Current State and Future Perspectives on Personalized Metabolomics
Abstract
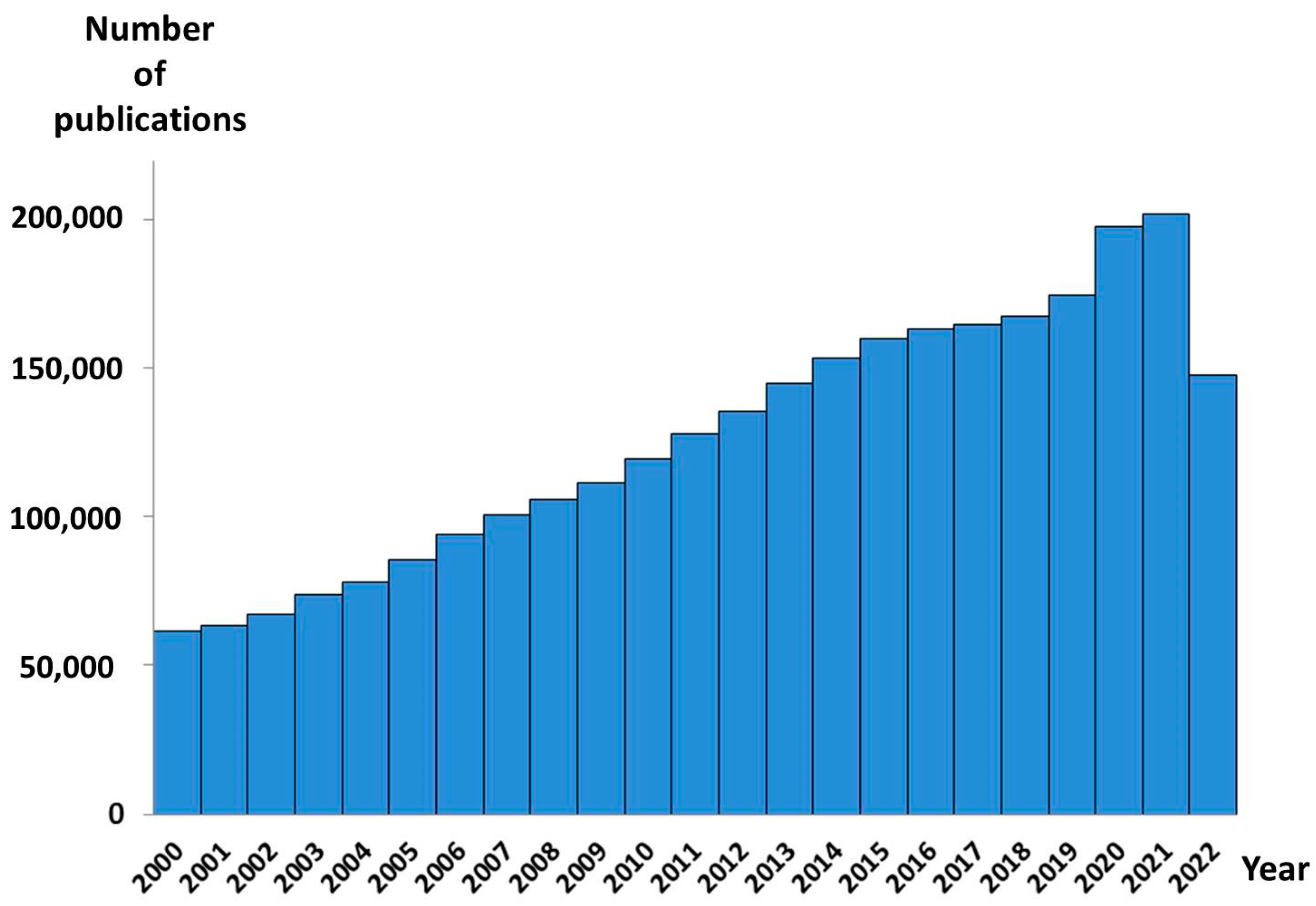
1. Introduction
2. The Bottle Necks of Personalized Metabolomics
2.1. Preanalitical and Analytical Methods
2.2. Data Processing and Interpretation
2.3. Data Interpretation for the End-Users
3. Possible Ways of a Personalized Metabolomics Implementation
3.1. Multi-Omics Tests
3.2. Laboratory Developed Tests
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sagner, M.; McNeil, A.; Puska, P.; Auffray, C.; Price, N.D.; Hood, L.; Lavie, C.J.; Han, Z.G.; Chen, Z.; Brahmachari, S.K.; et al. The P4 Health Spectrum—A Predictive, Preventive, Personalized and Participatory Continuum for Promoting Healthspan. Prog. Cardiovasc. Dis. 2017, 59, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Hood, L.; Flores, M. A personal view on systems medicine and the emergence of proactive P4 medicine: Predictive, preventive, personalized and participatory. New. Biotechnol. 2012, 29, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M. Where are all the new omics-based tests? Clin. Chem. 2014, 60, 1256–1257. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Macel, M.; Van Dam, N.M.; Keurentjes, J.J.B. Metabolomics: The chemistry between ecology and genetics. Mol. Ecol. Resour. 2010, 10, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Bar, N.; Korem, T.; Weissbrod, O.; Zeevi, D.; Rothschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weinberger, A.; Le Roy, C.I.; et al. A reference map of potential determinants for the human serum metabolome. Nature 2020, 588, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, Q.; Lu, L.; Wang, J.; Liu, D.; Liu, Z. Metabolomic Profiling of Amino Acids in Human Plasma Distinguishes Diabetic Kidney Disease From Type 2 Diabetes Mellitus. Front. Med. 2021, 8, 2342. [Google Scholar] [CrossRef]
- Di Minno, A.; Gelzo, M.; Caterino, M.; Costanzo, M.; Ruoppolo, M.; Castaldo, G. Challenges in Metabolomics-Based Tests, Biomarkers Revealed by Metabolomic Analysis, and the Promise of the Application of Metabolomics in Precision Medicine. Int. J. Mol. Sci. 2022, 23, 5213. [Google Scholar] [CrossRef] [PubMed]
- Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Mass spectrometry-based metabolomics diagnostics—Myth or reality? Expert Rev. Proteom. 2021, 18, 7–12. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Lokhov, P.G.; Trifonova, O.P.; Maslov, D.L.; Lichtenberg, S.; Balashova, E.E. Personal Metabolomics: A Global Challenge. Metabolites 2021, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhong, L. Applications of liquid chromatography-mass spectrometry based metabolomics in predictive and personalized medicine. Front. Mol. Biosci. 2022, 9, 1049016. [Google Scholar] [CrossRef] [PubMed]
- Castelli, F.A.; Rosati, G.; Moguet, C.; Fuentes, C.; Marrugo-Ramírez, J.; Lefebvre, T.; Volland, H.; Merkoçi, A.; Simon, S.; Fenaille, F.; et al. Metabolomics for personalized medicine: The input of analytical chemistry from biomarker discovery to point-of-care tests. Anal. Bioanal. Chem. 2021, 414, 759–789. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Hollywood, K.A.; Goodacre, R. Metabolomics for the masses: The future of metabolomics in a personalized world. New Horiz. Transl. Med. 2017, 3, 294. [Google Scholar] [CrossRef]
- Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational metabolomics: Current challenges and future opportunities. Metabolites 2019, 9, 108. [Google Scholar] [CrossRef]
- Zhang, X.W.; Li, Q.H.; Xu, Z.D.; Dou, J.J. Mass spectrometry-based metabolomics in health and medical science: A systematic review. RSC Adv. 2020, 10, 3092–3104. [Google Scholar] [CrossRef]
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.D.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef]
- López-López, Á.; López-Gonzálvez, Á.; Barker-Tejeda, T.C.; Barbas, C. A review of validated biomarkers obtained through metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef]
- Lichtenberg, S.; Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G.; Maslov, O.P.; Balashova, D.L.; Lokhov, E.E.; Metabolomic, P.G. Metabolomic Laboratory-Developed Tests: Current Status and Perspectives. Metabolites 2021, 11, 423. [Google Scholar] [CrossRef]
- Sansone, S.A.; Fan, T.; Goodacre, R.; Griffin, J.L.; Hardy, N.W.; Kaddurah-Daouk, R.; Kristal, B.S.; Lindon, J.; Mendes, P.; Morrison, N.; et al. The metabolomics standards initiative. Nat. Biotechnol. 2007, 25, 846–848. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Alexander, A.E.; Ae, A.; Ae, D.B.; Ae, M.H.B.; Beger, R.; Daykin, C.A.; Teresa, A.E.; Fan, W.-M.; Oliver, A.E.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, J.A.; Brennan, L.; Broadhurst, D.; Fiehn, O.; Cascante, M.; Dunn, W.B.; Schmidt, M.A.; Velagapudi, V. Preanalytical processing and biobanking procedures of biological samples for metabolomics research: A white paper, community perspective (for “Precision medicine and pharmacometabolomics task group”—The metabolomics society initiative). Clin. Chem. 2018, 64, 1158–1182. [Google Scholar] [CrossRef]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 4. [Google Scholar] [CrossRef]
- Brown, M.; Dunn, W.B.; Ellis, D.I.; Goodacre, R.; Handl, J.; Knowles, J.D.; O’Hagan, S.; Spasić, I.; Kell, D.B. A metabolome pipeline: From concept to data to knowledge. Metabolomics 2005, 1, 39–51. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Long, N.P.; Nghi, T.D.; Kang, Y.P.; Anh, N.H.; Kim, H.M.; Park, S.K.; Kwon, S.W. Toward a Standardized Strategy of Clinical Metabolomics for the Advancement of Precision Medicine. Metabolites 2020, 10, 51. [Google Scholar] [CrossRef]
- Lippi, G.; Betsou, F.; Cadamuro, J.; Cornes, M.; Fleischhacker, M.; Fruekilde, P.; Neumaier, M.; Nybo, M.; Padoan, A.; Plebani, M.; et al. Preanalytical challenges-time for solutions. Clin. Chem. Lab. Med. 2019, 57, 974–981. [Google Scholar] [CrossRef]
- Marciano, D.P.; Snyder, M.P. Personalized metabolomics. Methods Mol. Biol. 2019, 1978, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Desaire, H. How (Not) to Generate a Highly Predictive Biomarker Panel Using Machine Learning. J. Proteome Res. 2022, 21, 2071–2074. [Google Scholar] [CrossRef] [PubMed]
