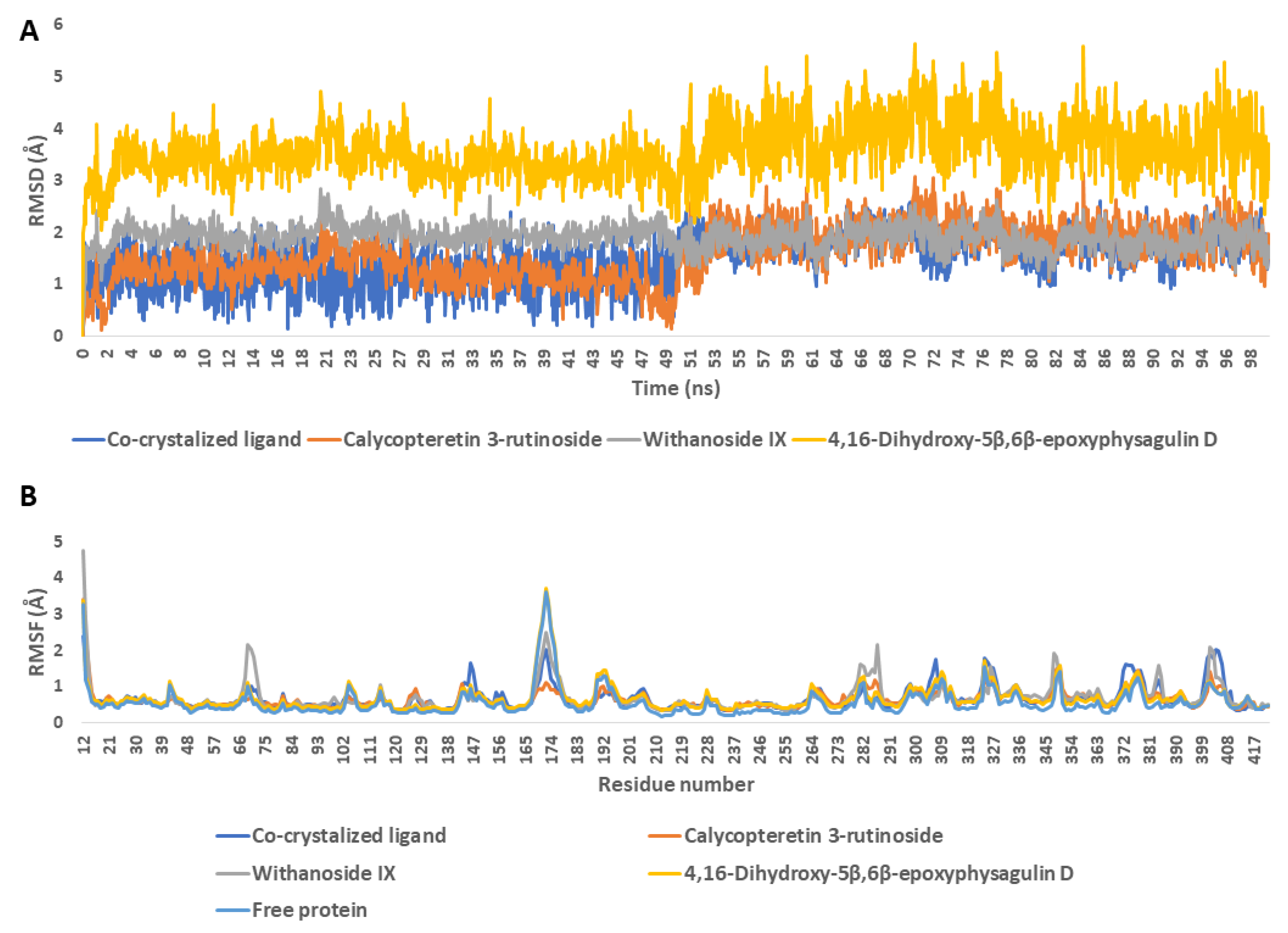
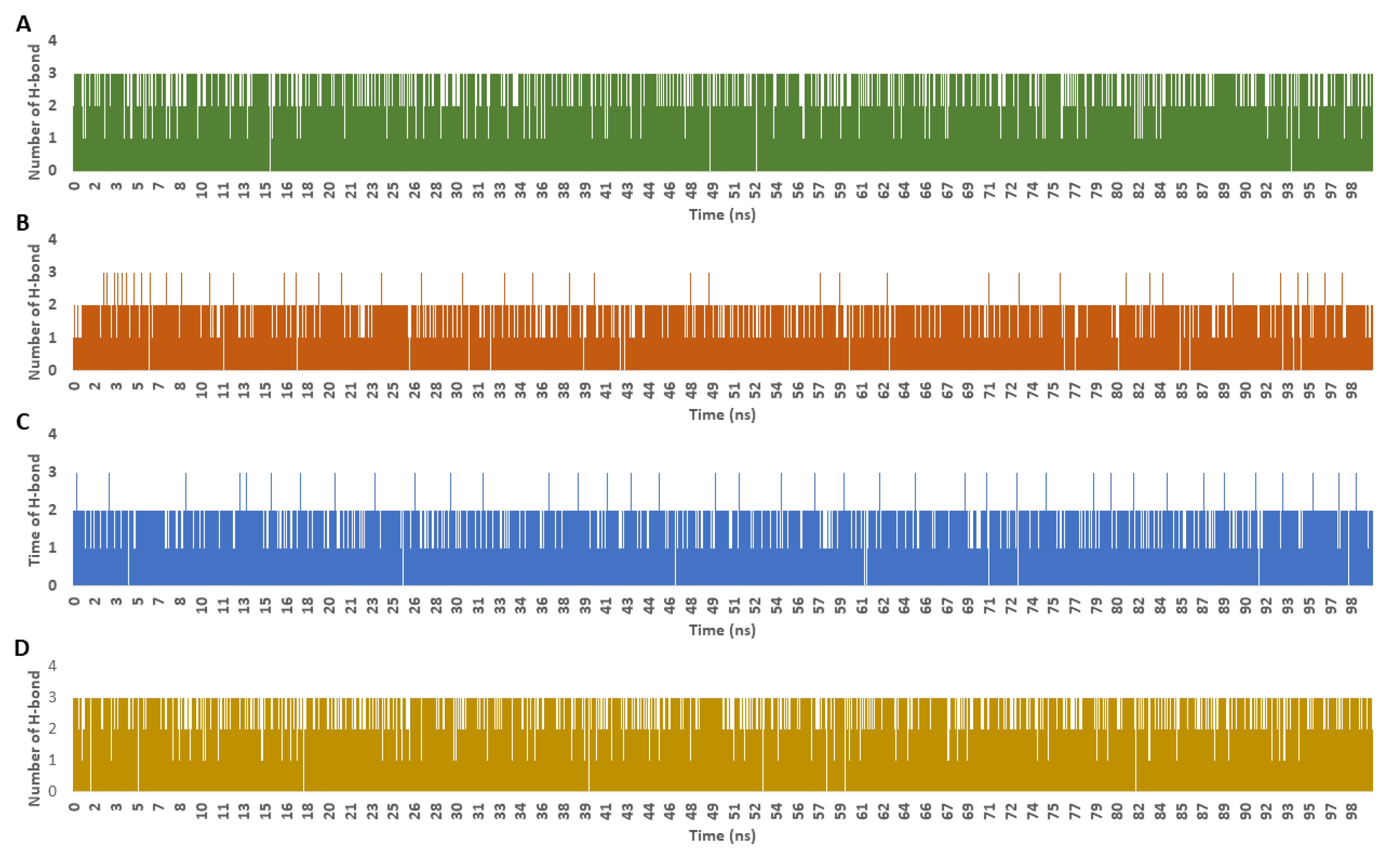
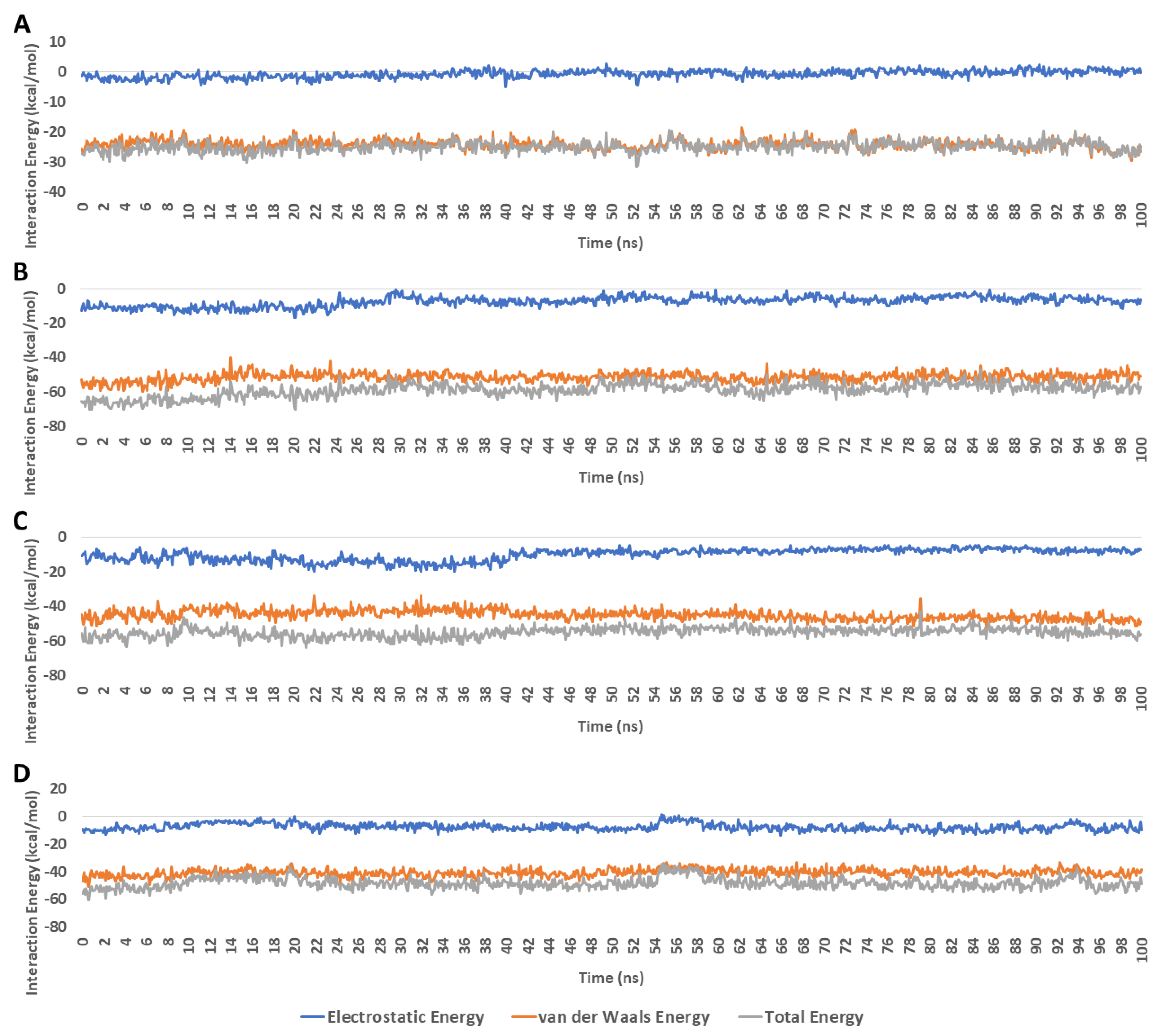
| Drug | Mode of Action | |
|---|---|---|
 Sodium stibogluconate Sodium stibogluconate | 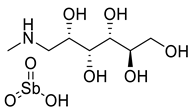 Meglumine antimoniate Meglumine antimoniate | Inhibit the parasite’s glycolysis and fatty acids β-oxidation |
Amphotericin B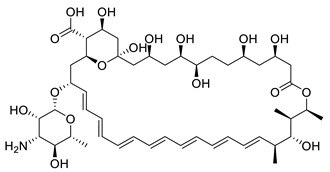 | Binds the membrane sterols of the parasite and alters its permeability to K+ and Mg2+ selectively | |
Pentamidine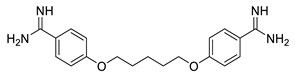 | Interferes with DNA synthesis and modifies the morphology of the kinetoplast | |
Miltefosine | Associated with leishmanial alkyl-lipid metabolism and phospholipid biosynthesis | |
Paromomycin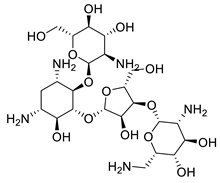 | Inhibits the protein biosynthesis in sensitive Leishmania parasites | |
| No. | Name | Pose Score (kcal/mol) | Reference for Compound Isolation |
|---|---|---|---|
| Alkaloids | |||
| 1 | Somniferine | −16.4 | [43] |
| 2 | D-α-Aminoadipic acid | −13.8 | [44] |
| 3 | Anaferine | −12.6 | [45] |
| 4 | Anahygrine | −11.2 | [45] |
| 5 | Tropine | −11.2 | [46] |
| 6 | Cuscohygrine | −10.7 | [45] |
| 7 | Isopelletierine | −10.4 | [47] |
| 8 | Putrescine | −10.0 | [48] |
| 9 | γ-Aminobutyric acid | −9.6 | [44] |
| 10 | Withasomnine | −9.0 | [49] |
| Phenolic compounds | |||
| 1 | Calycopteretin-3-rutinoside * | −23.3 | [50] |
| 2 | N-trans-feruloyl-methoxytyramine | −16.9 | [51] |
| 3 | Naringenin | −15.8 | [44] |
| 4 | Quercetin | −15.7 | [44] |
