Profiling of Volatile Organic Compounds from Four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A Metabolomics Study
Abstract
:1. Introduction
2. Materials and Methods
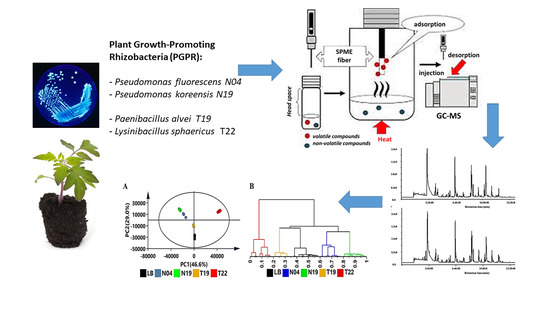
2.1. Bacterial Growth and Experimental Design
2.2. Volatile Collection and Headspace Sampling through SPME
2.3. Gas Chromatography—Time-of-Flight—Mass Spectrometric Analysis
2.4. Multivariate Data Analysis (MVDA)
3. Results
4. Discussion
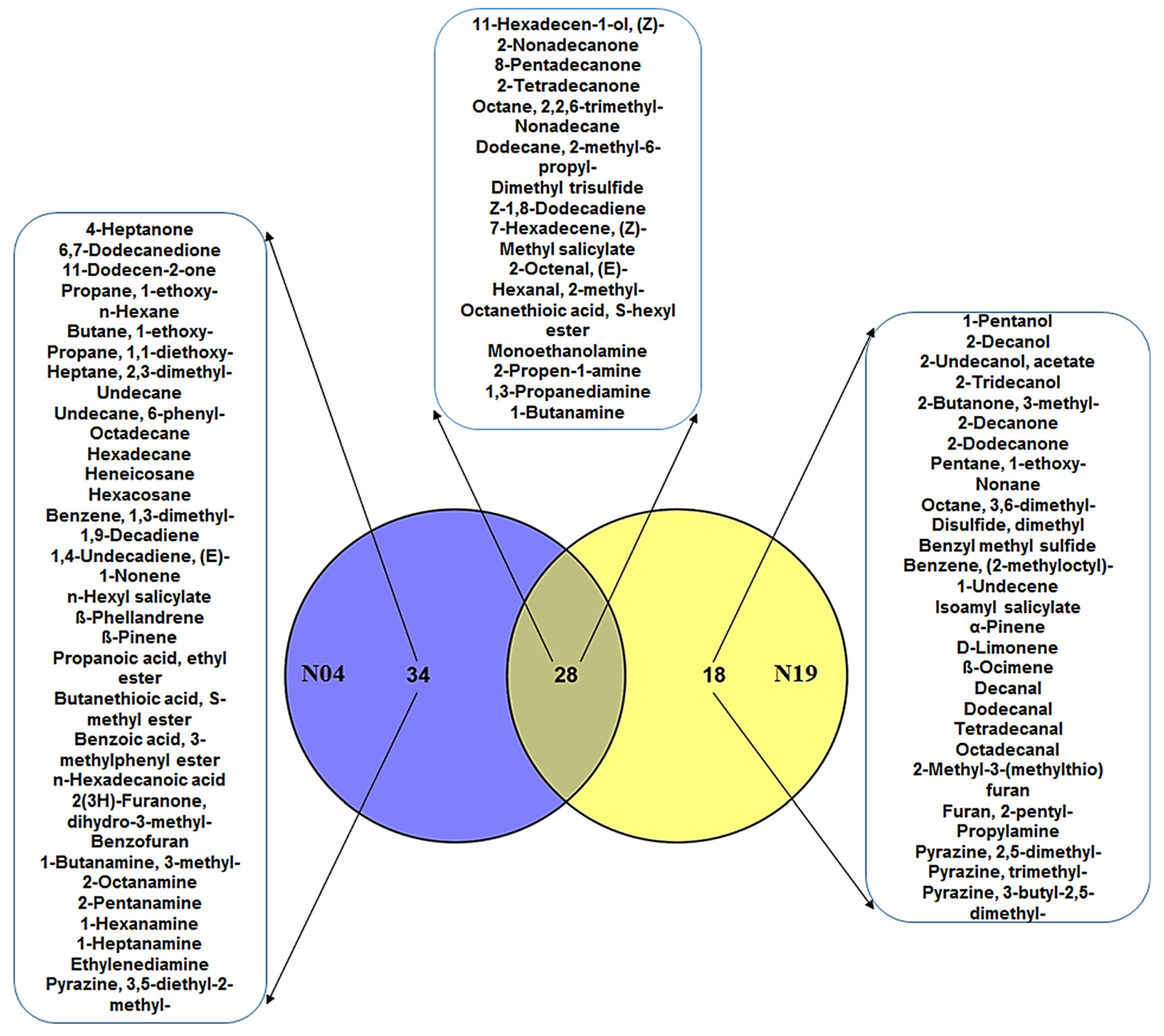
4.1. Comparison and Evaluation of Volatile Organic Compounds Secreted by the Two PGPR Pseudomonas Strains
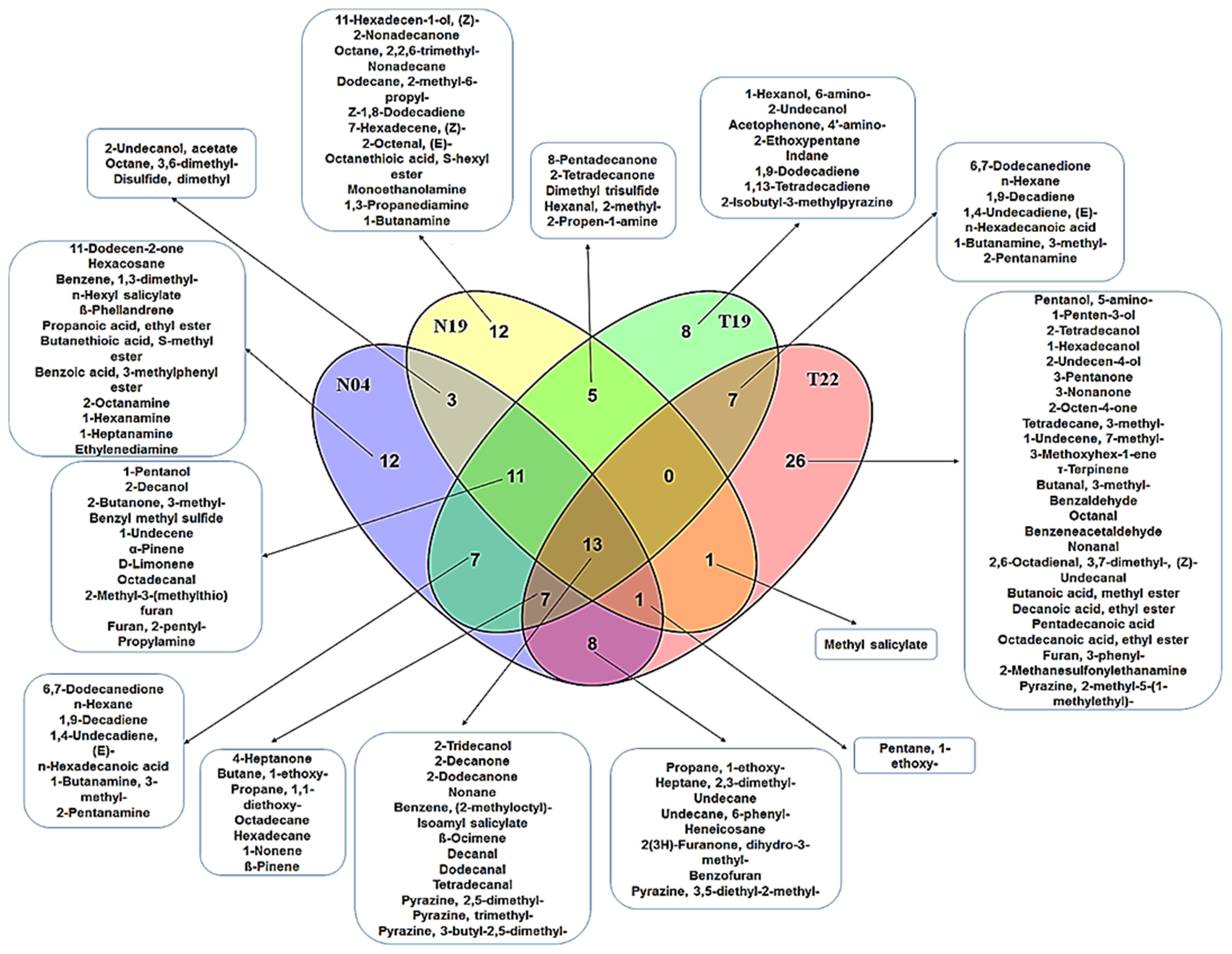
4.2. Comparison and Evaluation of Volatile Organic Compounds Secreted by the Two Pseudomonas Strains, Paenibacillus alvei (T22) and Lysinibacillus sphaericus (T19)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Ahmad, H.M.; Fiaz, S.; Hafeez, S.; Zahra, S.; Shah, A.N.; Gul, B.; Aziz, O.; Rahman, M.U.; Fakhar, A.; Rafique, M.; et al. Plant growth-promoting rhizobacteria eliminate the effect of drought stress in plants: A review. Front. Plant Sci. 2022, 13, 875774. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Piater, L.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Rhizosphere tripartite interactions and PGPR-Mediated metabolic reprogramming towards ISR and plant priming: A metabolomics review. Biology 2022, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Erb, M. Volatiles as inducers and suppressors of plant defense and immunity—Origins, specificity, perception and signaling. Curr. Opin. Plant Biol. 2018, 44, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bano, A.M.D.; Babar, M.D.A. Impacts of plant growth promoters and plant growth regulators on rainfed agriculture. PLoS ONE 2020, 15, e0232926. [Google Scholar] [CrossRef] [PubMed]
- Mommer, L.; Kirkegaard, J.; Van Ruijven, J. Root—Root interactions: Towards a rhizosphere framework. Trends Plant Sci. 2016, 21, 209–217. [Google Scholar] [CrossRef]
- Macías-Rodríguez, L.; Contreras-Cornejo, H.A.; Adame-Garnica, S.G.; Del-Val, E.; Larsen, J. The interactions of Trichoderma at multiple trophic levels: Inter-kingdom communication. Microbiol. Res. 2020, 240, 126552. [Google Scholar] [CrossRef]