- Bruno, C.; Patin, F.; Bocca, C.; Nadal-Desbarats, L.; Bonnier, F.; Reynier, P.; Emond, P.; Vourc’h, P.; Joseph-Delafont, K.; Corcia, P.; et al. The combination of four analytical methods to explore skeletal muscle metabolomics: Better coverage of metabolic pathways or a marketing argument? J. Pharm. Biomed. Anal. 2018, 148, 273–279. [Google Scholar] [CrossRef]
- Kim, Y.M.; Heyman, H.M. Mass Spectrometry-Based Metabolomics. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- González-Riano, C.; Dudzik, D.; Garcia, A.; Gil-De-La-Fuente, A.; Gradillas, A.; Godzien, J.; López-Gonzálvez, Á; Rey-Stolle, F.; Rojo, D.; Ruperez, F.J.; et al. Recent developments along the analytical process for metabolomics workflows. Anal. Chem. 2020, 92, 203–226. [Google Scholar] [CrossRef]
- Gika, H.G.; Wilson, I.D.; Theodoridis, G.A. Omics | Metabolomics: An analytical perspective. In Encyclopedia of Analytical Science; Elsevier: Amsterdam, The Netherlands, 2019; pp. 82–89. [Google Scholar] [CrossRef]
- Lodge, S.; Nitschke, P.; Loo, R.L.; Kimhofer, T.; Bong, S.-H.; Richards, T.; Begum, S.; Spraul, M.; Schaefer, H.; Lindon, J.C.; et al. Low Volume in Vitro Diagnostic Proton NMR Spectroscopy of Human Blood Plasma for Lipoprotein and Metabolite Analysis: Application to SARS-CoV-2 Biomarkers. J. Proteome Res. 2021, 20, 1415–1423. [Google Scholar] [CrossRef]
- Khodadadi, M.; Pourfarzam, M. A review of strategies for untargeted urinary metabolomic analysis using gas chromatography-mass spectrometry. Metabolomics 2020, 16, 66. [Google Scholar] [CrossRef]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC–MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef]
- Wang, S.; Blair, I.A.; Mesaros, C. Analytical Methods for Mass Spectrometry-Based Metabolomics Studies. Adv. Exp. Med. Biol. 2019, 1140, 635–647. [Google Scholar] [CrossRef]
- Bhatia, A.; Sarma, S.J.; Lei, Z.; Sumner, L.W. UHPLC-QTOF-MS/MS-SPE-NMR: A Solution to the Metabolomics Grand Challenge of Higher-Throughput, Confident Metabolite Identifications. Methods Mol. Biol. 2019, 2037, 113–133. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Sarvin, B.; Lagziel, S.; Sarvin, N.; Mukha, D.; Kumar, P.; Aizenshtein, E.; Shlomi, T. Fast and sensitive flow-injection mass spectrometry metabolomics by analyzing sample-specific ion distributions. Nat. Commun. 2020, 11, 3186. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Ponomarenko, E.A.; Archakov, A.I. Mass spectrometry-based metabolomics analysis of obese patients’ blood plasma. Int. J. Mol. Sci. 2020, 21, 568. [Google Scholar] [CrossRef] [PubMed]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef] [PubMed]
- Bartroff, J.; Song, J. Sequential Tests of Multiple Hypotheses Controlling False Discovery and Nondiscovery Rates. Seq. Anal. 2020, 39, 65–91. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Domingo-Almenara, X.; Siuzdak, G. Metabolomics Data Processing Using XCMS. Methods Mol. Biol. 2020, 2104, 11–24. [Google Scholar] [CrossRef]