| 5 | Kaempferol | −15.4 | [44] |
| 6 | Catechin | −14.4 | [44] |
| 7 | Withaninsam A | −13.5 | [52] |
| 8 | Butein | −13.1 | [53] |
| 9 | Withaninsam B | −12.6 | [52] |
| 10 | Vanillic acid | −12.4 | [54] |
| 11 | Syringic acid | −12.2 | [55] |
| 12 | Acetosyringone | −12.0 | [56] |
| 13 | Aesculetin | −11.7 | [57] |
| 14 | Podocarpic acid | −10.3 | [44] |
| 15 | P-Coumaric acid | −9.1 | [58] |
| Sterols | |||
| 1 | 3β-Stigmasta-5,24-dien-3-ol | −13.6 | [59] |
| 2 | Campesterol | −13.6 | [44] |
| 3 | Stigmasterol acetate | −13.5 | [60] |
| 4 | β-Sitosterol oleate | −13.3 | [60] |
| 5 | β-Sitosterol | −11.7 | [61] |
| 6 | Cholesterol | −11.6 | [44] |
| 7 | Stigmasterol | −11.3 | [61] |
| 8 | Brassicasterol | −11.1 | [44] |
| 9 | Crinosterol | −11.0 | [60] |
| 10 | 3β-Ergosta-5,24-dien-3-ol | −10.5 | [59] |
| Withanones | |||
| 1 | Isowithanone | −15.6 | [62] |
| 2 | 27-Hydroxywithanone | −15.1 | [63] |
| 3 | 4α-Hydroxywithanone | −15.1 | [57] |
| 4 | 2,3-Dihydro-3β-hydroxywithanone | −15.0 | [64] |
| 5 | Withanone | −14.1 | [65] |
| 6 | 14β-Hydroxywithanone | −13.8 | [66] |
| Chloride containing withanolides | |||
| 1 | Withanolide C | −15.7 | [67] |
| 2 | 4-Deoxyphysalolactone | −14.1 | [67] |
| 3 | (4β,5β,6α,22R) 5-Chloro-4,6,27-trihydroxy-1-oxowitha-2,24-dienolide 27-Acetate | −12.1 | [68] |
| 4 | Withanolide D chlorohydrin | −12.1 | [69] |
| 5 | 6α-Chloro-5β,17α-dihydroxywithaferin A | −11.8 | [70] |
| 6 | Withanolide Z | −11.7 | [28] |
| Sulfur-containing withanolides | |||
| 1 | Withanolide sulfoxide | −17.7 | [71] |