- Kong, Z.; Liu, H. Modification of rhizosphere microbial communities: A possible mechanism of plant growth promoting rhizobacteria enhancing plant growth and fitness. Front. Plant Sci. 2022, 13, 920813. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151. [Google Scholar] [CrossRef]
- Vlot, A.C.; Rosenkranz, M. Volatile compounds—The language of all kingdoms? J. Exp. Bot. 2022, 73, 445–448. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Bakker, P.A.H.M. Interactions between plants and beneficial Pseudomonas spp.: Exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 2007, 92, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Huang, J.; Lu, X.; Zhou, C. Development of plant systemic resistance by beneficial rhizobacteria: Recognition, initiation, elicitation and regulation. Front. Plant Sci. 2022, 13, 952397. [Google Scholar] [CrossRef]
- Li, W.; Lee, S.-Y.; Cho, Y.-J.; Ghim, S.-Y.; Jung, H.-Y. Mediation of induced systemic resistance by the plant growth-promoting rhizobacteria Bacillus pumilus S2-3-2. Mol. Biol. Rep. 2020, 47, 8429–8438. [Google Scholar] [CrossRef] [PubMed]
- Ortíz-Castro, R.; Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; López-Bucio, J. The role of microbial signals in plant growth and development. Plant Signal. Behav. 2009, 4, 701–712. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant—Microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Lemfack, M.C.; Nickel, J.; Dunkel, M.; Preissner, R.; Piechulla, B. mVOC: A database of microbial volatiles. Nucleic Acids Res. 2014, 42, 744–748. [Google Scholar] [CrossRef]
- Audrain, B.; Farag, M.A.; Ryu, C.-M.; Ghigo, J.-M. Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol. Rev. 2017, 39, 222–233. [Google Scholar] [CrossRef]
- Farag, M.A.; Zhang, H.; Ryu, C.-M. Dynamic chemical communication between plants and bacteria through airborne signals: Induced resistance by bacterial volatiles. J. Chem. Ecol. 2013, 39, 1007–1018. [Google Scholar] [CrossRef]
- Farag, M.A.; Ryu, C.-M.; Sumner, L.W.; Paré, P.W. GC–MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 2006, 67, 2262–2268. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Ryu, C.-M.; Zhang, S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef]
- Cho, S.M.; Kang, B.R.; Han, S.H.; Anderson, A.J.; Park, J.-Y.; Lee, Y.-H. 2R,3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2008, 21, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.S.; Ryu, C.-M.; Lee, S.; Park, H.B.; Han, K.S.; Lee, J.H.; Lee, K.; Chung, W.S.; Jeong, M.-J.; Kim, H.K.; et al. Proteome analysis of Arabidopsis seedlings exposed to bacterial volatiles. Planta 2010, 232, 1355–1370. [Google Scholar] [CrossRef] [PubMed]