- Hsu, Y.H.H.; Churchhouse, C.; Pers, T.H.; Mercader, J.M.; Metspalu, A.; Fischer, K.; Fortney, K.; Morgen, E.K.; Gonzalez, C.; Gonzalez, M.E.; et al. PAIRUP-MS: Pathway analysis and imputation to relate unknowns in profiles from mass spectrometry-based metabolite data. PLoS Comput. Biol. 2019, 15, e1006734. [Google Scholar] [CrossRef]
- Collins, S.L.; Koo, I.; Peters, J.M.; Smith, P.B.; Patterson, A.D. Current Challenges and Recent Developments in Mass Spectrometry-Based Metabolomics. Annu. Rev. Anal. Chem. 2021, 14, 467–487. [Google Scholar] [CrossRef]
- Wishart, D.; Arndt, D.; Pon, A.; Sajed, T.; Guo, A.C.; Djoumbou, Y.; Knox, C.; Wilson, M.; Liang, Y.; Grant, J.; et al. T3DB: The toxic exposome database. Nucleic Acids Res. 2015, 43, D928–D934. [Google Scholar] [CrossRef]
- Montenegro-Burke, J.R.; Guijas, C.; Siuzdak, G. METLIN: A Tandem Mass Spectral Library of Standards. Methods Mol. Biol. 2020, 2104, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Frainay, C.; Schymanski, E.L.; Neumann, S.; Merlet, B.; Salek, R.M.; Jourdan, F.; Yanes, O. Mind the Gap: Mapping Mass Spectral Databases in Genome-Scale Metabolic Networks Reveals Poorly Covered Areas. Metabolites 2018, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Damont, A.; Olivier, M.F.; Warnet, A.; Lyan, B.; Pujos-Guillot, E.; Jamin, E.L.; Debrauwer, L.; Bernillon, S.; Junot, C.; Tabet, J.C.; et al. Proposal for a chemically consistent way to annotate ions arising from the analysis of reference compounds under ESI conditions: A prerequisite to proper mass spectral database constitution in metabolomics. J. Mass Spectrom. 2019, 54, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Böcker, S. Searching molecular structure databases using tandem MS data: Are we there yet? Curr. Opin. Chem. Biol. 2017, 36, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nash, W.J.; Dunn, W.B. From mass to metabolite in human untargeted metabolomics: Recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data. TrAC Trends Anal. Chem. 2019, 120, 115324. [Google Scholar] [CrossRef]
- Yi, Z.; Zhu, Z.J. Overview of Tandem Mass Spectral and Metabolite Databases for Metabolite Identification in Metabolomics. Methods Mol. Biol. 2020, 2104, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 2018, 37, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Yurkovich, J.T.; Hood, L. Blood Is a Window into Health and Disease. Clin. Chem. 2019, 65, 1204–1206. [Google Scholar] [CrossRef]
- Zukunft, S.; Prehn, C.; Röhring, C.; Möller, G.; Hrabě de Angelis, M.; Adamski, J.; Tokarz, J. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics 2018, 14, 18. [Google Scholar] [CrossRef]
- Saoi, M.; Britz-Mckibbin, P. New Advances in Tissue Metabolomics: A Review. Metabolites 2021, 11, 672. [Google Scholar] [CrossRef]
- Considine, E.C. The search for clinically useful biomarkers of complex disease: A data analysis perspective. Metabolites 2019, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [PubMed]
- Lilley, L.M.; Sanche, S.; Moore, S.C.; Salemi, M.R.; Vu, D.; Iyer, S.; Hengartner, N.W.; Mukundan, H. Methods to capture proteomic and metabolomic signatures from cerebrospinal fluid and serum of healthy individuals. Sci. Rep. 2022, 12, 1–12. [Google Scholar] [CrossRef]