| 2 | 5α,17α-Dihydroxy-6α,7α-epoxy-1-oxo-3β-O-sulfate-witha-24-enolide | −15.4 | [72] |
| 3 | 2,3-Dihydrowithanone-3β-O-sulfate | −14.6 | [73] |
| 4 | 2,3-Dihydrowithaferin A-3β-O-sulfate | −14.2 | [73] |
| 5 | Ashwagandhanolide | −14.2 | [74] |
| Withanamides | |||
| 1 | Withanamide F * | −18.4 | [75] |
| 2 | Withanamide H | −17.3 | [75] |
| 3 | Withanamide E | −16.5 | [75] |
| 4 | Withanamide C | −15.7 | [75] |
| 5 | Withanamide G | −15.5 | [75] |
| 6 | Withanamide B | −15.4 | [75] |
| 7 | Withanamide A | −15.3 | [75] |
| 8 | Withanamide D | −15.0 | [75] |
| 9 | Withanamide I | −14.2 | [75] |
| Withanolides | |||
| 1 | Withanolide A * | −18.7 | [66] |
| 2 | (4β,5β,6β,17α,22R) 5,6-Epoxy-4,17,27-trihydroxy-1-oxowitha-2,24-dienolide * | −18.5 | [66] |
| 3 | Somniferanolide | −18.0 | [76] |
| 4 | Withanolide H | −17.7 | [77] |
| 5 | 17-Isowithanolide E | −17.6 | [78] |
| 6 | Withanolide K | −17.4 | [79] |
| 7 | (20R,22R)14,20-Dihydroxy-1-oxowitha-2,4,6,24-tetraenolide | −17.4 | [66] |
| 8 | Withacoagulin I | −17.3 | [44] |
| 9 | 3α-(Uracil-1-yl)-2,3-dihydrowithaferin A | −17.1 | [80] |
| 10 | 14,17-Dihydroxywithanolide R | −17.0 | [57] |
| 11 | 27-Hydroxywithanolide D | −16.6 | [81] |
| 12 | Withanolide D | −16.6 | [82] |
| 13 | 24,25-Dihydro-27-desoxywithaferin A | −16.4 | [83] |
| 14 | Somniwithanolide | −16.3 | [76] |
| 15 | Withanolide S | −16.1 | [82] |
| 16 | 5,6:14,15-Diepoxy-4,27-dihydroxy-1-oxowitha-2,24-dienolide | −16.0 | [68] |
| 17 | (3α,4β,5β,6α,22R) 3,6-Epoxy-4,5,27-trihydroxy-1-oxowith-24-enolide | −15.9 | [84] |
