Bariatric Surgery Associates with Nonalcoholic Steatohepatitis/Hepatocellular Carcinoma Amelioration via SPP1 Suppression
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source and Analysis
2.2. Function Enrichment Analysis and Protein-Protein Interaction (PPI) Network
2.3. Gene Profiling in Liver Hepatic Carcinoma (LIHC)
2.4. Cell Enrichment Analysis
2.5. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.6. Mouse Models
2.7. RNA Quantification
2.8. Statistical Analysis
3. Results
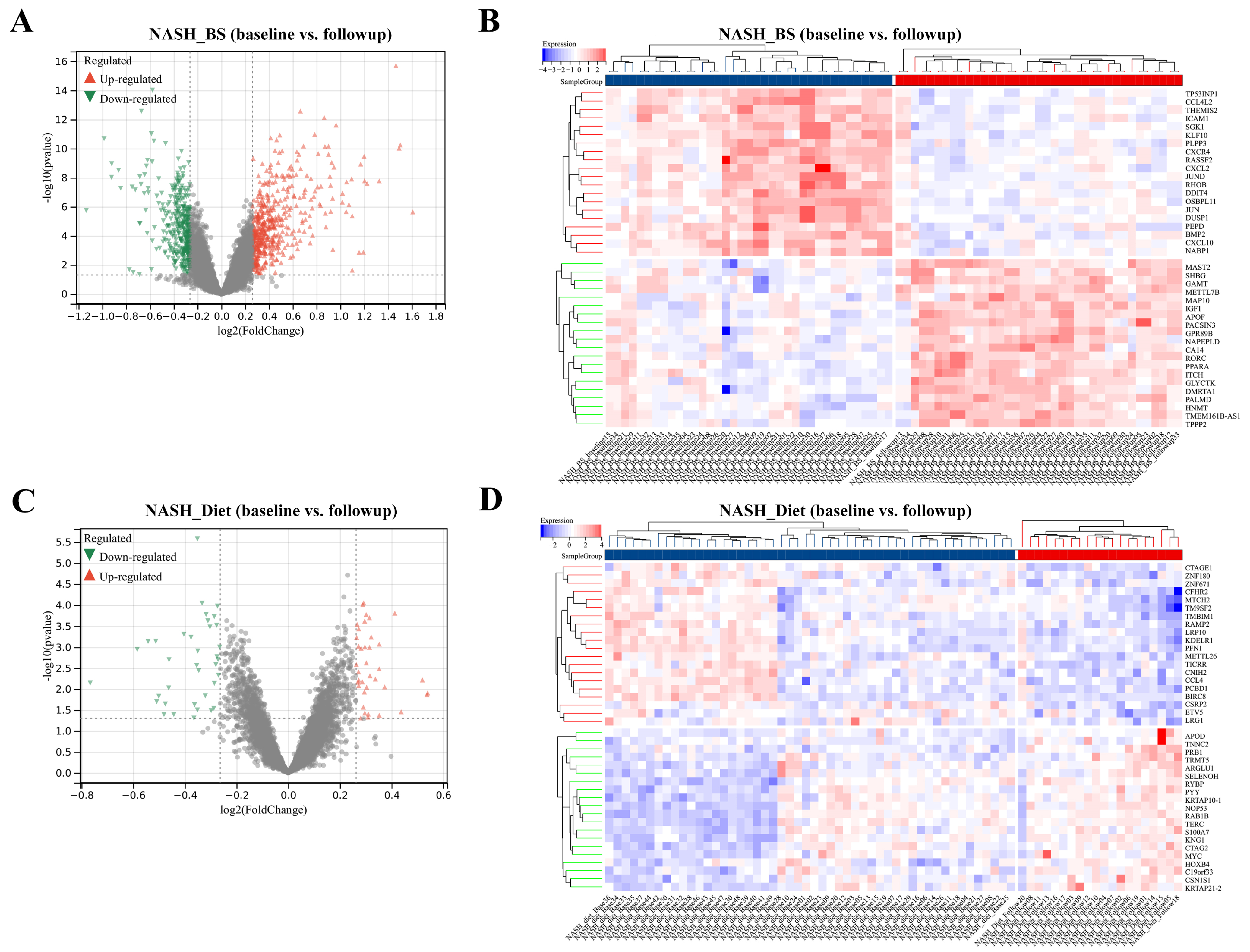
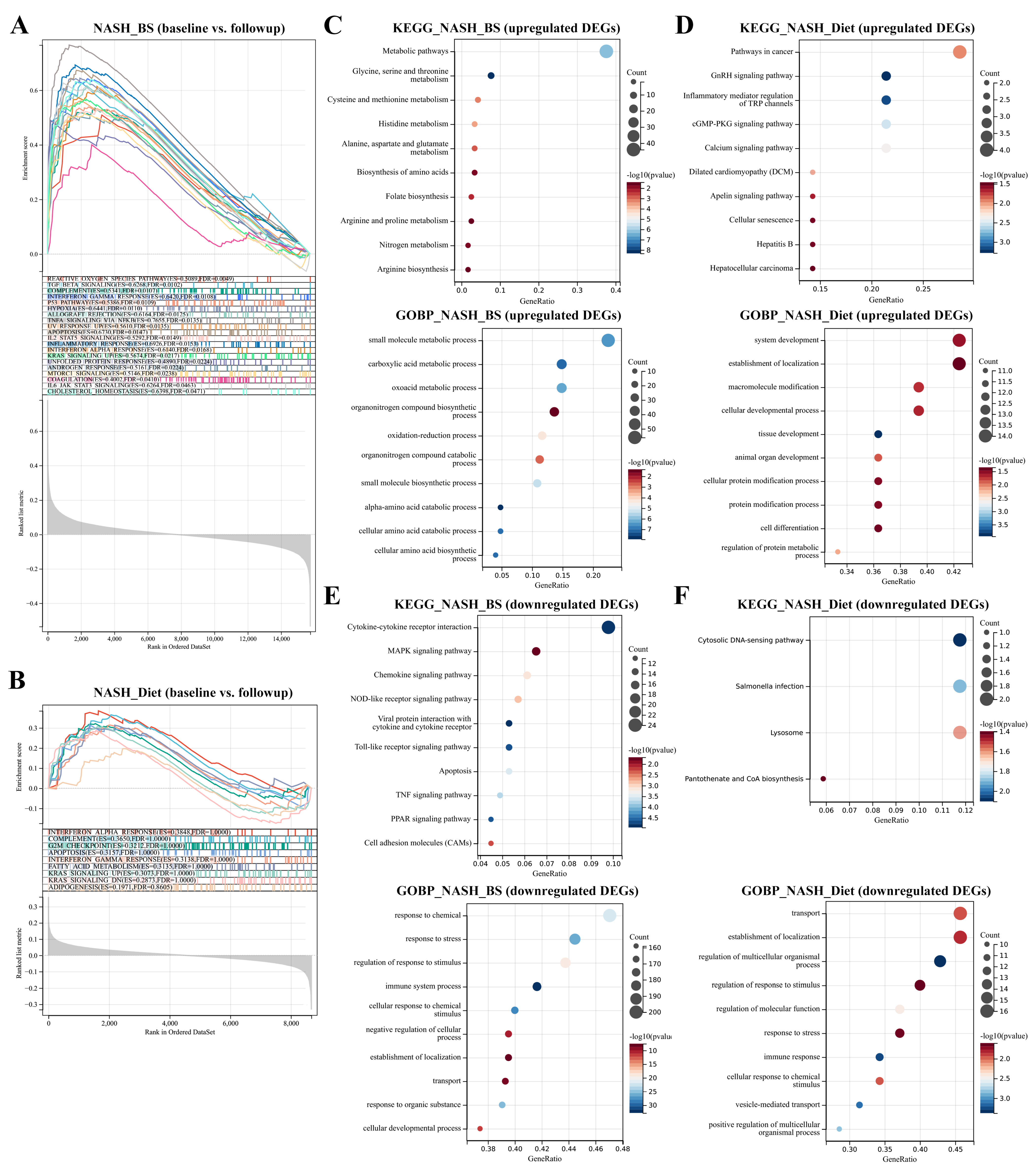
3.1. DEGs Mining and Function Enrichment
3.2. BS Attenuates Risks of Inflammation, Steatosis, and Fibrogenesis in NASH Liver, Superior to Diet
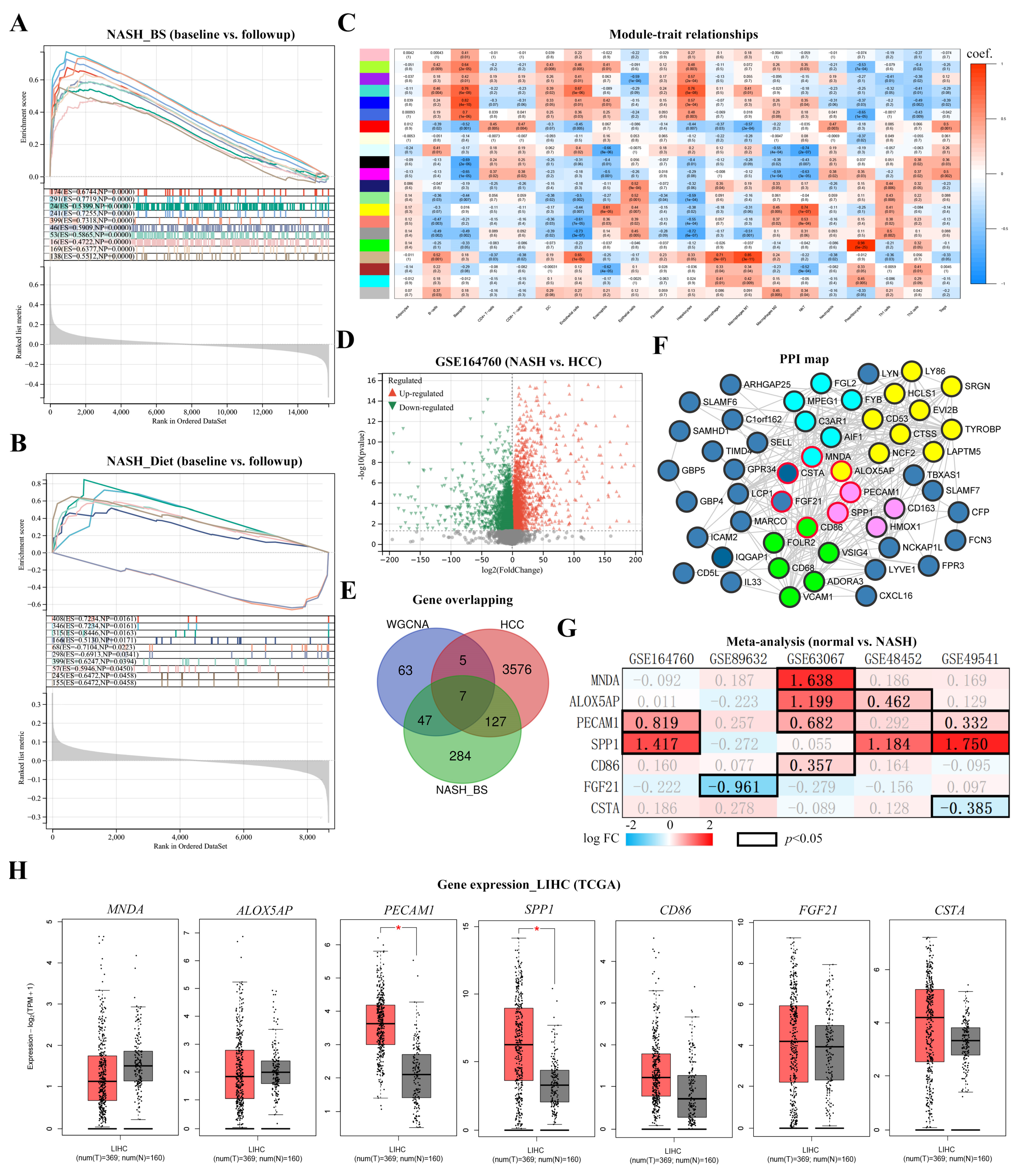
3.3. Key Factor Identification in Post-BS NASH Livers
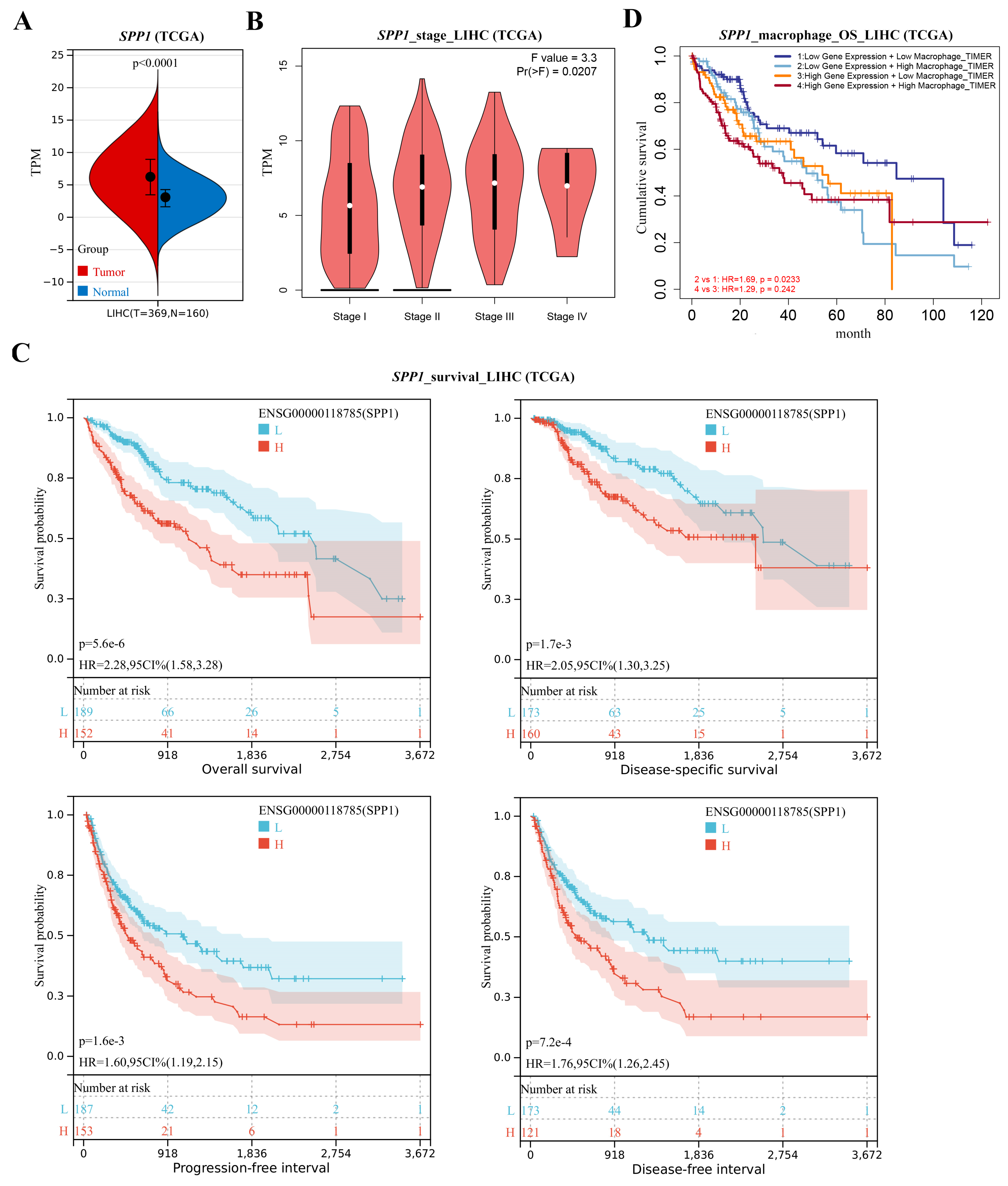
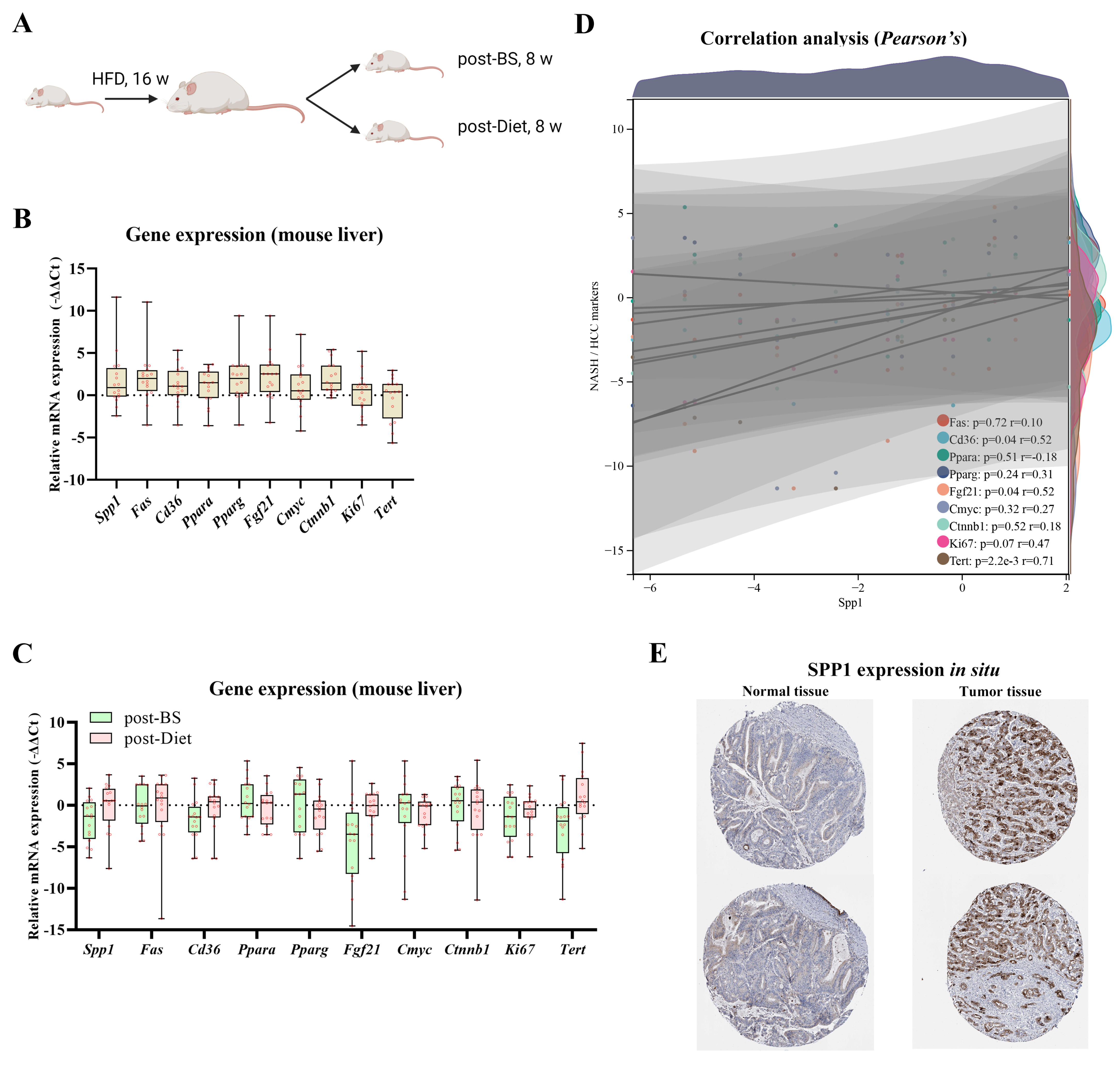
3.4. Spp1 Expression Was Attenuated after BS and Associated with NASH/HCC Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the american association for the study of liver diseases, american college of gastroenterology, and the american gastroenterological association. Hepatology 2012, 55, 2005–2023. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, F.; Shubrook, J.H.; Younossi, Z.; Natarajan, Y.; Bugianesi, E.; Rinella, M.E.; Harrison, S.A.; Mantzoros, C.; Pfotenhauer, K.; Klein, S.; et al. Preparing for the nash epidemic: A call to action. Gastroenterology 2021, 161, 1030–1042.e1038. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, D.; Ditah, I.C.; Saeian, K.; Lalehzari, M.; Aronsohn, A.; Gorospe, E.C.; Charlton, M. Changes in the prevalence of hepatitis c virus infection, nonalcoholic steatohepatitis, and alcoholic liver disease among patients with cirrhosis or liver failure on the waitlist for liver transplantation. Gastroenterology 2017, 152, 1090–1099.e1091. [Google Scholar] [CrossRef] [PubMed]
- Oseini, A.M.; Sanyal, A.J. Therapies in non-alcoholic steatohepatitis (nash). Liver Int. 2017, 37 (Suppl. S1), 97–103. [Google Scholar] [CrossRef]
- Machado, M.; Marques-Vidal, P.; Cortez-Pinto, H. Hepatic histology in obese patients undergoing bariatric surgery. J. Hepatol. 2006, 45, 600–606. [Google Scholar] [CrossRef]
- Boza, C.; Riquelme, A.; Ibañez, L.; Duarte, I.; Norero, E.; Viviani, P.; Soza, A.; Fernandez, J.I.; Raddatz, A.; Guzman, S.; et al. Predictors of nonalcoholic steatohepatitis (nash) in obese patients undergoing gastric bypass. Obes. Surg. 2005, 15, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Hannah, W.N., Jr.; Harrison, S.A. Effect of weight loss, diet, exercise, and bariatric surgery on nonalcoholic fatty liver disease. Clin. Liver. Dis. 2016, 20, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Romero-Gómez, M.; Zelber-Sagi, S.; Trenell, M. Treatment of nafld with diet, physical activity and exercise. J. Hepatol. 2017, 67, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.C.; Abbasi, F.; Lamendola, C.; Carter, S.; McLaughlin, T.L. Serum alanine aminotransferase levels decrease further with carbohydrate than fat restriction in insulin-resistant adults. Diabetes Care 2007, 30, 1075–1080. [Google Scholar] [CrossRef]