- Planchamp, C.; Glauser, G.; Mauch-Mani, B. Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 2014, 5, 719. [Google Scholar] [CrossRef]
- Insam, H.; Seewald, M.S.A. Volatile organic compounds (VOCs) in soils. Biol. Fertil. Soils 2010, 46, 199–213. [Google Scholar] [CrossRef]
- Garbeva, P.; Hordijk, C.; Gerards, S.; de Boer, W. Volatiles produced by the mycophagous soil bacterium Collimonas. FEMS Microbiol. Ecol. 2014, 87, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1846. [Google Scholar] [CrossRef]
- Rudolph, N.; Labuschagne, N.; Aveling, T. The effect of plant growth promoting rhizobacteria on seed germination and seedling growth of maize. Seed Sci. Technol. 2015, 43, 507–518. [Google Scholar] [CrossRef]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Labuschagne, N. Differential metabolic reprogramming in Paenibacillus alvei-primed Sorghum bicolor seedlings in response to Fusarium pseudograminearum infection. Metabolites 2019, 9, 150. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Labuschagne, N.; Dubery, I.A. Unravelling the metabolic reconfiguration of the post-challenge primed state in Sorghum bicolor responding to Colletotrichum sublineolum infection. Metabolites 2019, 9, 194. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic profiling of PGPR-treated tomato plants reveal priming-related adaptations of secondary metabolites and aromatic amino acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef]
- Badoil, L.; Benanou, D. Characterization of volatile and semivolatile compounds in waste landfill leachates using stir bar sorptive extraction—GC/MS. Anal. Bioanal. Chem. 2009, 393, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Iizuka, Y.; Kobayashi, M.; Fukushima, A.; Saito, K. Development of a direct headspace collection method from Ara-bidopsis seedlings using HS-SPME-GC-TOF-MS analysis. Metabolites 2013, 3, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Rasanen, I.; Viinamäki, J.; Vuori, E.; Ojanperä, I. Headspace in-tube extraction gas chromatography—Mass spectrometry for the analysis of hydroxylic methyl-derivatized and volatile organic compounds in blood and urine. J. Anal. Toxicol. 2017, 34, 113–121. [Google Scholar] [CrossRef]