| 18 | 3β-(Uracil-1-yl)-2,3-dihydrowithaferin A | −15.7 | [80] |
| 19 | Tubocapsanolide F | −15.6 | [85] |
| 20 | 4-Hydroxywithanolide E | −15.6 | [86] |
| 21 | Withanolide E | −15.4 | [79] |
| 22 | (3β,5α,6α,7α,17α,22R) 6,7-Epoxy-3,5,17-trihydroxy-1-oxowith-24-enolide | −15.4 | [72] |
| 23 | Quresimine B | −15.3 | [87] |
| 24 | Sominolide | −15.3 | [88] |
| 25 | Withanolide Ws 1 | −15.3 | [89] |
| 26 | Withanolide L | −15.2 | [79] |
| 27 | Withasomniferol C | −15.2 | [90] |
| 28 | Withacoagin | −15.2 | [91] |
| 29 | Somniferawithanolide | −15. 2 | [76] |
| 30 | 6,7-Epoxy-5,23-dihydroxy-1-oxowitha-2,24-Dienolide | −15.0 | [92] |
| 31 | Withaoxylactone | −14.8 | [93] |
| 32 | Dihydrowithaferin A | −14.8 | [63] |
| 33 | Withanolide J | −14.7 | [79] |
| 34 | 5-Deoxywithanolide R | −14.7 | [94] |
| 35 | Withanolide I | −14.7 | [79] |
| 36 | 17α-Hydroxywithanolide D | −14.5 | [81] |
| 37 | Withasomnilide | −14.4 | [76] |
| 38 | (3β,5α,6α,7α,20R,22R) 6,7-Epoxy-3,5,20-trihydroxy-1-oxowith-24-enolide | −14.4 | [95] |
| 39 | Withanolide G | −14.3 | [79] |
| 40 | 3β-O-Butyl-2,3-dihydrowithaferin A | −14. 3 | [80] |
| 41 | 27-Deoxywithaferin A | −14.3 | [96] |
| 42 | Pubesenolide (sominone) | −14.3 | [97] |
| 43 | Quresimine A | −14.2 | [93] |
| 44 | 27-Hydroxywithanolide B | −14.2 | [28] |
| 45 | Withanolide B | −14.2 | [98] |
| 46 | Withasomniferanolide | −14.1 | [76] |
| 47 | Somnifericin | −14.1 | [99] |
| 48 | 20-Deoxywithanolide D | −14.1 | [100] |
| 49 | (5α,6α,7α,16β,17(20)E) 6,7-Epoxy-5,16-dihydroxy-1-oxowitha-2,17(20),24-trienolide 16-acetate | −14.1 | [98] |
