Dietary Polyphenols in Relation to Gut Microbiota Composition in Saudi Arabian Females
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Sample Size Calculation
2.3. Anthropometric Measurements
2.4. Gut Microbiota Composition
2.4.1. DNA Extraction
2.4.2. Library Preparation and Sequencing
2.4.3. Characterization of Microbial Composition
2.5. Dietary Intake
2.6. Assessment of Polyphenol Intake
2.7. Statistical Analysis
3. Results
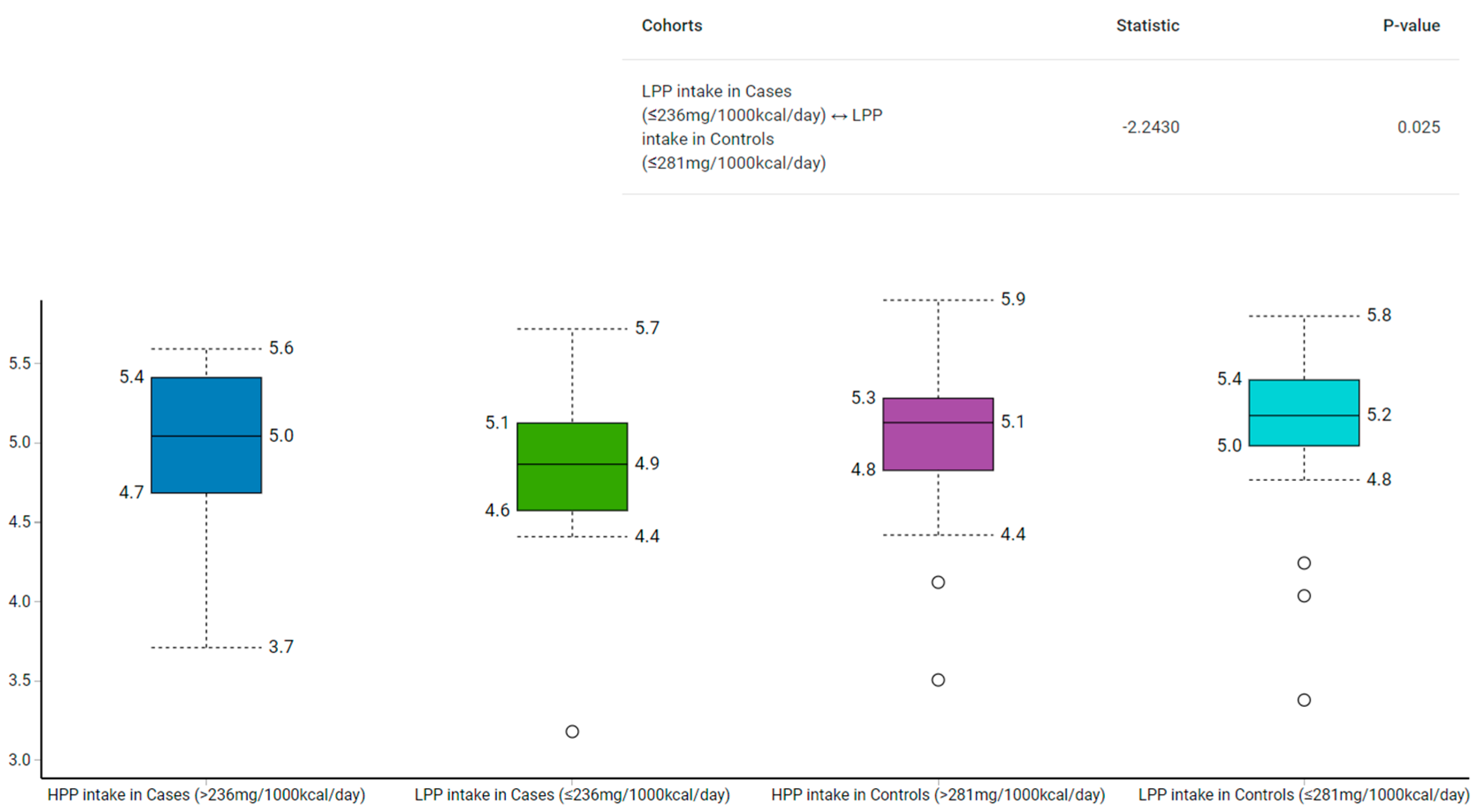
3.1. Dietary Polyphenols Intake
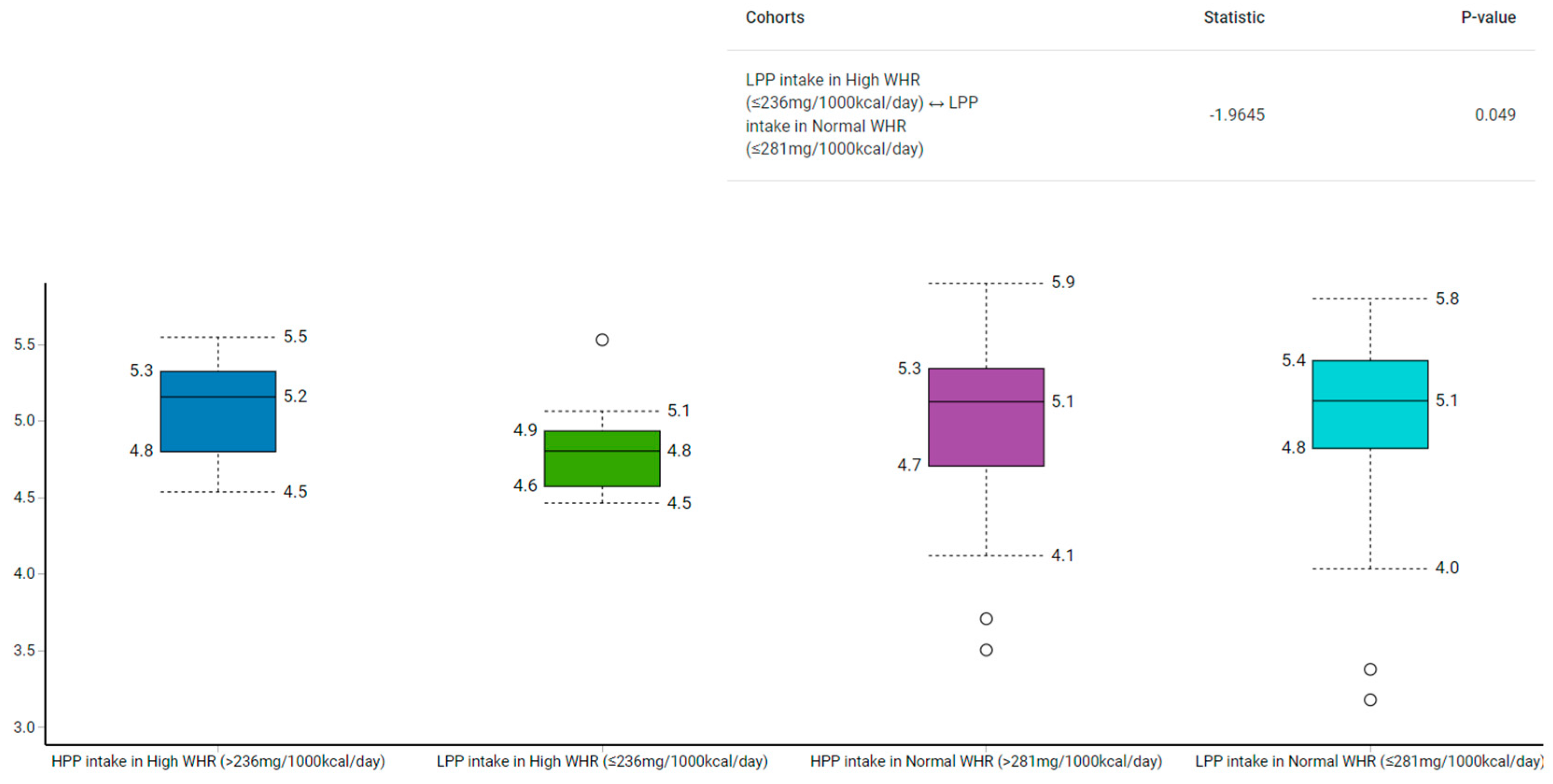
3.2. Anthropometrics Measurements
3.3. Gut Microbiota Composition
3.4. Correlations between Gut Microbiota, Dietary Polyphenol Intake, and BMI
3.5. Gut Microbiota Diversity Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- World Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 16 March 2020).
