Similar 5F-APINACA Metabolism between CD-1 Mouse and Human Liver Microsomes Involves Different P450 Cytochromes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Steady-State Kinetics for 5F-APINACA Metabolism by Human and Mouse Liver Microsomes
2.3. Kinetic Analyses with Explicit Equations and Numerical Methods
2.4. Chemical Inhibitor Phenotyping of Reactions for CYP Contributions
2.5. HPLC Resolution and Analysis of 5F-APINACA Analytes from Reactions
2.6. Statistical Analyses
3. Results
3.1. Conditions Were Optimized to Improve Accuracy of Kinetic Measures for Microsomal 5F-APINACA Metabolism
3.2. Human 5F-APINACA Metabolism Favored Two Competing Pathways Similarly
3.3. Human 5F-APINACA Microsomal Metabolism Impacted by Substrate Effector Site
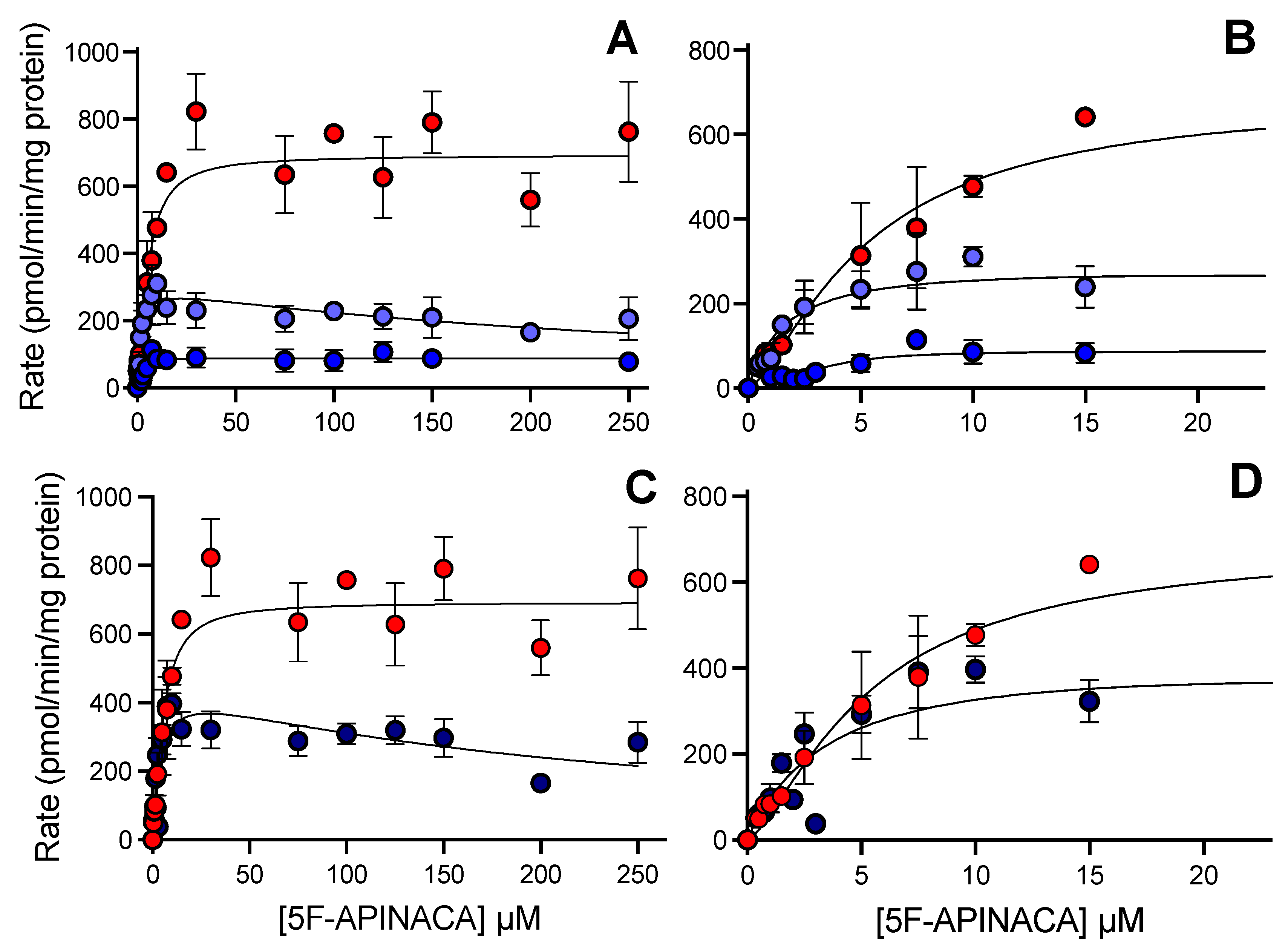
3.4. CD-1 Mouse 5F-APINACA Microsomal Metabolism Favored the Same Two Competing Pathways
3.5. CD-1 Mouse 5F-APINACA Microsomal Metabolism Impacted Similarly by Substrate Effector Site
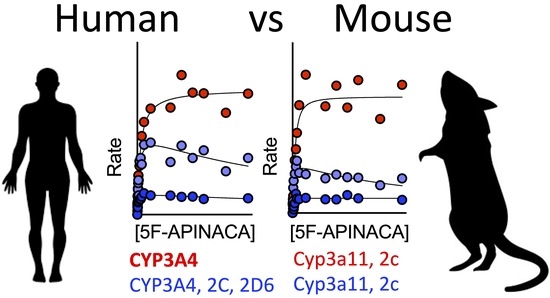
3.6. CYP Selectivity and Specificity toward 5F-APINACA Differ between Humans and CD-1 Mice
4. Discussion
4.1. Experimental Design Impacts Metabolism Observations and Conclusions
4.2. Human and CD-1 Mice Share Common Competing Metabolic Pathways for 5F-APINACA
4.3. Similarities and Differences in CYPs Responsible for 5F-APINACA Metabolism Exist between Species
4.4. Limitations of the Current Findings
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- News: January 2020—UNODC-SMART: Share of NPS Stimulants and Synthetic Cannabinoids Large but Stable While Opioids Increase. Available online: https://www.unodc.org/LSS/Announcement/Details/0096a6ec-c5d0-404f-a559-d15fb84ca75c (accessed on 22 September 2020).
- Van Hout, M.C.; Hearne, E. User Experiences of Development of Dependence on the Synthetic Cannabinoids, 5f-AKB48 and 5F-PB-22, and Subsequent Withdrawal Syndromes. Int. J. Ment. Health Addict. 2017, 15, 565–579. [Google Scholar] [CrossRef]