| 50 | Withanolide M | −14.0 | [79] |
| 51 | Withanolide O | −14.0 | [101] |
| 52 | Dunawithagenin | −13.9 | [102] |
| 53 | 5,6-Epoxy-4-hydroxy-1-oxowitha-2,16,24-trienolide | −13.9 | [72] |
| 54 | Withanolide U | −13.9 | [101] |
| 55 | (5α,17αOH,22R) 5,17-Dihydroxy-1-oxowitha-2,6,24-trienolide | −13.7 | [103] |
| 56 | 4-Deoxywithaperuvin | −13.7 | [104] |
| 57 | 5,6-Epoxy-20-hydroxy-1,4-dioxowitha-2,24-dienolide | −13.7 | [105] |
| 58 | 17-Hydroxywithaferin A | −13.6 | [106] |
| 59 | 2,3-Dehydrosomnifericin | −13.5 | [107] |
| 60 | 5,6-Epoxy-4-hydroxy-1-oxowitha-2,14,24-trienolide | −13.5 | [108] |
| 61 | Withanolide Q | −13.5 | [92] |
| 62 | Withasomniferin A | −13.5 | [94] |
| 63 | (14α,20R,22R)14,20-Dihydroxy-1-oxowitha-2,5,16,24-tetraenolide | −13.5 | [109] |
| 64 | 3β-(Adenin-9-yl)-2,3-dihydrowithaferin A | −13.4 | [80] |
| 65 | Withanolide F | −13.4 | [79] |
| 66 | Withanolide T | −13.3 | [66] |
| 67 | Withanolide Y | −13.3 | [110] |
| 68 | Withanolide N | −13.3 | [82] |
| 69 | Withacoagulin G | −13.2 | [44] |
| 70 | (4β,5β,6β,20R,22R) 5,6-Epoxy-4,20-dihydroxy-1-oxowith-24-enolide | −13.2 | [105] |
| 71 | 14α-Hydroxywithanolide D | −13.2 | [111] |
| 72 | 4-Dimethyloxocyclopropyl-2,3-dihydrowithaferin A | −13.1 | [83] |
| 73 | 24,25-Dihydrowithanolide D | −13.1 | [71] |
| 74 | Withanolide P | −12.8 | [81] |
| 75 | 5,6-Epoxy-20-hydroxy-1,4-dioxowith-2-Enolide | −12.6 | [112] |
| 76 | Withasomniferol B | −12.5 | [113] |
| 77 | Withaferin A | −12.5 | [114] |
| 78 | Withasomniferol A | −12.5 | [113] |