- Brito, F.; Curcio, H.F.Q.; da Silva Fidalgo, T.K. Periodontal disease metabolomics signatures from different biofluids: A systematic review. Metabolomics 2022, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.A.; Bossuyt, P.M.M. Waste, leaks, and failures in the biomarker pipeline. Clin. Chem. 2017, 63, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Perez De Souza, L.; Alseekh, S.; Brotman, Y.; Fernie, A.R. Network-based strategies in metabolomics data analysis and interpretation: From molecular networking to biological interpretation. Expert Rev. Proteom. 2020, 17, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Gummesson, A.; Zhong, W.; Koistinen, I.S.; Lakshmikanth, T.; Olsson, L.M.; Boulund, F.; Neiman, M.; Stenlund, H.; Hellström, C.; et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat. Commun. 2020, 11, 4487. [Google Scholar] [CrossRef]
- Gurke, R.; Bendes, A.; Bowes, J.; Koehm, M.; Twyman, R.M.; Barton, A.; Elewaut, D.; Goodyear, C.; Hahnefeld, L.; Hillenbrand, R.; et al. Omics and Multi-Omics Analysis for the Early Identification and Improved Outcome of Patients with Psoriatic Arthritis. Biomedicines 2022, 10, 2387. [Google Scholar] [CrossRef]
- Zheng, M.; Piermarocchi, C.; Mias, G.I. Temporal response characterization across individual multiomics profiles of prediabetic and diabetic subjects. Sci. Rep. 2022, 12, 1–13. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Chen, R.; Mias, G.I.; Li-Pook-Than, J.; Jiang, L.; Lam, H.Y.K.; Chen, R.; Miriami, E.; Karczewski, K.J.; Hariharan, M.; Dewey, F.E.; et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Schüssler-Fiorenza Rose, S.M.; Contrepois, K.; Moneghetti, K.J.; Zhou, W.; Mishra, T.; Mataraso, S.; Dagan-Rosenfeld, O.; Ganz, A.B.; Dunn, J.; Hornburg, D.; et al. A longitudinal big data approach for precision health. Nat. Med. 2019, 25, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.; Perelman, D.; Breschi, A.; Limcaoco, P.; Kellogg, R.; McLaughlin, T.; Snyder, M. Glucotypes reveal new patterns of glucose dysregulation. PLoS Biol. 2018, 16, e2005143. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, S.; Zhou, W.; Schüssler-Fiorenza Rose, S.M.; Sailani, M.R.; Contrepois, K.; Avina, M.; Ashland, M.; Brunet, A.; Snyder, M. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 2020, 26, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Hood, L.; Price, N.D. Promoting Wellness & Demystifying Disease: The 100K Project. Clin. Omi. 2015, 1, 20–23. [Google Scholar] [CrossRef]
- Price, N.D.; Magis, A.T.; Earls, J.C.; Glusman, G.; Levy, R.; Lausted, C.; McDonald, D.T.; Kusebauch, U.; Moss, C.L.; Zhou, Y.; et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 2017, 35, 747–756. [Google Scholar] [CrossRef]
- Magis, A.T.; Rappaport, N.; Conomos, M.P.; Omenn, G.S.; Lovejoy, J.C.; Hood, L.; Price, N.D. Untargeted longitudinal analysis of a wellness cohort identifies markers of metastatic cancer years prior to diagnosis. Sci. Rep. 2020, 10, 16275. [Google Scholar] [CrossRef]