- Browning, J.D.; Baker, J.A.; Rogers, T.; Davis, J.; Satapati, S.; Burgess, S.C. Short-term weight loss and hepatic triglyceride reduction: Evidence of a metabolic advantage with dietary carbohydrate restriction. Am. J. Clin. Nutr. 2011, 93, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, H.; Oien, D.M. Metabolic/bariatric surgery worldwide 2011. Obes. Surg. 2013, 23, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Castagneto-Gissey, L.; Mingrone, G. Insulin sensitivity and secretion modifications after bariatric surgery. J. Endocrinol. Invest. 2012, 35, 692–698. [Google Scholar] [PubMed]
- Mingrone, G.; Panunzi, S.; De Gaetano, A.; Guidone, C.; Iaconelli, A.; Leccesi, L.; Nanni, G.; Pomp, A.; Castagneto, M.; Ghirlanda, G.; et al. Bariatric surgery versus conventional medical therapy for type 2 diabetes. N. Engl. J. Med. 2012, 366, 1577–1585. [Google Scholar] [CrossRef]
- Pais, R.; Aron-Wisnewsky, J.; Bedossa, P.; Ponnaiah, M.; Oppert, J.M.; Siksik, J.M.; Genser, L.; Charlotte, F.; Thabut, D.; Clement, K.; et al. Persistence of severe liver fibrosis despite substantial weight loss with bariatric surgery. Hepatology 2022, 76, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, H.; Buchwald, J.N. Metabolic (bariatric and nonbariatric) surgery for type 2 diabetes: A personal perspective review. Diabetes Care 2019, 42, 331–340. [Google Scholar] [CrossRef]
- Rustgi, V.K.; Li, Y.; Gupta, K.; Minacapelli, C.D.; Bhurwal, A.; Catalano, C.; Elsaid, M.I. Bariatric surgery reduces cancer risk in adults with nonalcoholic fatty liver disease and severe obesity. Gastroenterology 2021, 161, 171–184.e110. [Google Scholar] [CrossRef]