- Winstock, A.; Lynskey, M.; Borschmann, R.; Waldron, J. Risk of Emergency Medical Treatment Following Consumption of Cannabis or Synthetic Cannabinoids in a Large Global Sample. J. Psychopharmacol. 2015, 29, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.; Mukaratirwa, S.; Petersen-Jones, M. Incidences and Range of Spontaneous Findings in the Lymphoid and Haemopoietic System of Control Charles River CD-1 Mice (Crl: CD-1(ICR) BR) Used in Chronic Toxicity Studies. Toxicol. Pathol. 2012, 40, 375–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alleva, E.; Laviola, G. Short-Term and Delayed Behavioral Effects of Pre- and Post-Weaning Morphine in Mice. Pharmacol. Biochem. Behav. 1987, 26, 539–542. [Google Scholar] [CrossRef]
- Perez, V.J.; Gonzalez, G.E.; Smith, C.J. Exposure to Ethanol during Pregnancy in Mice: Potential Importance of Dose for the Development of Tolerance in Offspring. Physiol. Behav. 1983, 30, 485–488. [Google Scholar] [CrossRef]
- File, S.E. Strain Differences in Mice in the Development of Tolerance to the Anti-Pentylenetetrazole Effects of Diazepam. Neurosci. Lett. 1983, 42, 95–98. [Google Scholar] [CrossRef]
- Moser, V.C.; Balster, R.L. The Effects of Acute and Repeated Toluene Exposure on Operant Behavior in Mice. Neurobehav. Toxicol. Teratol. 1981, 3, 471–475. [Google Scholar]
- Kaplan, G.B.; Greenblatt, D.J.; Kent, M.A.; Cotreau-Bibbo, M.M. Caffeine Treatment and Withdrawal in Mice: Relationships between Dosage, Concentrations, Locomotor Activity and A1 Adenosine Receptor Binding. J. Pharmacol. Exp. Ther. 1993, 266, 1563–1572. [Google Scholar]
- Milne, B.; Cervenko, F.W.; Jhamandas, K.H. Physical Dependence on Nitrous Oxide in Mice: Resemblance to Alcohol but Not to Opiate Withdrawal. Can. Anaesth. Soc. J. 1981, 28, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Sansone, M.; Battaglia, M.; Ekblom, J.; Nalepa, I.; Vetulani, J. Avoidance Learning during Antidepressant Withdrawal in Mice. J. Pharm. Pharmacol. 1991, 43, 51–53. [Google Scholar] [CrossRef]
- Kaplan, G.B.; Bharmal, N.H.; Leite-Morris, K.A.; Adams, W.R. Role of Adenosine A1 and A2A Receptors in the Alcohol Withdrawal Syndrome. Alcohol 1999, 19, 157–162. [Google Scholar] [CrossRef]