- WHO GHO | Global Health Observatory Data Repository (South-East Asia Region) | Prevalence of Obesity among Adults, BMI ≥ 30, Crude—Estimates by WHO Region. Available online: https://apps.who.int/gho/data/view.main-searo.BMI30CREGv?lang=en (accessed on 25 April 2021).
- WHO GHO | Global Health Observatory Data Repository (South-East Asia Region) | Prevalence of Overweight among Adults, BMI ≥ 25, Crude—Estimates by WHO Region. Available online: https://apps.who.int/gho/data/view.main-searo.BMI25CREGv?lang=en (accessed on 25 April 2021).
- MoH World Health Survey Saudi Arabia (KSAWHS) 2019 Final Report. 2019. Available online: https://www.moh.gov.sa/en/Ministry/Statistics/Population-Health-Indicators/Documents/World-Health-Survey-Saudi-Arabia.pdf (accessed on 1 November 2022).
- Hanson, M.A.; Gluckman, P.D. Early Developmental Conditioning of Later Health and Disease: Physiology or Pathophysiology? Physiol. Rev. 2014, 94, 1027–1076. [Google Scholar] [CrossRef] [PubMed]
- CDC. Adult Obesity Causes & Consequences | Overweight & Obesity | CDC. Available online: https://www.cdc.gov/obesity/adult/causes.html (accessed on 17 March 2020).
- Naukkarinen, J.; Rissanen, A.; Kaprio, J.; Pietiläinen, K.H. Causes and consequences of obesity: The contribution of recent twin studies. Int. J. Obes. 2011, 36, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.J.; West, N.P.; Cripps, A.W. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015, 3, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Pott-Junior, H.; Nascimento, C.M.C.; Costa-Guarisco, L.P.; Gomes, G.A.D.O.; Gramani-Say, K.; Orlandi, F.D.S.; Gratão, A.C.M.; Orlandi, A.A.D.S.; Pavarini, S.C.I.; Vasilceac, F.A.; et al. Vitamin D Deficient Older Adults Are More Prone to Have Metabolic Syndrome, but Not to a Greater Number of Metabolic Syndrome Parameters. Nutrients 2020, 12, 748. [Google Scholar] [CrossRef] [PubMed]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2019, 287, 833–855. [Google Scholar] [CrossRef]
- Wilkins, L.J.; Monga, M.; Miller, A.W. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci. Rep. 2019, 9, 12918. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, Q.; Balakrishnan, B.; Belobrajdic, D.P.; Feng, Q.-J.; Zhang, W. Role of Dietary Nutrients in the Modulation of Gut Microbiota: A Narrative Review. Nutrients 2020, 12, 381. [Google Scholar] [CrossRef]
- Plamada, D.; Vodnar, D.C. Polyphenols—Gut Microbiota Interrelationship: A Transition to a New Generation of Prebiotics. Nutrients 2021, 14, 137. [Google Scholar] [CrossRef]
- Fang, C.; Kim, H.; Yanagisawa, L.; Bennett, W.; Sirven, M.A.; Alaniz, R.C.; Talcott, S.T.; Mertens-Talcott, S.U. Gallotannins and Lactobacillus plantarum WCFS1 Mitigate High-Fat Diet-Induced Inflammation and Induce Biomarkers for Thermogenesis in Adipose Tissue in Gnotobiotic Mice. Mol. Nutr. Food Res. 2019, 63, e1800937. [Google Scholar] [CrossRef]