- Taminau, J.; Meganck, S.; Lazar, C.; Steenhoff, D.; Coletta, A.; Molter, C.; Duque, R.; de Schaetzen, V.; Weiss Solís, D.Y.; Bersini, H.; et al. Unlocking the potential of publicly available microarray data using insilicodb and insilicomerging r/bioconductor packages. BMC Bioinform. 2012, 13, 335. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Jia, A.; Xu, L.; Wang, Y. Venn diagrams in bioinformatics. Brief Bioinform. 2021, 22, bbab108. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tang, Z.; Zhang, W.; Ye, Z.; Liu, F. Gepia2021: Integrating multiple deconvolution-based analysis into gepia. Nucleic Acids Res. 2021, 49, W242–W246. [Google Scholar] [CrossRef]
- The genotype-tissue expression (gtex) project. Nat. Genet. 2013, 45, 580–585. [CrossRef]
- Gu, Z.; Hübschmann, D. Make interactive complex heatmaps in r. Bioinformatics 2021, 38, 1460–1462. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. Xcell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. Timer2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–w514. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Wgcna: An r package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Qi, X.; Li, X.; Jiao, Y.; Chen, S.; Song, P.; Qian, Z.; Tang, L. The effects of antral preservation and antral resection on body composition, glycemic control and bone mineral density following vertical sleeve gastrectomy in c57bl/6j mice with obesity and type 2 diabetes. Diabetes Metab. Syndr. Obes. 2022, 15, 297–305. [Google Scholar] [CrossRef]