- Wu, H.; MacDougall, R.S.; Clithero, A.D.; Leitermann, R.J.; Terashvili, M.; Tseng, L.F. Opposite Conditioned Place Preference Responses to Endomorphin-1 and Endomorphin-2 in the Mouse. Neurosci. Lett. 2004, 365, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Salzmann, J.; Marie-Claire, C.; Le Guen, S.; Roques, B.P.; Noble, F. Importance of ERK Activation in Behavioral and Biochemical Effects Induced by MDMA in Mice. Br. J. Pharmacol. 2003, 140, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Rauhut, A.S.; Hawrylak, M.; Mardekian, S.K. Bupropion Differentially Alters the Aversive, Locomotor and Rewarding Properties of Nicotine in CD-1 Mice. Pharmacol. Biochem. Behav. 2008, 90, 598–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plasterer, M.R.; Bradshaw, W.S.; Booth, G.M.; Carter, M.W.; Schuler, R.L.; Hardin, B.D. Developmental Toxicity of Nine Selected Compounds Following Prenatal Exposure in the Mouse: Naphthalene, p-Nitrophenol, Sodium Selenite, Dimethyl Phthalate, Ethylenethiourea, and Four Glycol Ether Derivatives. J. Toxicol. Environ. Health Part A Curr. Issues 1985, 15, 25–38. [Google Scholar] [CrossRef]
- Mutluay, D.; Güngör, Ş.; Tenekeci, G.Y.; Köksoy, S.; Çoban, C.S. Effects of Synthetic (JWH-018) Cannabinoids Treatment on Spermatogenesis and Sperm Function. Drug Chem. Toxicol. 2022, 45, 215–222. [Google Scholar] [CrossRef]
- Ossato, A.; Uccelli, L.; Bilel, S.; Canazza, I.; Di Domenico, G.; Pasquali, M.; Pupillo, G.; De Luca, M.A.; Boschi, A.; Vincenzi, F.; et al. Psychostimulant Effect of the Synthetic Cannabinoid JWH-018 and AKB48: Behavioral, Neurochemical, and Dopamine Transporter Scan Imaging Studies in Mice. Front. Psychiatry 2017, 8, 130. [Google Scholar] [CrossRef] [Green Version]
- Canazza, I.; Ossato, A.; Vincenzi, F.; Gregori, A.; Di Rosa, F.; Nigro, F.; Rimessi, A.; Pinton, P.; Varani, K.; Borea, P.A.; et al. Pharmaco-Toxicological Effects of the Novel Third-Generation Fluorinate Synthetic Cannabinoids, 5F-ADBINACA, AB-FUBINACA, and STS-135 in Mice. In Vitro and in Vivo Studies. Hum. Psychopharmacol. Clin. Exp. 2017, 32, e2601. [Google Scholar] [CrossRef] [Green Version]
- Canazza, I.; Ossato, A.; Trapella, C.; Fantinati, A.; De Luca, M.A.; Margiani, G.; Vincenzi, F.; Rimondo, C.; Di Rosa, F.; Gregori, A.; et al. Effect of the Novel Synthetic Cannabinoids AKB48 and 5F-AKB48 on “Tetrad”, Sensorimotor, Neurological and Neurochemical Responses in Mice. In Vitro and in Vivo Pharmacological Studies. Psychopharmacology 2016, 233, 3685–3709. [Google Scholar] [CrossRef]
