Novel Regulators of Macropinocytosis-Dependent Growth Revealed by Informer Set Library Screening in Pancreatic Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell culture and Reagents
2.2. Dextran Uptake Assay
2.3. Cell Viability Assay
2.4. Generation of NHE1 Knockdown Cells
2.5. Quantitative Real-Time PCR
2.6. Informer Set Library Screening
2.7. Statistics
3. Results
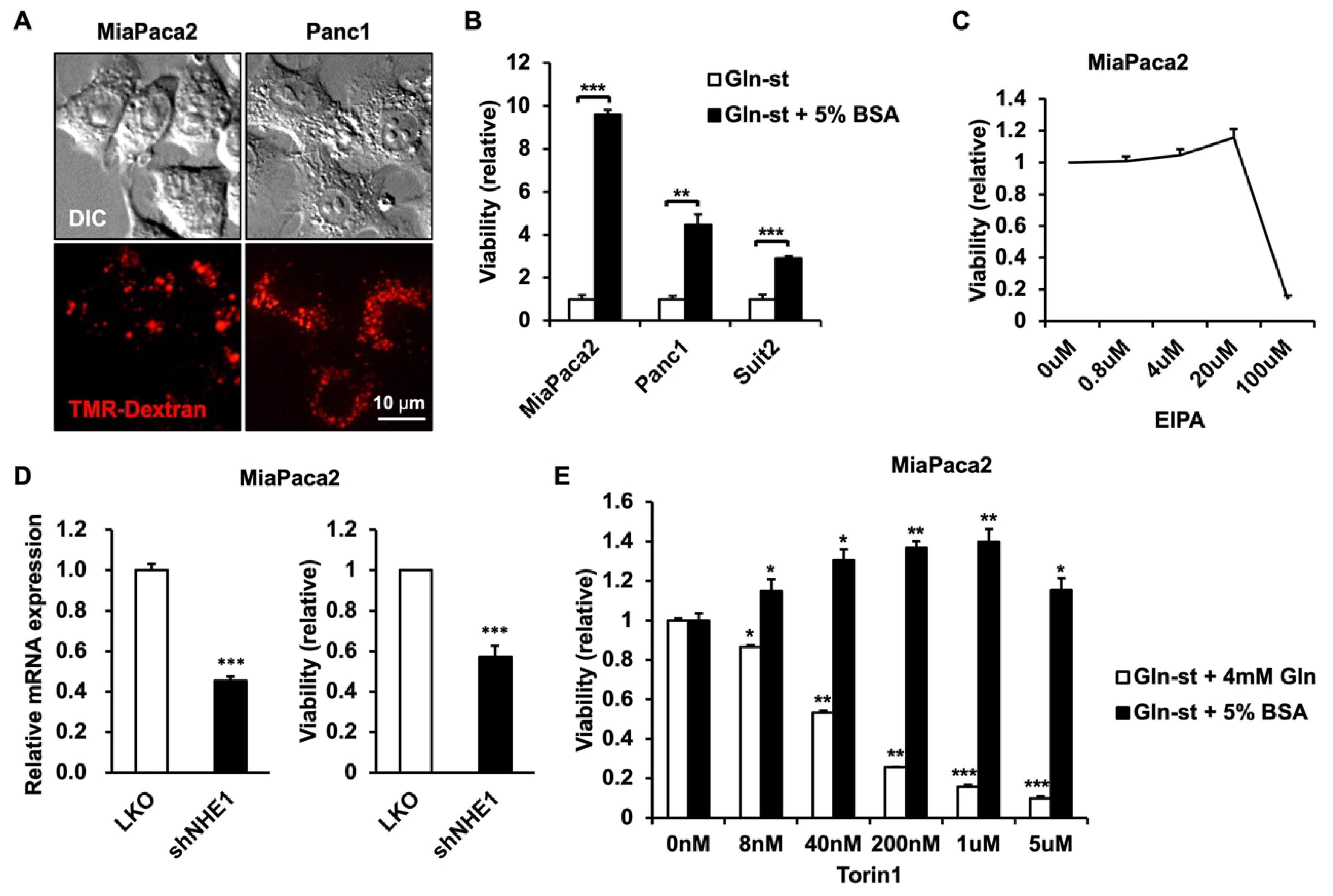
3.1. PDAC Cells Utilize Extracellular Protein to Enhance Survival in Glutamine-Deprived Conditions
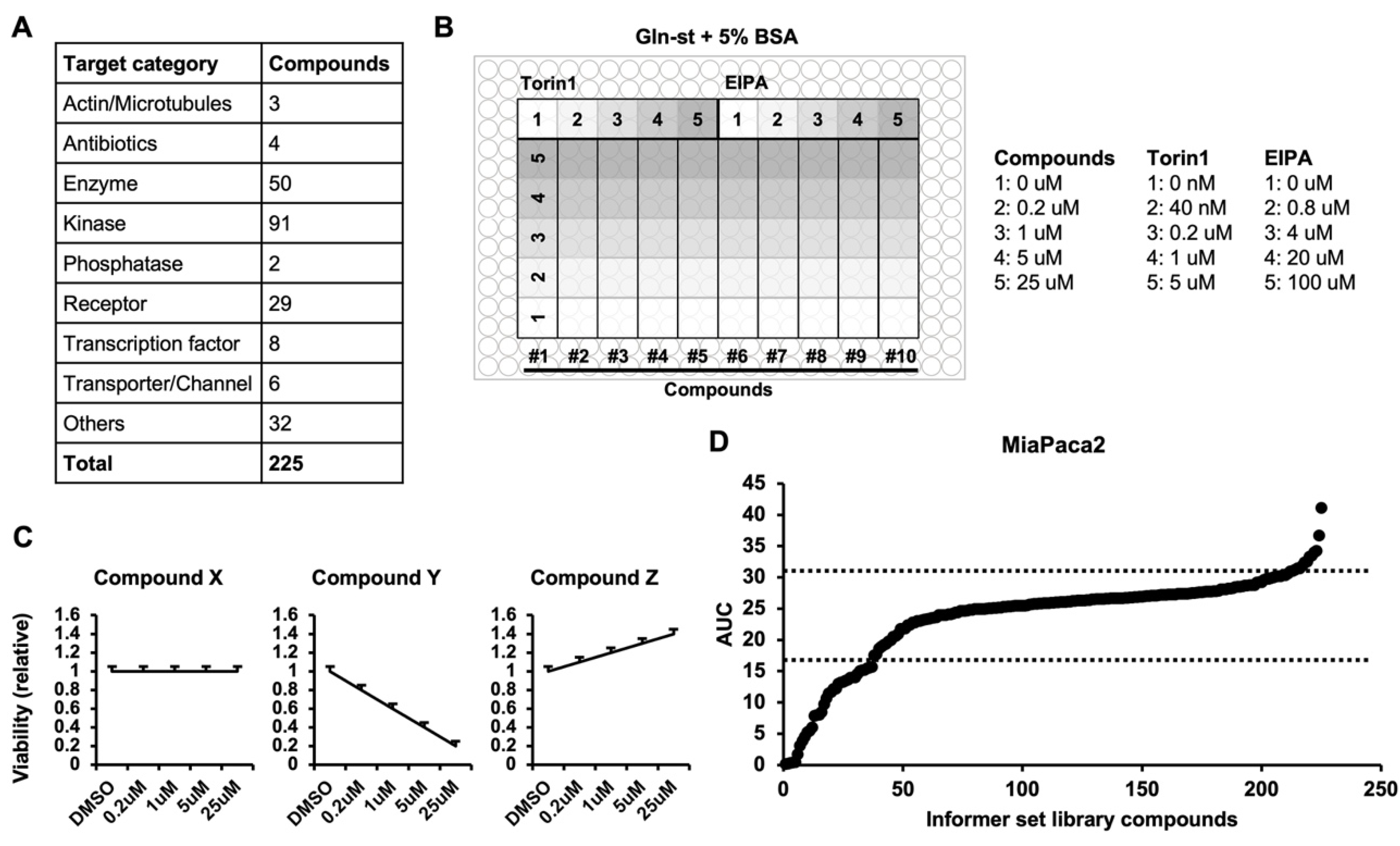
3.2. Macropinocytosis Regulator Screen using the Informer Set Library
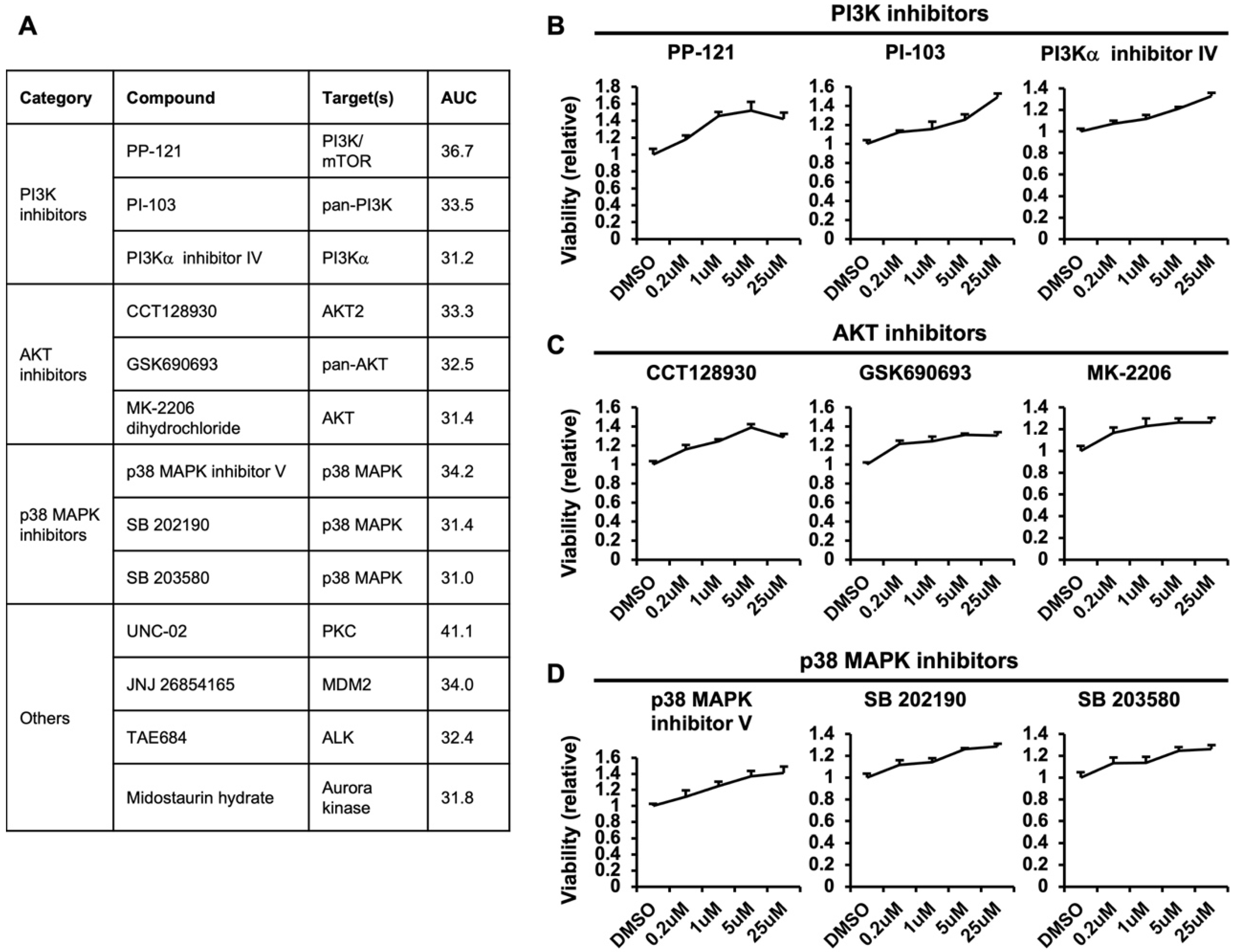
3.3. Positive Regulators of Macropinocytosis-Dependent Growth
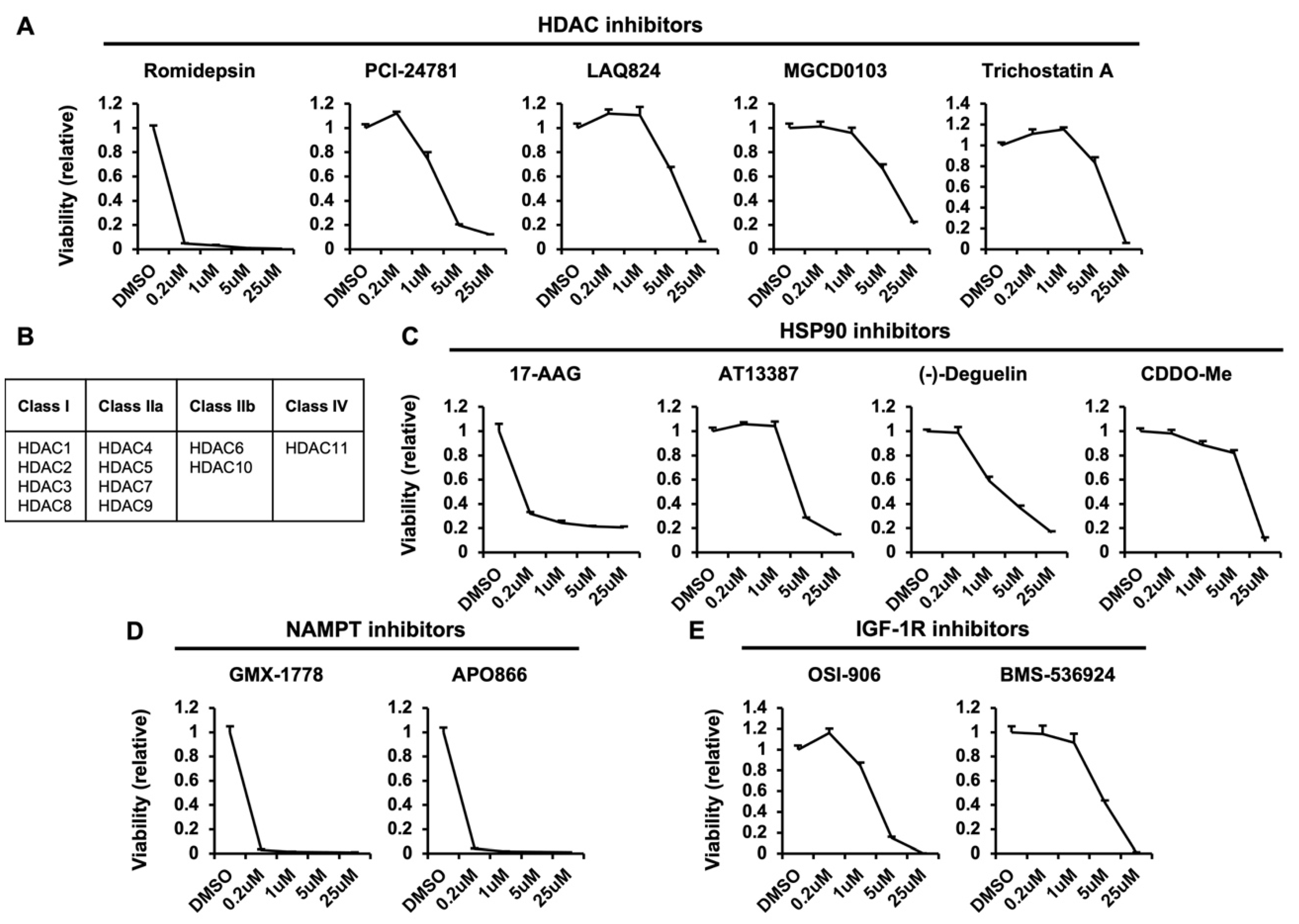
3.4. Negative Regulators of Macropinocytosis-Dependent Growth
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Kamphorst, J.J.; Nofal, M.; Commisso, C.; Hackett, S.R.; Lu, W.; Grabocka, E.; Vander Heiden, M.G.; Miller, G.; Drebin, J.A.; Bar-Sagi, D.; et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015, 75, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef]
- Davidson, S.M.; Jonas, O.; Keibler, M.A.; Hou, H.W.; Luengo, A.; Mayers, J.R.; Wyckoff, J.; Del Rosario, A.M.; Whitman, M.; Chin, C.R.; et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat. Med. 2017, 23, 235–241. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Renschler, M.F. Increased survival in pancreatic cancer with nab-Paclitaxel plus Gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Pelossof, R.; Fairchild, L.; Huang, C.-H.; Widmer, C.; Sreedharan, V.T.; Sinha, N.; Lai, D.-Y.; Guan, Y.; Premsrirut, P.K.; Tschaharganeh, D.F.; et al. Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol. 2017, 35, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Fellmann, C.; Hoffmann, T.; Sridhar, V.; Hopfgartner, B.; Muhar, M.; Roth, M.; Lai, D.Y.; Barbosa, I.A.; Kwon, J.S.; Guan, Y.; et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013, 5, 1704–1713. [Google Scholar] [CrossRef] [PubMed]
- Jayashankar, V.; Edinger, A.L. Macropinocytosis confers resistance to therapies targeting cancer anabolism. Nat. Commun. 2020, 11, 1121. [Google Scholar] [CrossRef]
- Palm, W.; Park, Y.; Wright, K.; Pavlova, N.N.; Tuveson, D.A.; Thompson, C.B. The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell 2015, 162, 259–270. [Google Scholar] [CrossRef]
- Li, L.; Wan, T.; Wan, M.; Liu, B.; Cheng, R.; Zhang, R. The effect of the size of fluorescent dextran on its endocytic pathway: Size-based endocytic entry for fluid cargoes. Cell. Biol. Int. 2015, 39, 531–539. [Google Scholar] [CrossRef]
- Fennell, M.; Commisso, C.; Ramirez, C.; Garippa, R.; Bar-Sagi, D. High-content, full genome siRNA screen for regulators of oncogenic HRAS -driven Macropinocytosis. ASSAY Drug Dev. Technol. 