| 79 | 5,6-Epoxy-4,20-Dihydroxy-3-methoxy-1-oxowithanolide | −12.2 | [115] |
| 80 | 27-Deoxy-14-hydroxywithaferin A | −12.2 | [116] |
| 81 | Withanolide R | −11.9 | [92] |
| 82 | 27-Hydroxywithanolide I | −11.7 | [102] |
| 83 | Viscosalactone B | −11.6 | [117] |
| 84 | 5-Ethoxy-6,14,17,20-tetrahydroxy-1-oxowitha-2,24-dienolide | −11.6 | [102] |
| 85 | Withalactone | −11.5 | [93] |
| 86 | (1α,3β,5α,6α,7α,20S,22R) 6,7-Epoxy-1,3,5-trihydroxywith-24-enolide | −11.3 | [118] |
| 87 | Withasomidienone | −10.1 | [119] |
| Withanosides | |||
| 1 | 4,16-Dihydroxy-5β,6β-epoxyphysagulin D * | −24.0 | [83] |
| 2 | Withanoside IX * | −22.2 | [84] |
| 3 | Physagulin D (1→6)-β-D-glucopyranosyl-(1→4)-β-D-glucopyranoside * | −22.1 | [83] |
| 4 | Withanoside VIII | −20.5 | [84] |
| 5 | Withanoside X * | −19.9 | [84] |
| 6 | Withanoside II * | −19.6 | [120] |
| 7 | Withanoside IV | −19.2 | [120] |
| 8 | 24,25-Dihydrowithanoside VI * | −18.5 | [75] |
| 9 | Sitoindoside IX * | −18.1 | [121] |
| 10 | Withanoside III | −17.8 | [120] |
| 11 | Withanoside VII | −17.5 | [120] |
| 12 | Withanoside V | −16.6 | [120] |
| 13 | Withanoside VI | −16.3 | [120] |
| 14 | Glucosomniferanolide | −16.1 | [90] |
| 15 | Withanoside XI | −15.7 | [84] |
| 16 | Sitoindoside VII | −14.4 | [122] |
| 17 | Sitoindoside VIII | −14.0 | [122] |
| 18 | Withanoside I | −14.0 | [120] |
| 19 | Sitoindoside X | −12.9 | [121] |
| Energy Component | Calycopteretin 3-Rutinoside | Withanoside IX | 4,16-Dihydroxy-5,6- epoxyphysagulin D | Co-Crystalized Inhibitor |
|---|---|---|---|---|
| ΔGgas | −17.98 | −24.63 | −21.45 | −28.73 |