- Ossato, A.; Canazza, I.; Trapella, C.; Vincenzi, F.; De Luca, M.A.; Rimondo, C.; Varani, K.; Borea, P.A.; Serpelloni, G.; Marti, M. Effect of JWH-250, JWH-073 and Their Interaction on “Tetrad”, Sensorimotor, Neurological and Neurochemical Responses in Mice. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 67, 31–50. [Google Scholar] [CrossRef]
- Little, P.J.; Compton, D.R.; Johnson, M.R.; Melvin, L.S.; Martin, B.R. Pharmacology and Stereoselectivity of Structurally Novel Cannabinoids in Mice. J. Pharmacol. Exp. Ther. 1988, 247, 1046–1051. [Google Scholar]
- Banister, S.D.; Moir, M.; Stuart, J.; Kevin, R.C.; Wood, K.E.; Longworth, M.; Wilkinson, S.M.; Beinat, C.; Buchanan, A.S.; Glass, M.; et al. Pharmacology of Indole and Indazole Synthetic Cannabinoid Designer Drugs AB-FUBINACA, ADB-FUBINACA, AB-PINACA, ADB-PINACA, 5F-AB-PINACA, 5F-ADB-PINACA, ADBICA, and 5F-ADBICA. ACS Chem. Neurosci. 2015, 6, 1546–1559. [Google Scholar] [CrossRef]
- Schindler, C.W.; Gramling, B.R.; Justinova, Z.; Thorndike, E.B.; Baumann, M.H. Synthetic Cannabinoids Found in “Spice” Products Alter Body Temperature and Cardiovascular Parameters in Conscious Male Rats. Drug Alcohol Depend. 2017, 179, 387–394. [Google Scholar] [CrossRef]
- Tai, S.; Hyatt, W.S.; Gu, C.; Franks, L.N.; Vasiljevik, T.; Brents, L.K.; Prather, P.L.; Fantegrossi, W.E. Repeated Administration of Phytocannabinoid Δ(9)-THC or Synthetic Cannabinoids JWH-018 and JWH-073 Induces Tolerance to Hypothermia but Not Locomotor Suppression in Mice, and Reduces CB1 Receptor Expression and Function in a Brain Region-Specific Manner. Pharmacol. Res. 2015, 102, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Wilson, C.D.; Tai, S.; Ewing, L.; Crane, J.; Lockhart, T.; Fujiwara, R.; Radominska-Pandya, A.; Fantegrossi, W.E. Convulsant Effects of Abused Synthetic Cannabinoids JWH-018 and 5F-AB-PINACA Are Mediated by Agonist Actions at CB1 Receptors in Mice. J. Pharmacol. Exp. Ther. 2019, 368, 146–156. [Google Scholar] [CrossRef] [Green Version]