- Younossi, Z.; Tacke, F.; Arrese, M.; Chander Sharma, B.; Mostafa, I.; Bugianesi, E.; Wai-Sun Wong, V.; Yilmaz, Y.; George, J.; Fan, J.; et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 2019, 69, 2672–2682. [Google Scholar] [CrossRef] [PubMed]
- Puengel, T.; Liu, H.; Guillot, A.; Heymann, F.; Tacke, F.; Peiseler, M. Nuclear receptors linking metabolism, inflammation, and fibrosis in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2022, 23, 2668. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From nash to hcc: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of nafld-related hcc: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Lefere, S.; Onghena, L.; Vanlander, A.; van Nieuwenhove, Y.; Devisscher, L.; Geerts, A. Bariatric surgery and the liver-mechanisms, benefits, and risks. Obes. Rev. 2021, 22, e13294. [Google Scholar] [CrossRef] [PubMed]
- Arterburn, D.E.; Telem, D.A.; Kushner, R.F.; Courcoulas, A.P. Benefits and risks of bariatric surgery in adults: A review. JAMA 2020, 324, 879–887. [Google Scholar] [CrossRef]
- Adams, T.D.; Davidson, L.E.; Litwin, S.E.; Kim, J.; Kolotkin, R.L.; Nanjee, M.N.; Gutierrez, J.M.; Frogley, S.J.; Ibele, A.R.; Brinton, E.A.; et al. Weight and metabolic outcomes 12 years after gastric bypass. N. Engl. J. Med. 2017, 377, 1143–1155. [Google Scholar] [CrossRef]
- Mechanick, J.I.; Apovian, C.; Brethauer, S.; Timothy Garvey, W.; Joffe, A.M.; Kim, J.; Kushner, R.F.; Lindquist, R.; Pessah-Pollack, R.; Seger, J.; et al. Clinical practice guidelines for the perioperative nutrition, metabolic, and nonsurgical support of patients undergoing bariatric procedures—2019 update: Cosponsored by american association of