2015, 13, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Singla, B.; Ghoshal, P.; Faulkner, J.L.; Cherian-Shaw, M.; O’Connor, P.M.; She, J.X.; de Chantemele, E.J.B.; Csányi, G. Identification of novel macropinocytosis inhibitors using a rational screen of Food and Drug Administration-approved drugs: Identification of novel macropinocytosis inhibitors. Br. J. Pharmacol. 2018, 175, 3640–3655. [Google Scholar] [CrossRef] [PubMed]
- Redelman-Sidi, G.; Binyamin, A.; Gaeta, I.; Palm, W.; Thompson, C.B.; Romesser, P.B.; Lowe, S.W.; Bagul, M.; Doench, J.G.; Root, D.E.; et al. The canonical Wnt pathway drives macropinocytosis in cancer. Cancer Res. 2018, 78, 4658–4670. [Google Scholar] [CrossRef]
- Gao, Y.S.; Hubbert, C.C.; Lu, J.; Lee, Y.S.; Lee, J.Y.; Yao, T.P. Histone Deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol. Cell Biol. 2007, 27, 8637–8647. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.Y.; Shang, B.Y.; Du, Y.; Li, Y.; Li, L.; Xu, X.D.; Zhen, Y.S. A new HDAC inhibitor cinnamoylphenazine shows antitumor activity in association with intensive macropinocytosis. Oncotarget 2017, 8, 14748–14758. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Bae, S.-C. Histone deacetylase inhibitors: Molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Ju, H.-Q.; Zhuang, Z.-N.; Li, H.; Tian, T.; Lu, Y.-X.; Fan, X.-Q.; Zhou, H.-J.; Mo, H.-Y.; Sheng, H.; Chiao, P.J.; et al. Regulation of the Nampt-mediated NAD salvage pathway and its therapeutic implications in pancreatic cancer. Cancer Lett. 2016, 379, 1–11. [Google Scholar] [CrossRef]