- Alves-Santos, A.M.; Sugizaki, C.S.A.; Lima, G.C.; Naves, M.M.V. Prebiotic effect of dietary polyphenols: A systematic review. J. Funct. Foods 2020, 74, 104169. [Google Scholar] [CrossRef]
- Corrêa, T.A.F.; Rogero, M.M.; Hassimotto, N.M.A.; Lajolo, F.M. The Two-Way Polyphenols-Microbiota Interactions and Their Effects on Obesity and Related Metabolic Diseases. Front. Nutr. 2019, 6, 188. [Google Scholar] [CrossRef] [PubMed]
- Duarte, L.; Gasaly, N.; Poblete-Aro, C.; Uribe, D.; Echeverria, F.; Gotteland, M.; Garcia-Diaz, D.F. Polyphenols and their anti-obesity role mediated by the gut microbiota: A comprehensive review. Rev. Endocr. Metab. Disord. 2021, 22, 367–388. [Google Scholar] [CrossRef]
- Queipo-Ortuño, M.I.; Boto-Ordóñez, M.; Murri, M.; Gomez-Zumaquero, J.M.; Clemente-Postigo, M.; Estruch, R.; Cardona Diaz, F.; Andrés-Lacueva, C.; Tinahones, F.J. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012, 95, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Eid, N.; Osmanova, H.; Natchez, C.; Walton, G.; Costabile, A.; Gibson, G.; Rowland, I.; Spencer, J.P.E. Impact of palm date consumption on microbiota growth and large intestinal health: A randomised, controlled, cross-over, human intervention study. Br. J. Nutr. 2015, 114, 1226–1236. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Indias, I.; Sánchez-Alcoholado, L.; Pérez-Martínez, P.; Andrés-Lacueva, C.; Cardona, F.; Tinahones, F.J.; Queipo-Ortuño, M.I. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct. 2016, 7, 1775–1787. [Google Scholar] [CrossRef]
- Most, J.; Penders, J.; Lucchesi, M.; Goossens, G.H.; Blaak, E.E. Gut microbiota composition in relation to the metabolic response to 12-week combined polyphenol supplementation in overweight men and women. Eur. J. Clin. Nutr. 2017, 71, 1040–1045. [Google Scholar] [CrossRef]
- Aljazairy, E.A.; Al-Musharaf, S.; Abudawood, M.; Almaarik, B.; Hussain, S.D.; Alnaami, A.M.; Sabico, S.; Al-Daghri, N.M.; Clerici, M.; Aljuraiban, G.S. Influence of Adiposity on the Gut Microbiota Composition of Arab Women: A Case-Control Study. Biology 2022, 11, 1586. [Google Scholar] [CrossRef] [PubMed]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in Lean and Overweight Healthy Subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Fernandes, J.J.D.R.; Su, W.; Rahat-Rozenbloom, S.; Wolever, T.M.S.; Comelli, E. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes 2014, 4, e121. [Google Scholar] [CrossRef]
- WHO. Waist Circumference and Waist-Hip Ratio: Report of a WHO Expert Consultation; World Health Organization: Geneva, Switzerland, 2008. [Google Scholar]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2017, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Leo, L.; Sudarshan, S.; Felix, E. Microbiome Diversity. Orchestrating Microbiome Analysis. Available online: https://microbiome.github.io/OMA/index.html (accessed on 28 July 2021).