clinical endocrinologists/american college of endocrinology, the obesity society, american society for metabolic and bariatric surgery, obesity medicine association, and american society of anesthesiologists. Obesity 2020, 28, O1–O58. [Google Scholar]
- Cherla, D.V.; Rodriguez, N.A.; Vangoitsenhoven, R.; Singh, T.; Mehta, N.; McCullough, A.J.; Brethauer, S.A.; Schauer, P.R.; Aminian, A. Impact of sleeve gastrectomy and roux-en-y gastric bypass on biopsy-proven non-alcoholic fatty liver disease. Surg. Endosc. 2020, 34, 2266–2272. [Google Scholar] [CrossRef]
- Miras, A.D.; le Roux, C.W. Mechanisms underlying weight loss after bariatric surgery. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 575–584. [Google Scholar] [CrossRef]
- Sinclair, P.; Brennan, D.J.; le Roux, C.W. Gut adaptation after metabolic surgery and its influences on the brain, liver and cancer. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 606–624. [Google Scholar] [CrossRef]
- Chambers, A.P.; Jessen, L.; Ryan, K.K.; Sisley, S.; Wilson-Pérez, H.E.; Stefater, M.A.; Gaitonde, S.G.; Sorrell, J.E.; Toure, M.; Berger, J.; et al. Weight-independent changes in blood glucose homeostasis after gastric bypass or vertical sleeve gastrectomy in rats. Gastroenterology 2011, 141, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Caiazzo, R.; Lassailly, G.; Leteurtre, E.; Baud, G.; Verkindt, H.; Raverdy, V.; Buob, D.; Pigeyre, M.; Mathurin, P.; Pattou, F. Roux-en-y gastric bypass versus adjustable gastric banding to reduce nonalcoholic fatty liver disease: A 5-year controlled longitudinal study. Ann. Surg. 2014, 260, 893–898; discussion 898–899. [Google Scholar] [CrossRef] [PubMed]