- Chini, C.; Guerrico AM, G.; Nin, V.; Camacho-Pereira, J.; Escande, C.; Barbosa, M.T.; Chini, E.N. Targeting of NAD metabolism in pancreatic cancer cells: Potential novel therapy for pancreatic tumors. Clin. Cancer Res. 2014, 20, 120–130. [Google Scholar] [CrossRef]
- Yuan, J.; Yin, Z.; Tao, K.; Wang, G.; Gao, J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy (Review). Oncol. Lett. 2017, 15, 41–47. [Google Scholar] [CrossRef]
- Bar-Sagi, D.; Feramisco, J.R. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 1986, 233, 1061–1068. [Google Scholar] [CrossRef]
- Johnson, J.C.; Martinez, O.; Honko, A.N.; Hensley, L.E.; Olinger, G.G.; Basler, C.F. Pyridinyl imidazole inhibitors of p38 MAP kinase impair viral entry and reduce cytokine induction by Zaire ebolavirus in human dendritic cells. Antivir. Res. 2014, 107, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhu, Z.; Tong, B.C.-K.; Iyaswamy, A.; Xie, W.-J.; Zhu, Y.; Sreenivasmurthy, S.G.; Senthilkumar, K.; Cheung, K.-H.; Song, J.-X.; et al. A stress response p38 MAP kinase inhibitor SB202190 promoted TFEB/TFE3-dependent autophagy and lysosomal biogenesis independent of p38. Redox Biol. 2020, 32, 101445. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; She, H.; Zhang, T.; Xu, H.; Cheng, L.; Yepes, M.; Mao, Z. p38 MAPK inhibits autophagy and promotes microglial inflammatory responses by phosphorylating ULK1. J. Cell Biol. 2018, 217, 315–328. [Google Scholar] [CrossRef] [PubMed]