- Pinson, A.; Yarbrough, A.L.; Bush, J.M.; Cabanlong, C.V.; Shoeib, A.; Jackson, B.K.; Fukuda, S.; Gogoi, J.; Fantegrossi, W.E.; McCain, K.; et al. Metabolism, CB1 Cannabinoid Receptor Binding and in Vivo Activity of Synthetic Cannabinoid 5F-AKB48: Implications for Toxicity. Pharmacol. Biochem. Behav. 2020, 195, 172949. [Google Scholar] [CrossRef]
- Pinson, A.O.; Pouncey, D.L.; Schleiff, M.A.; Fantegrossi, W.E.; Prather, P.L.; Radominska-Pandya, A.; Boysen, G.; Miller, G.P. Significance of Competing Metabolic Pathways for 5F-APINACA Based on Quantitative Kinetics. Molecules 2020, 25, 4820. [Google Scholar] [CrossRef]
- Holm, N.B.; Pedersen, A.J.; Dalsgaard, P.W.; Linnet, K. Metabolites of 5F-AKB-48, a Synthetic Cannabinoid Receptor Agonist, Identified in Human Urine and Liver Microsomal Preparations Using Liquid Chromatography High-Resolution Mass Spectrometry. Drug Test. Anal. 2015, 7, 199–206. [Google Scholar] [CrossRef]
- Vikingsson, S.; Josefsson, M.; Gréen, H. Identification of AKB-48 and 5F-AKB-48 Metabolites in Authentic Human Urine Samples Using Human Liver Microsomes and Time of Flight Mass Spectrometry. J. Anal. Toxicol. 2015, 39, 426–435. [Google Scholar] [CrossRef]
- Wang, Z.; Paragas, E.M.; Nagar, S.; Korzekwa, K. Complex Cytochrome P450 Kinetics Due to Multisubstrate Binding and Sequential Metabolism. Part 1. Theoretical Considerations. Drug Metab. Dispos. 2021, 49, 1090–1099. [Google Scholar] [CrossRef]
- Paragas, E.M.; Wang, Z.; Korzekwa, K.; Nagar, S. Complex Cytochrome P450 Kinetics Due to Multisubstrate Binding and Sequential Metabolism. Part 2. Modeling of Experimental Data. Drug Metab. Dispos. 2021, 49, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Schellinger, A.P.; Carr, P.W. Solubility of Buffers in Aqueous–Organic Eluents for Reversed-Phase Liquid Chromatography. Lc Gc N. Am. 2004, 22, 544–549. [Google Scholar]
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Davis, M.A.; Barnette, D.A.; Flynn, N.R.; Pidugu, A.S.; Swamidass, S.J.; Boysen, G.; Miller, G.P. CYP2C19 and 3A4 Dominate Metabolic Clearance and Bioactivation of Terbinafine Based on Computational and Experimental Approaches. Chem. Res. Toxicol. 2019, 32, 1151–1164. [Google Scholar] [CrossRef]