- Lassailly, G.; Caiazzo, R.; Buob, D.; Pigeyre, M.; Verkindt, H.; Labreuche, J.; Raverdy, V.; Leteurtre, E.; Dharancy, S.; Louvet, A.; et al. Bariatric surgery reduces features of nonalcoholic steatohepatitis in morbidly obese patients. Gastroenterology 2015, 149, 379–388; quiz e15–16. [Google Scholar] [CrossRef] [PubMed]
- Peiseler, M.; Schwabe, R.; Hampe, J.; Kubes, P.; Heikenwälder, M.; Tacke, F. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease—Novel insights into cellular communication circuits. J. Hepatol. 2022, 77, 1136–1160. [Google Scholar] [CrossRef]
- Song, Z.; Chen, W.; Athavale, D.; Ge, X.; Desert, R.; Das, S.; Han, H.; Nieto, N. Osteopontin takes center stage in chronic liver disease. Hepatology 2021, 73, 1594–1608. [Google Scholar] [CrossRef]
- Zhu, C.; Kim, K.; Wang, X.; Bartolome, A.; Salomao, M.; Dongiovanni, P.; Meroni, M.; Graham, M.J.; Yates, K.P.; Diehl, A.M.; et al. Hepatocyte notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 2018, 10, eaat0344. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, N.; Sawada, Y.; Endo, I.; Saito, K.; Uemura, Y.; Nakatsura, T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 10573–10583. [Google Scholar] [CrossRef] [PubMed]
- Guillot, A.; Kohlhepp, M.S.; Bruneau, A.; Heymann, F.; Tacke, F. Deciphering the immune microenvironment on a single archival formalin-fixed paraffin-embedded tissue section by an immediately implementable multiplex fluorescence immunostaining protocol. Cancers 2020, 12, 2449. [Google Scholar] [CrossRef]