- Zubair, N.; Conomos, M.P.; Hood, L.; Omenn, G.S.; Price, N.D.; Spring, B.J.; Magis, A.T.; Lovejoy, J.C. Genetic Predisposition Impacts Clinical Changes in a Lifestyle Coaching Program. Sci. Rep. 2019, 9, 6805. [Google Scholar] [CrossRef]
- Diener, C.; Dai, C.L.; Wilmanski, T.; Baloni, P.; Smith, B.; Rappaport, N.; Hood, L.; Magis, A.T.; Gibbons, S.M. Genome–microbiome interplay provides insight into the determinants of the human blood metabolome. Nat. Metab. 2022, 4, 1560–1572. [Google Scholar] [CrossRef]
- Earls, J.C.; Rappaport, N.; Heath, L.; Wilmanski, T.; Magis, A.T.; Schork, N.J.; Omenn, G.S.; Lovejoy, J.; Hood, L.; Price, N.D. Multi-Omic Biological Age Estimation and Its Correlation with Wellness and Disease Phenotypes: A Longitudinal Study of 3558 Individuals. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2019, 74, S52. [Google Scholar] [CrossRef]
- Oldoni, E.; Saunders, G.; Bietrix, F.; Garcia Bermejo, M.L.; Niehues, A.; ’t Hoen, P.A.C.; Nordlund, J.; Hajduch, M.; Scherer, A.; Kivinen, K.; et al. Tackling the translational challenges of multi-omics research in the realm of European personalised medicine: A workshop report. Front. Mol. Biosci. 2022, 9, 974799. [Google Scholar] [CrossRef] [PubMed]
- Krassowski, M.; Das, V.; Sahu, S.K.; Misra, B.B. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 2020, 11, 610798. [Google Scholar] [CrossRef] [PubMed]
- EATRIS-Plus—Flagship in Personalised Medicine—EATRIS. Available online: https://eatris.eu/projects/eatris-plus/ (accessed on 5 December 2022).
- Schreier, J.; Feeney, R.; Keeling, P. Diagnostics Reform and Harmonization of Clinical Laboratory Testing. J. Mol. Diagn. 2019, 21, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Spitzenberger, F.; Patel, J.; Gebuhr, I.; Kruttwig, K.; Safi, A.; Meisel, C. Laboratory-Developed Tests: Design of a Regulatory Strategy in Compliance with the International State-of-the-Art and the Regulation (EU) 2017/746 (EU IVDR [In Vitro Diagnostic Medical Device Regulation]). Ther. Innov. Regul. Sci. 2022, 56, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Graden, K.C.; Bennett, S.A.; Delaney, S.R.; Gill, H.E.; Willrich, M.A.V. A High-Level Overview of the Regulations Surrounding a Clinical Laboratory and Upcoming Regulatory Challenges for Laboratory Developed Tests. Lab. Med. 2021, 52, 315–328. [Google Scholar] [CrossRef]
- Genzen, J.R. Regulation of Laboratory-Developed Tests. Am. J. Clin. Pathol. 2019, 152, 122–131. [Google Scholar] [CrossRef]
- FDA Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs). Draft Guidance. Available online: https://www.fda.gov/media/89841/download (accessed on 24 November 2022).
- Centers for Medicare and Medicaid Services. Background Document on CLIA Oversight of LDTs. Available online: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/Downloads/LDT-and-CLIA_FAQs.pdf (accessed on 24 November 2022).
- FDA. The Public Health Evidence for FDA Oversight of Laboratory Developed Tests: 20 Case Studies—The Real and Potential Harms to Patients and to Public Health from Certain Laboratory Developed Tests (LDTs). Available online: http://wayback.archive-it.org/7993/20171115144712/ (accessed on 24 November 2022).