- De Grazia, S.; Gionfriddo, E.; Pawliszyn, J. A new and efficient solid phase microextraction approach for analysis of high fat content food samples using a matrix-compatible coating. Talanta 2017, 167, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.-H.; Chen, G.-S.; Xiong, Z.-H.; Fan, Y.-X.; Wang, X.-C.; Liu, Y. Applications of solid-phase microextraction in food analysis. TrAC Trends Anal. Chem. 2016, 80, 12–29. [Google Scholar] [CrossRef]
- Zhang, Q.-H.; Zhou, L.-D.; Chen, H.; Wang, C.-Z.; Xia, Z.-N.; Yuan, C.-S. Solid-phase microextraction technology for in vitro and in vivo metabolite analysis. TrAC Trends Anal. Chem. 2016, 80, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Jelen, H.H.; Obuchowska, M.; Zawirska-Wojtasiak, R.; Wasowicz, E. Headspace solid-phase microextraction use for the characterization of volatile compounds in vegetable oils of different sensory quality. J. Agric. Food Chem. 2000, 48, 2360–2367. [Google Scholar] [CrossRef]
- Doleschall, F.; Recseg, K.; Kemény, Z.; Kővári, K. Comparison of differently coated SPME fibres applied for monitoring volatile substances in vegetable oils. Eur. J. Lipid Sci. Technol. 2003, 105, 333–338. [Google Scholar] [CrossRef]
- Szekely Gabor, J. Hierarchical clustering via joint-within diastances: Extending ward’s minimum variance and method. J. Classif. 2005, 22, 158–183. [Google Scholar]
- Fouedjio, F. A hierarchical clustering method for multivariate geostatistical data. Spat. Stat. 2016, 18, 333–351. [Google Scholar] [CrossRef]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical per-spective. Trends Food Sci. Technol. 2018, 7, 83–90. [Google Scholar] [CrossRef]
- Homayoun, S.B.; Shrikant, I.B.; Kazem, M.; Hemmat, M.; Reza, M.; Sadeghi-Bazargani, H.; Bangdiwala, S.I.; Mohammad, K.; Maghsoudi, H.; Mohammadi, R. Compared application of the new OPLS-DA statistical model versus partial least squares regression to manage large numbers of variables in an injury case-control study. Sci. Res. Essays 2011, 6, 4369–4377. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for significance testing of PLS and OPLS models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2013, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- NIST Research Library. Available online: www.nist.gov/nist-research-library (accessed on 4 April 2020).