- Johnson, K.A. New Standards for Collecting and Fitting Steady State Kinetic Data. Beilstein J. Org. Chem. 2019, 15, 16–29. [Google Scholar] [CrossRef]
- Martignoni, M.; Groothuis, G.M.M.; de Kanter, R. Species Differences between Mouse, Rat, Dog, Monkey and Human CYP-Mediated Drug Metabolism, Inhibition and Induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef]
- Davidson, C.; Opacka-Juffry, J.; Arevalo-Martin, A.; Garcia-Ovejero, D.; Molina-Holgado, E.; Molina-Holgado, F. Spicing Up Pharmacology: A Review of Synthetic Cannabinoids from Structure to Adverse Events. Adv. Pharmacol. 2017, 80, 135–168. [Google Scholar] [CrossRef]
- Miwa, G.T.; Lu, A.Y. Kinetic Isotope Effects and “metabolic Switching” in Cytochrome P450-Catalyzed Reactions. Bioessays 1987, 7, 215–219. [Google Scholar] [CrossRef]


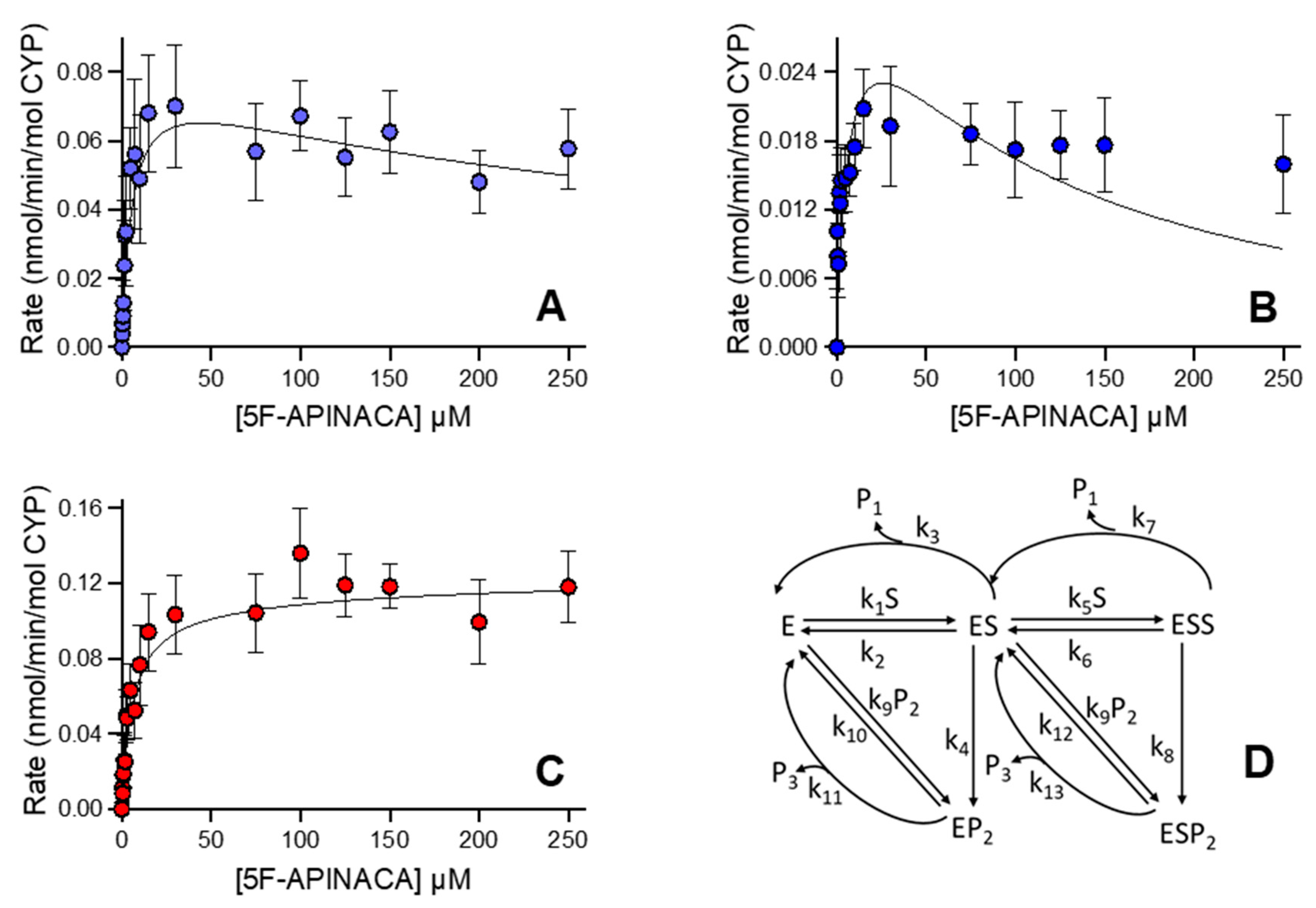


| Metabolite | Model | Vmax, pmol min−1 mg protein−1 | Km, µM | Ki, µM | Vmax/Km, pmol min−1 mg protein−1 µM substrate−1 |
|---|---|---|---|---|---|
| 5OH-APINACA | Michaelis–Menten | 465 (446–485) | 6.7 (5.6–7.9) | 69 | |
| 5F-APINACA-OH | Substrate inhibition | 310 (280–350) | 3.8 (2.9–5.1) | 360 (230–600) | 82 |
| 5F-APINACA-(OH)2 | Substrate inhibition | 77 (70–86) | 1.3 (0.9–1.9) | 810 (420–2800) | 59 |
| Combined | Substrate inhibition | 400 (330–510) | 3.3 (1.9–6.0) | 300 (150–790) | 120 |
| Metabolite | kcat1 min−1 | Km1 µM | kcat1/Km1 mL min−1 nmol−1 | kcat2 min−1 | Km2 µM | kcat2/Km2 mL min−1 nmol−1 |
|---|---|---|---|---|---|---|
| 5OH-APINACA (P1) | k3 1.10 (0.35) | ((k2 + k3 + k4)/k1) 5.69 (0.35) | 0.193 (0.015) | k7 1.23 (0.1) | ((k6 + k7 + k8)/k5) 181 (10) | 0.0065 (0.0013) |
| 5F-APINACA-OH (P2) | k4 1.18 (0.05) | ((k2 + k3 + k4)/k1) 5.69 (0.35) | 0.207 (0.016) | k8 0.17 (0.08) | ((k6 + k7 + k8)/k5) 85 (5) | 0.0009 (0.0001) |
| 5F-APINACA-(OH)2 (P3) | k11 40 b | ((k10 + k11)/k9) 82 (20) | 0.49 (0.12) | k13 40 b | ((k12 + k13)/k9) 58 (6) | 0.69 (0.07) |
| Metabolite | Model | Vmax pmol min−1 mg protein−1 | Km or Kh µM | h | Ki µM | Vmax/Km or Vmax/Kh pmol min−1 mg protein−1 µM substrate−1 |
|---|---|---|---|---|---|---|
| 5OH-APINACA | Hill | 690 (650–730) | 5.4 (4.4–6.7) | 1.4 (1.0–1.8) | 128 | |