| ΔGsolv | 9.76 | 16.15 | 11.64 | 15.44 |
| ΔGTotal | −8.21 | −8.47 | −9.80 | −13.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orabi, M.A.A.; Alshahrani, M.M.; Sayed, A.M.; Abouelela, M.E.; Shaaban, K.A.; Abdel-Sattar, E.-S. Identification of Potential Leishmania N-Myristoyltransferase Inhibitors from Withania somnifera (L.) Dunal: A Molecular Docking and Molecular Dynamics Investigation. Metabolites 2023, 13, 93. https://doi.org/10.3390/metabo13010093
Orabi MAA, Alshahrani MM, Sayed AM, Abouelela ME, Shaaban KA, Abdel-Sattar E-S. Identification of Potential Leishmania N-Myristoyltransferase Inhibitors from Withania somnifera (L.) Dunal: A Molecular Docking and Molecular Dynamics Investigation. Metabolites. 2023; 13(1):93. https://doi.org/10.3390/metabo13010093
Chicago/Turabian StyleOrabi, Mohamed A. A., Mohammed Merae Alshahrani, Ahmed M. Sayed, Mohamed E. Abouelela, Khaled A. Shaaban, and El-Shaymaa Abdel-Sattar. 2023. "Identification of Potential Leishmania N-Myristoyltransferase Inhibitors from Withania somnifera (L.) Dunal: A Molecular Docking and Molecular Dynamics Investigation" Metabolites 13, no. 1: 93. https://doi.org/10.3390/metabo13010093
APA StyleOrabi, M. A. A., Alshahrani, M. M., Sayed, A. M., Abouelela, M. E., Shaaban, K. A., & Abdel-Sattar, E.-S. (2023). Identification of Potential Leishmania N-Myristoyltransferase Inhibitors from Withania somnifera (L.) Dunal: A Molecular Docking and Molecular Dynamics Investigation. Metabolites, 13(1), 93. https://doi.org/10.3390/metabo13010093