- King, J.S.; Kay, R.R. The origins and evolution of macropinocytosis. Philos. Trans. R. Soc. B Biol. Sci. 2018, 374, 20180158. [Google Scholar] [CrossRef] [PubMed]
- Akishiba, M.; Takeuchi, T.; Kawaguchi, Y.; Sakamoto, K.; Yu, H.H.; Nakase, I.; Futaki, S. Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat. Chem. 2017, 9, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Tejeda-Muñoz, N.; Albrecht, L.V.; Bui, M.H.; De Robertis, E.M. Wnt canonical pathway activates macropinocytosis and lysosomal degradation of extracellular proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 10402–10411. [Google Scholar] [CrossRef]
- Albrecht, L.V.; Tejeda-Muñoz, N.; Bui, M.H.; Cicchetto, A.C.; Di Biagio, D.; Colozza, G.; Schmid, E.; Piccolo, S.; Christofk, H.R.; De Robertis, E.M. GSK3 Inhibits Macropinocytosis and Lysosomal Activity through the Wnt Destruction Complex Machinery. Cell Rep. 2020, 32, 107973. [Google Scholar] [CrossRef]
- Ramaiah, M.J.; Tangutur, A.D.; Manyam, R.R. Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 2021, 277, 119504. [Google Scholar] [CrossRef]
- Li, L.; Chen, N.-N.; You, Q.-D.; Xu, X.-L. An updated patent review of anticancer Hsp90 inhibitors (2013-present). Expert Opin. Ther. Pat. 2020, 31, 67–80. [Google Scholar] [CrossRef]
- Winn, B.J.; Kersten, R.C. Teprotumumab. Ophthalmology 2021, 128, 1627–1651. [Google Scholar] [CrossRef]