- Alkhalaf, M.; Edwards, C.; Combet, E. Validation of a Food Frequency Questionnaire Specific for Salt Intake in Saudi Arabian Adults Using Urinary Biomarker and Repeated Multiple Pass 24-Hour Dietary Recall. Proc. Nutr. Soc. 2015, 74, E337. [Google Scholar] [CrossRef]
- Bingham, S.A.; Gill, C.; Welch, A.; Cassidy, A.; Runswick, S.A.; Oakes, S.; Lubin, R.; Thurnham, D.I.; Key, T.J.; Roe, L.; et al. Validation of dietary assessment methods in the UK arm of EPIC using weighed records, and 24-hour urinary nitrogen and potassium and serum vitamin C and carotenoids as biomarkers. Int. J. Epidemiol. 1997, 26, S137–S151. [Google Scholar] [CrossRef]
- Perezjimenez, J.; Neveu, V.; Vos, F.; Scalbert, A. Identification of the 100 richest dietary sources of polyphenols: An application of the Phenol-Explorer database. Eur. J. Clin. Nutr. 2010, 64, S112–S120. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, J.; Liang, H.; Ye, L.; Lan, L.; Lu, F.; Wang, Q.; Lei, T.; Yang, X.; Cui, P.; et al. Differences in Alpha Diversity of Gut Microbiota in Neurological Diseases. Front. Neurosci. 2022, 16, 892. [Google Scholar] [CrossRef]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Ma, G.; Chen, Y. Polyphenol supplementation benefits human health via gut microbiota: A systematic review via meta-analysis. J. Funct. Foods 2020, 66, 103829. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; del Pugar, E.M.G.; López-Almela, I.; Moya-Pérez, Á.; Codoñer-Franch, P.; Sanz, Y. Depletion of Blautia Species in the Microbiota of Obese Children Relates to Intestinal Inflammation and Metabolic Phenotype Worsening. Msystems 2020, 5, e00857-19. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, A.; Sandhu, A.K.; Edirisinghe, I.; Burton-Freeman, B.M. Functional Deficits in Gut Microbiome of Young and Middle-Aged Adults with Prediabetes Apparent in Metabolizing Bioactive (Poly)phenols. Nutrients 2020, 12, 3595. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Imlay, J.A. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol. Microbiol. 2013, 90, 1356–1371. [Google Scholar] [CrossRef]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Dehoux, P.; Marvaud, J.C.; Abouelleil, A.; Earl, A.M.; Lambert, T.; Dauga, C. Comparative genomics of Clostridium bolteae and Clostridium clostridioforme reveals species-specific genomic properties and numerous putative antibiotic resistance determinants. BMC Genom. 2016, 17, 819. [Google Scholar] [CrossRef]
- Esfandiar, Z.; Hosseini-Esfahani, F.; Mirmiran, P.; Yuzbashian, E.; Azizi, F. The Association of Dietary Polyphenol Intake with the Risk of Type 2 Diabetes: Tehran Lipid and Glucose Study. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, ume 13, 1643–1652. [Google Scholar] [CrossRef]