- Calvert, J.; Saber, N.; Hoffman, J.; Das, R. Machine-Learning-Based Laboratory Developed Test for the Diagnosis of Sepsis in High-Risk Patients. Diagnostics 2019, 9, 20. [Google Scholar] [CrossRef]
- Kulis-Horn, R.K.; Tiemann, C. Evaluation of a laboratory-developed test for simultaneous detection of norovirus and rotavirus by real-time RT-PCR on the Panther Fusion® system. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 103–112. [Google Scholar] [CrossRef]
- Brukner, I.; Eintracht, S.; Forgetta, V.; Papadakis, A.I.; Spatz, A.; Oughton, M. Laboratory-developed test for detection of acute Clostridium difficile infections with the capacity for quantitative sample normalization. Diagn. Microbiol. Infect. Dis. 2019, 95, 113–118. [Google Scholar] [CrossRef]
- Tinawi-Aljundi, R.; King, L.; Knuth, S.T.; Gildea, M.; Ng, C.; Kahl, J.; Dion, J.; Young, C.; Schervish, E.W.; Frontera, J.R.; et al. One-year monitoring of an oligonucleotide fluorescence in situ hybridization probe panel laboratory-developed test for bladder cancer detection. Res. Rep. Urol. 2015, 7, 49. [Google Scholar] [CrossRef][Green Version]
- Fiset, P.O.; Labbé, C.; Young, K.; Craddock, K.J.; Smith, A.C.; Tanguay, J.; Pintilie, M.; Wang, R.; Torlakovic, E.; Cheung, C.; et al. Anaplastic lymphoma kinase 5A4 immunohistochemistry as a diagnostic assay in lung cancer: A Canadian reference testing center’s results in population-based reflex testing. Cancer 2019, 125, 4043–4051. [Google Scholar] [CrossRef] [PubMed]
- Munari, E.; Zamboni, G.; Lunardi, G.; Marconi, M.; Brunelli, M.; Martignoni, G.; Netto, G.J.; Quatrini, L.; Vacca, P.; Moretta, L.; et al. PD-L1 expression in non–small cell lung cancer: Evaluation of the diagnostic accuracy of a laboratory-developed test using clone E1L3N in comparison with 22C3 and SP263 assays. Hum. Pathol. 2019, 90, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Burchard, P.R.; Abou Tayoun, A.N.; Lefferts, J.A.; Lewis, L.D.; Tsongalis, G.J.; Cervinski, M.A. Development of a rapid clinical TPMT genotyping assay. Clin. Biochem. 2014, 47, 126–129. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Maslov, D.L.; Lichtenberg, S.; Trifonova, O.P.; Balashova, E.E. Holistic Metabolomic Laboratory-Developed Test (LDT): Development and Use for the Diagnosis of Early-Stage Parkinson’s Disease. Metabolites 2020, 11, 14. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Trifonova, O.P.; Maslov, D.L.; Lichtenberg, S.; Balashova, E.E. Diagnosis of Parkinson’s Disease by A Metabolomics-Based Laboratory-Developed Test (LDT). Diagnostics 2020, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Metabolon Launches Meta UDx™ Test to Speed Diagnosis of Rare and Undiagnosed Diseases in Children and Adults—Metabolon. Available online: https://www.metabolon.com/news/meta-udx-launch/ (accessed on 24 November 2022).
- Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Evaluation of Dried Blood Spot Sampling for Clinical Metabolomics: Effects of Different Papers and Sample Storage Stability. Metabolites 2019, 9, 277. [Google Scholar] [CrossRef] [PubMed]
- The Preventative Health Company. Nightingale Health. Available online: https://nightingalehealth.com/ (accessed on 22 December 2022).
- AminoIndex®. Innovation in Action. Innovation. Ajinomoto Group Global Website—Eat Well, Live Well. Available online: https://www.ajinomoto.com/innovation/action/aminoindex (accessed on 22 December 2022).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Current State and Future Perspectives on Personalized Metabolomics. Metabolites 2023, 13, 67. https://doi.org/10.3390/metabo13010067
Trifonova OP, Maslov DL, Balashova EE, Lokhov PG. Current State and Future Perspectives on Personalized Metabolomics. Metabolites. 2023; 13(1):67. https://doi.org/10.3390/metabo13010067
Chicago/Turabian StyleTrifonova, Oxana P., Dmitry L. Maslov, Elena E. Balashova, and Petr G. Lokhov. 2023. "Current State and Future Perspectives on Personalized Metabolomics" Metabolites 13, no. 1: 67. https://doi.org/10.3390/metabo13010067
APA StyleTrifonova, O. P., Maslov, D. L., Balashova, E. E., & Lokhov, P. G. (2023). Current State and Future Perspectives on Personalized Metabolomics. Metabolites, 13(1), 67. https://doi.org/10.3390/metabo13010067