- Available online: https://metabolomics.ucdavis.edu/compound-identification (accessed on 7 October 2020).
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A conversation on data mining strategies in LC-MS untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 18–20. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjo, L.; Shockcor, J.P.; Gottfries, J.; Moritz, T. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Van Velzen, E.J.J.; Van Duijnhoven, J.P.M.; Van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. BioSyst. 2014, 11, 13–19. [Google Scholar] [CrossRef]
- Gutiérrez-Luna, F.M.; López-Bucio, J.; Altamirano-Hernández, J.; Valencia-Cantero, E.; De La Cruz, H.R.; Macías-Rodríguez, L. Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound emission. Symbiosis 2010, 51, 75–83. [Google Scholar] [CrossRef]
- Finkel, O.M.; Castrillo, G.; Paredes, S.H.; González, I.S.; Dangl, J.L.; Hill, C. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 2018, 38, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Silverman, P.; Raskin, I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 1997, 385, 718–721. [Google Scholar] [CrossRef]
- Hunziker, L.; Bönisch, D.; Groenhagen, U.; Bailly, A.; Schulz, S.; Weisskopf, L. Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl. Environ. Microbiol. 2015, 81, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Lee, S.J.; Moon, J.H.; Park, K.H.; Yang, K.Y.; Cho, B.H. GacS-dependent production of 2R, 3R-butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Mol. Plant Microbe Interact. 2006, 19, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Massalha, H.; Korenblum, E.; Tholl, D.; Aharoni, A. Small molecules below-ground: The role of specialized metabolites in the rhizosphere. Plant J. 2017, 90, 788–807. [Google Scholar] [CrossRef]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-W.; Mu, W.; Zhu, B.-Y.; DU, Y.-C.; Liu, F. Antagonistic activities of volatiles from four strains of Bacillus spp. and Paenibacillus spp. against soil-borne plant pathogens. Agric. Sci. China 2008, 7, 1104–1114. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolomic evaluation of tissue-specific defense responses in tomato plants modulated by PGPR-priming against Phytophthora capsici infection. Plants 2021, 10, 1530. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Profiling of Volatile Organic Compounds from Four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A Metabolomics Study. Metabolites 2022, 12, 763. https://doi.org/10.3390/metabo12080763
Mhlongo MI, Piater LA, Dubery IA. Profiling of Volatile Organic Compounds from Four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A Metabolomics Study. Metabolites. 2022; 12(8):763. https://doi.org/10.3390/metabo12080763
Chicago/Turabian StyleMhlongo, Msizi I., Lizelle A. Piater, and Ian A. Dubery. 2022. "Profiling of Volatile Organic Compounds from Four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A Metabolomics Study" Metabolites 12, no. 8: 763. https://doi.org/10.3390/metabo12080763
APA StyleMhlongo, M. I., Piater, L. A., & Dubery, I. A. (2022). Profiling of Volatile Organic Compounds from Four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A Metabolomics Study. Metabolites, 12(8), 763. https://doi.org/10.3390/metabo12080763