- Del Bo’, C.; Bernardi, S.; Marino, M.; Porrini, M.; Tucci, M.; Guglielmetti, S.; Cherubini, A.; Carrieri, B.; Kirkup, B.; Kroon, P.; et al. Systematic Review on Polyphenol Intake and Health Outcomes: Is there Sufficient Evidence to Define a Health-Promoting Polyphenol-Rich Dietary Pattern? Nutrients 2019, 11, 1355. [Google Scholar] [CrossRef]
- Williamson, G.; Holst, B. Dietary reference intake (DRI) value for dietary polyphenols: Are we heading in the right direction? Br. J. Nutr. 2008, 99, S55–S58. [Google Scholar] [CrossRef] [PubMed]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The Role of Polyphenols in Human Health and Food Systems: A Mini-Review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef]
- He, Y.; Wu, W.; Zheng, H.-M.; Li, P.; McDonald, D.; Sheng, H.-F.; Chen, M.-X.; Chen, Z.-H.; Ji, G.-Y.; Zheng, Z.-D.; et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 2018, 24, 1532–1535. [Google Scholar] [CrossRef]
- Costea, P.I.; Zeller, G.; Sunagawa, S.; Pelletier, E.; Alberti, A.; Levenez, F.; Tramontano, M.; Driessen, M.; Hercog, R.; Jung, F.-E.; et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017, 35, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Vincent, C.; Stephens, D.A.; Loo, V.G.; Edens, T.J.; A Behr, M.; Dewar, K.; Manges, A.R. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome 2013, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Mosca, A.; Leclerc, M.; Hugot, J.-P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Concha, F.; Prado, G.; Quezada, J.; Ramirez, A.; Bravo, N.; Flores, C.; Herrera, J.J.; Lopez, N.; Uribe, D.; Duarte-Silva, L.; et al. Nutritional and non-nutritional agents that stimulate white adipose tissue browning. Rev. Endocr. Metab. Disord. 2019, 20, 161–171. [Google Scholar] [CrossRef]
- Cani, P.D.; Van Hul, M.; Lefort, C.; Depommier, C.; Rastelli, M.; Everard, A. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar] [CrossRef]
- Hillmann, B.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Zhu, Q.; Gohl, D.; Beckman, K.B.; Knight, R.; Knights, D. Evaluating the Information Content of Shallow Shotgun Metagenomics. Msystems 2018, 3, e00069-18. [Google Scholar] [CrossRef]
- Freedman, L.S.; Midthune, D.; Arab, L.; Prentice, R.L.; Subar, A.F.; Willett, W.; Neuhouser, M.L.; Tinker, L.F.; Kipnis, V. Combining a Food Frequency Questionnaire With 24-Hour Recalls to Increase the Precision of Estimation of Usual Dietary Intakes—Evidence From the Validation Studies Pooling Project. Am. J. Epidemiology 2018, 187, 2227–2232. [Google Scholar] [CrossRef] [PubMed]

| Variables | Cases | Controls | ||||
|---|---|---|---|---|---|---|
| Low Polyphenols (≤236 mg/1000 kcal/Day) | High Polyphenols (>236 mg/1000 kcal/Day) | p-Value | Low Polyphenols (≤281 mg/1000 kcal/Day) | High Polyphenols (>281 mg/1000 kcal/Day) | p-Value | |
| Anthropometric Measurements | ||||||
| BMI (kg/m2) | 36.1 ± 4.4 | 36.0 ± 5.0 | 0.10 | 22.0 ± 2.0 | 21.4 ± 1.7 | 0.30 |