- Rao, R.S.; Rao, V.; Kini, S. Animal models in bariatric surgery—A review of the surgical techniques and postsurgical physiology. Obes. Surg. 2010, 20, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Lutz, T.A.; Bueter, M. The use of rat and mouse models in bariatric surgery experiments. Front. Nutr. 2016, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. Rna sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef] [PubMed]
- Guillot, A.; Tacke, F. Location, location, location—Spatial insight into hepatic macrophage populations. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 281–282. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Tang, L.; Guillot, A.; Liu, H. Bariatric Surgery Associates with Nonalcoholic Steatohepatitis/Hepatocellular Carcinoma Amelioration via SPP1 Suppression. Metabolites 2023, 13, 11. https://doi.org/10.3390/metabo13010011
Chen S, Tang L, Guillot A, Liu H. Bariatric Surgery Associates with Nonalcoholic Steatohepatitis/Hepatocellular Carcinoma Amelioration via SPP1 Suppression. Metabolites. 2023; 13(1):11. https://doi.org/10.3390/metabo13010011
Chicago/Turabian StyleChen, Shuai, Liming Tang, Adrien Guillot, and Hanyang Liu. 2023. "Bariatric Surgery Associates with Nonalcoholic Steatohepatitis/Hepatocellular Carcinoma Amelioration via SPP1 Suppression" Metabolites 13, no. 1: 11. https://doi.org/10.3390/metabo13010011
APA StyleChen, S., Tang, L., Guillot, A., & Liu, H. (2023). Bariatric Surgery Associates with Nonalcoholic Steatohepatitis/Hepatocellular Carcinoma Amelioration via SPP1 Suppression. Metabolites, 13(1), 11. https://doi.org/10.3390/metabo13010011