- Deleyto-Seldas, N.; Efeyan, A. The mTOR–autophagy axis and the control of metabolism. Front. Cell Dev. Biol. 2021, 9, 655731. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Song, J.H.; Kim, M.J.; Song, M.G.; Ku, A.A.; Bandyopadhyay, S.; McCormick, F.; Kim, S.E. Novel Regulators of Macropinocytosis-Dependent Growth Revealed by Informer Set Library Screening in Pancreatic Cancer Cells. Metabolites 2022, 12, 831. https://doi.org/10.3390/metabo12090831
Kim SH, Song JH, Kim MJ, Song MG, Ku AA, Bandyopadhyay S, McCormick F, Kim SE. Novel Regulators of Macropinocytosis-Dependent Growth Revealed by Informer Set Library Screening in Pancreatic Cancer Cells. Metabolites. 2022; 12(9):831. https://doi.org/10.3390/metabo12090831
Chicago/Turabian StyleKim, Sang Hoon, Jae Ho Song, Min Ji Kim, Mun Gu Song, Angel A. Ku, Sourav Bandyopadhyay, Frank McCormick, and Sung Eun Kim. 2022. "Novel Regulators of Macropinocytosis-Dependent Growth Revealed by Informer Set Library Screening in Pancreatic Cancer Cells" Metabolites 12, no. 9: 831. https://doi.org/10.3390/metabo12090831
APA StyleKim, S. H., Song, J. H., Kim, M. J., Song, M. G., Ku, A. A., Bandyopadhyay, S., McCormick, F., & Kim, S. E. (2022). Novel Regulators of Macropinocytosis-Dependent Growth Revealed by Informer Set Library Screening in Pancreatic Cancer Cells. Metabolites, 12(9), 831. https://doi.org/10.3390/metabo12090831