| WHR (ratio) | 0.8 ± 0.1 | 0.7 ± 0.1 | 0.60 | 0.7 ± 0.0 | 0.7 ± 0.1 | 0.66 |
| Body fat (%) | 51.0 ± 3.3 | 51.1 ± 3.3 | 0.94 | 35.1 ± 4.6 | 34.4 ± 6.3 | 0.67 |
| Muscle mass (%) | 26.6 ± 2.0 | 26.6 ± 1.9 | 0.94 | 26.6 ± 10.4 | 32.5 ± 7.2 | 0.03 |
| Gut Microbiota | ||||||
| Firmicutes | 0.21 (0.15–0.32) | 0.23 (0.16–0.30) | 0.98 | 0.21 (0.17–0.32) | 0.20 (0.17–0.27) | 0.46 |
| Blautia wexlerae | 0.01 (0.0038–0.01) | 0.01 (0.0034–0.01) | 0.30 | 0.0045 (0.0025–0.01) | 0.01 (0.0038–0.01) | 0.29 |
| Flavonifractor plautii | 0.0008 (0.0003–0.0018) | 0.0007 (0.0004–0.0011) | 0.49 | 0.0005 (0.0003–0.0008) | 0.0008 (0.0004–0.0018) | 0.07 |
| Clostridium bolteae | 0.0004 (0.0002–0.0013) | 0.0000 (0.0000–0.0005) | 0.03 | 0.0003 (0.0000–0.0006) | 0.0005 (0.0001–0.0007) | 0.29 |
| Faecalibacterium prausnitzii | 0.02 (0.02–0.03) | 0.02 (0.01–0.02) | 0.08 | 0.026 (0.01–0.03) | 0.02 (0.01–0.02) | 0.16 |
| Clostridioides difficile§ | 0.01 ± 0.05 | 0.0001 ± 0.0001 | 0.04 | 0.0001 ± 0.0001 | 0.01 ± 0.04 | 0.14 |
| Bacteroidetes | 0.74 (0.63–0.83) | 0.72 (0.66–0.81) | 0.98 | 0.72 (0.55–0.77) | 0.74 (0.66–0.79) | 0.35 |
| Bacteroides faecichinchillae§ | 0.004 ± 0.02 | 0.0001 ± 0.0001 | 0.97 | 0.01 ± 0.03 | 0.0001 ± 0.0001 | 0.02 |
| Bacteroides thetaiotaomicron | 0.01 (0.0023–0.01) | 0.01 (0.0035–0.01) | 0.73 | 0.01 (0.0038–0.01) | 0.01 (0.0033–0.01) | 0.61 |
| Actinobacteria | 0.02 (0.02–0.05) | 0.02 (0.01–0.04) | 0.50 | 0.03 (0.02–0.06) | 0.04 (0.01–0.07) | 0.85 |
| Bifidobacterium pseudocatenulatum | 0.0017 (0.0000–0.0057) | 0.0001 (0.0000–0.0015) | 0.04 | 0.0000 (0.0000–0.0048) | 0.0005 (0.0000–0.0031) | 0.33 |
| Verrucomicrobia | 0.0005 (0.0000–0.0043) | 0.0003 (0.0000–0.0015) | 0.59 | 0.0019 (0.0004–0.0128) | 0.0003 (0.0000–0.0038) | 0.03 |
| Akkermansia muciniphila | 0.0005 (0.0000–0.0043) | 0.0003 (0.0000–0.0015) | 0.76 | 0.0029 (0.0004–0.01) | 0.0003 (0.0000–0.0038) | 0.02 |
| Proteobacteria | 0.01 (0.01–0.02) | 0.01 (0.01–0.02) | 0.45 | 0.01 (0.01–0.02) | 0.01 (0.01–0.02) | 0.42 |
| Fusobacteria | 0.0000 (0.0000–0.0000) | 0.0000 (0.0000–0.0000) | 0.15 | 0.0000 (0.0000–0.0000) | 0.0000 (0.0000–0.0000) | 1.00 |
| F/B (ratio) | 0.29 (0.19–0.49) | 0.32 (0.20–0.44) | 0.96 | 0.28 (0.22–0.59) | 0.28 (0.21–0.41) | 0.40 |
| Variables | Polyphenol Intake (mg/1000 kcal/Day) | p-Value |
|---|---|---|
| Firmicutes | −0.02 | 0.86 |
| Blautia wexlerae | −0.07 | 0.49 |
| Flavonifractor plautii | 0.22 | 0.03 |
| Clostridium bolteae | 0.28 | <0.01 |
| Faecalibacterium prausnitzii | −0.18 | 0.10 |
| Clostridioides difficile | 0.01 | 0.95 |
| Bacteroidetes | 0.06 | 0.59 |
| Bacteroides faecichinchillae | −0.10 | 0.36 |
| Bacteroides thetaiotaomicron | −0.08 | 0.43 |
| Actinobacteria | −0.08 | 0.46 |
| Verrucomicrobia | −0.11 | 0.30 |
| Akkermansia muciniphila | −0.12 | 0.26 |
| Proteobacteria | −0.07 | 0.53 |