| 5F-APINACA-OH | Substrate inhibition | 310 (275–345) | 1.7 (1.2–2.4) | 280 (180–480) | 180 | |
| 5F-APINACA-(OH)2 | Hill | 88 (81–95) | 3.1 (2.1–4.0) | 2.3 (1.3–4.8) | 28 | |
| Combined | Substrate inhibition | 465 (330–730) | 3.8 (1.5–9.6) | 220 (74–1300) | 122 |
| Metabolite | kcat1 min−1 | Km1 µM | kcat1/Km1 mL min−1 nmol−1 | kcat2 min−1 | Km2 µM | kcat2/Km2, mL min−1 nmol−1 |
|---|---|---|---|---|---|---|
| 5OH-APINACA (P1) | k3 0.89 (0.04) | ((k2 + k3 + k4)/k1) 3.75 (0.21) | 0.238 (0.017) | k7 1.17 (0.05) | ((k6 + k7 + k8)/k5) 122 (6) | 0.0096 (0.0006) |
| 5F-APINACA-OH (P2) | k4 0.738 (0.017) | ((k2 + k3 + k4)/k1) 3.75 (21) | 0.197 (0.012) | k8 0.010 (0.04) | ((k6 + k7 + k8)/k5) 122 (6) | 0.0008 (0.00033) |
| 5F-APINACA-(OH)2 (P3) | k11 5 b | ((k10 + k11)/k9) 199 (4) | 0.0251 (0.0005) | k13 5 b | ((k12 + k13)/k9) 9.9 (0.7) | 0.504 (0.034) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crosby, S.V.; Ahmed, I.Y.; Osborn, L.R.; Wang, Z.; Schleiff, M.A.; Fantegrossi, W.E.; Nagar, S.; Prather, P.L.; Boysen, G.; Miller, G.P. Similar 5F-APINACA Metabolism between CD-1 Mouse and Human Liver Microsomes Involves Different P450 Cytochromes. Metabolites 2022, 12, 773. https://doi.org/10.3390/metabo12080773
Crosby SV, Ahmed IY, Osborn LR, Wang Z, Schleiff MA, Fantegrossi WE, Nagar S, Prather PL, Boysen G, Miller GP. Similar 5F-APINACA Metabolism between CD-1 Mouse and Human Liver Microsomes Involves Different P450 Cytochromes. Metabolites. 2022; 12(8):773. https://doi.org/10.3390/metabo12080773
Chicago/Turabian StyleCrosby, Samantha V., Izzeldin Y. Ahmed, Laura R. Osborn, Zeyuan Wang, Mary A. Schleiff, William E. Fantegrossi, Swati Nagar, Paul L. Prather, Gunnar Boysen, and Grover P. Miller. 2022. "Similar 5F-APINACA Metabolism between CD-1 Mouse and Human Liver Microsomes Involves Different P450 Cytochromes" Metabolites 12, no. 8: 773. https://doi.org/10.3390/metabo12080773
APA StyleCrosby, S. V., Ahmed, I. Y., Osborn, L. R., Wang, Z., Schleiff, M. A., Fantegrossi, W. E., Nagar, S., Prather, P. L., Boysen, G., & Miller, G. P. (2022). Similar 5F-APINACA Metabolism between CD-1 Mouse and Human Liver Microsomes Involves Different P450 Cytochromes. Metabolites, 12(8), 773. https://doi.org/10.3390/metabo12080773