| Fusobacteria | −0.10 | 0.35 |
| F/B (ratio) | 0.02 | 0.83 |
| Variables | Cases | Controls | ||||||
|---|---|---|---|---|---|---|---|---|
| Low Polyphenols (≤236 mg/1000 kcal/Day) | High Polyphenols (>236 mg/1000 kcal/Day) | Low Polyphenols (≤281 mg/1000 kcal/Day) | High Polyphenols (>281 mg/1000 kcal/Day) | |||||
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | |
| Firmicutes | −0.29 | 0.20 | 0.36 | 0.11 | 0.04 | 0.87 | −0.37 | 0.09 |
| Blautia wexlerae | −0.56 | <0.01 | 0.15 | 0.52 | 0.28 | 0.20 | −0.28 | 0.20 |
| Flavonifractor plautii | −0.04 | 0.88 | 0.29 | 0.20 | 0.14 | 0.52 | 0.15 | 0.50 |
| Clostridium bolteae | −0.16 | 0.49 | 0.28 | 0.22 | −0.11 | 0.60 | 0.54 | <0.01 |
| Faecalibacterium prausnitzii | 0.39 | 0.08 | −0.12 | 0.60 | −0.28 | 0.19 | −0.19 | 0.39 |
| Clostridioides difficile | 0.14 | 0.54 | − | − | 0.18 | 0.40 | 0.00 | 0.99 |
| Bacteroidetes | 0.29 | 0.20 | −0.31 | 0.16 | −0.19 | 0.38 | 0.37 | 0.08 |
| Bacteroides faecichinchillae | 0.17 | 0.46 | −0.11 | 0.63 | 0.26 | 0.24 | − | − |
| Bacteroides thetaiotaomicron | 0.19 | 0.42 | −0.29 | 0.20 | −0.45 | 0.03 | 0.04 | 0.86 |
| Actinobacteria | −0.11 | 0.63 | −0.02 | 0.94 | 0.29 | 0.18 | −0.24 | 0.27 |
| Verrucomicrobia | 0.11 | 0.63 | −0.21 | 0.36 | 0.31 | 0.15 | −0.27 | 0.22 |
| Akkermansia muciniphila | 0.11 | 0.63 | −0.22 | 0.35 | 0.27 | 0.22 | −0.27 | 0.22 |
| Proteobacteria | −0.17 | 0.47 | −0.04 | 0.86 | 0.35 | 0.10 | −0.04 | 0.87 |
| Fusobacteria | −0.35 | 0.12 | − | − | − | − | − | − |
| F/B (ratio) | −0.26 | 0.26 | 0.36 | 0.11 | 0.10 | 0.66 | −0.26 | 0.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsuhaibani, M.N.; Aljuraiban, G.S.; Aljazairy, E.A.; Abudawood, M.; Hussain, S.D.; Alnaami, A.; Sabico, S.; Al-Daghri, N.M.; Al-Musharaf, S. Dietary Polyphenols in Relation to Gut Microbiota Composition in Saudi Arabian Females. Metabolites 2023, 13, 6. https://doi.org/10.3390/metabo13010006
Alsuhaibani MN, Aljuraiban GS, Aljazairy EA, Abudawood M, Hussain SD, Alnaami A, Sabico S, Al-Daghri NM, Al-Musharaf S. Dietary Polyphenols in Relation to Gut Microbiota Composition in Saudi Arabian Females. Metabolites. 2023; 13(1):6. https://doi.org/10.3390/metabo13010006
Chicago/Turabian StyleAlsuhaibani, Munirah N., Ghadeer S. Aljuraiban, Esra’a A. Aljazairy, Manal Abudawood, Syed D. Hussain, Abdullah Alnaami, Shaun Sabico, Nasser M. Al-Daghri, and Sara Al-Musharaf. 2023. "Dietary Polyphenols in Relation to Gut Microbiota Composition in Saudi Arabian Females" Metabolites 13, no. 1: 6. https://doi.org/10.3390/metabo13010006
APA StyleAlsuhaibani, M. N., Aljuraiban, G. S., Aljazairy, E. A., Abudawood, M., Hussain, S. D., Alnaami, A., Sabico, S., Al-Daghri, N. M., & Al-Musharaf, S. (2023). Dietary Polyphenols in Relation to Gut Microbiota Composition in Saudi Arabian Females. Metabolites, 13(1), 6. https://doi.org/10.3390/metabo13010006






