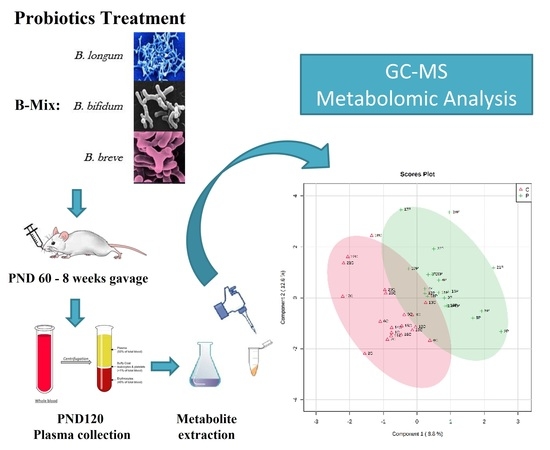
Effects of Chronic Bifidobacteria Administration in Adult Male Rats on Plasma Metabolites: A Preliminary Metabolomic Study
Abstract
:1. Introduction
2. Experimental Design
3. Material and Methods
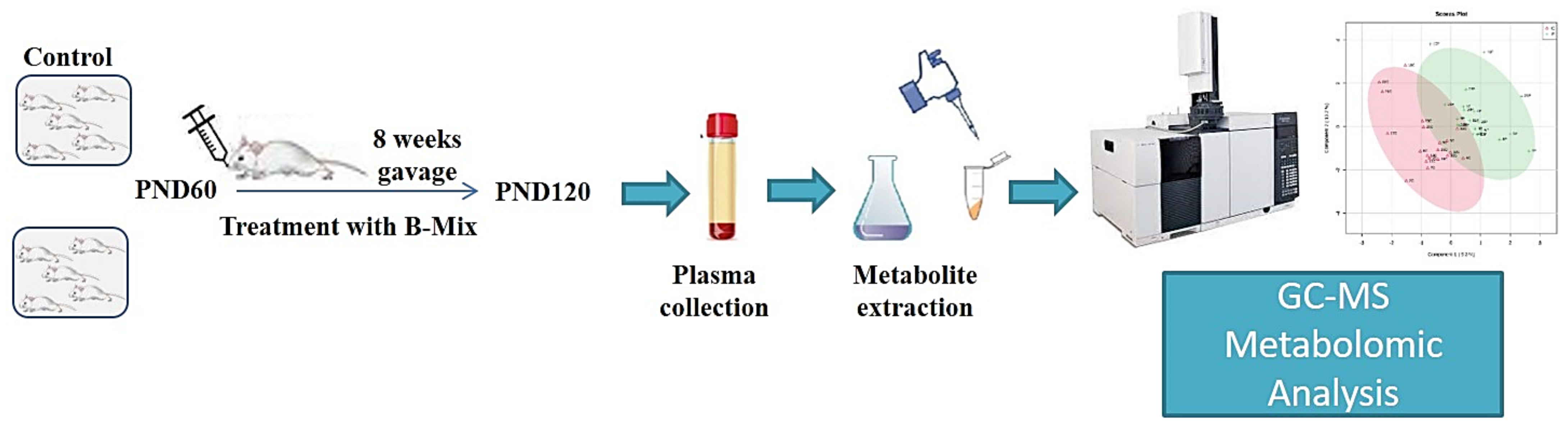
3.1. Animals
3.2. Bifidobacteria Treatment
3.3. Plasma Collection
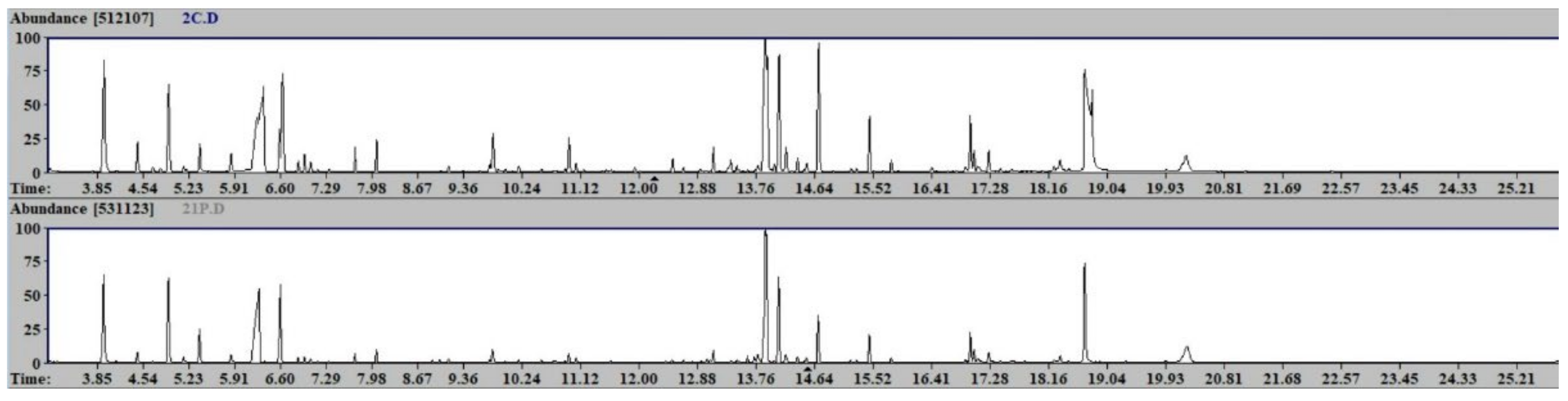
3.4. Sample Preparation and GC-MS Analysis
3.5. Statistical Analysis
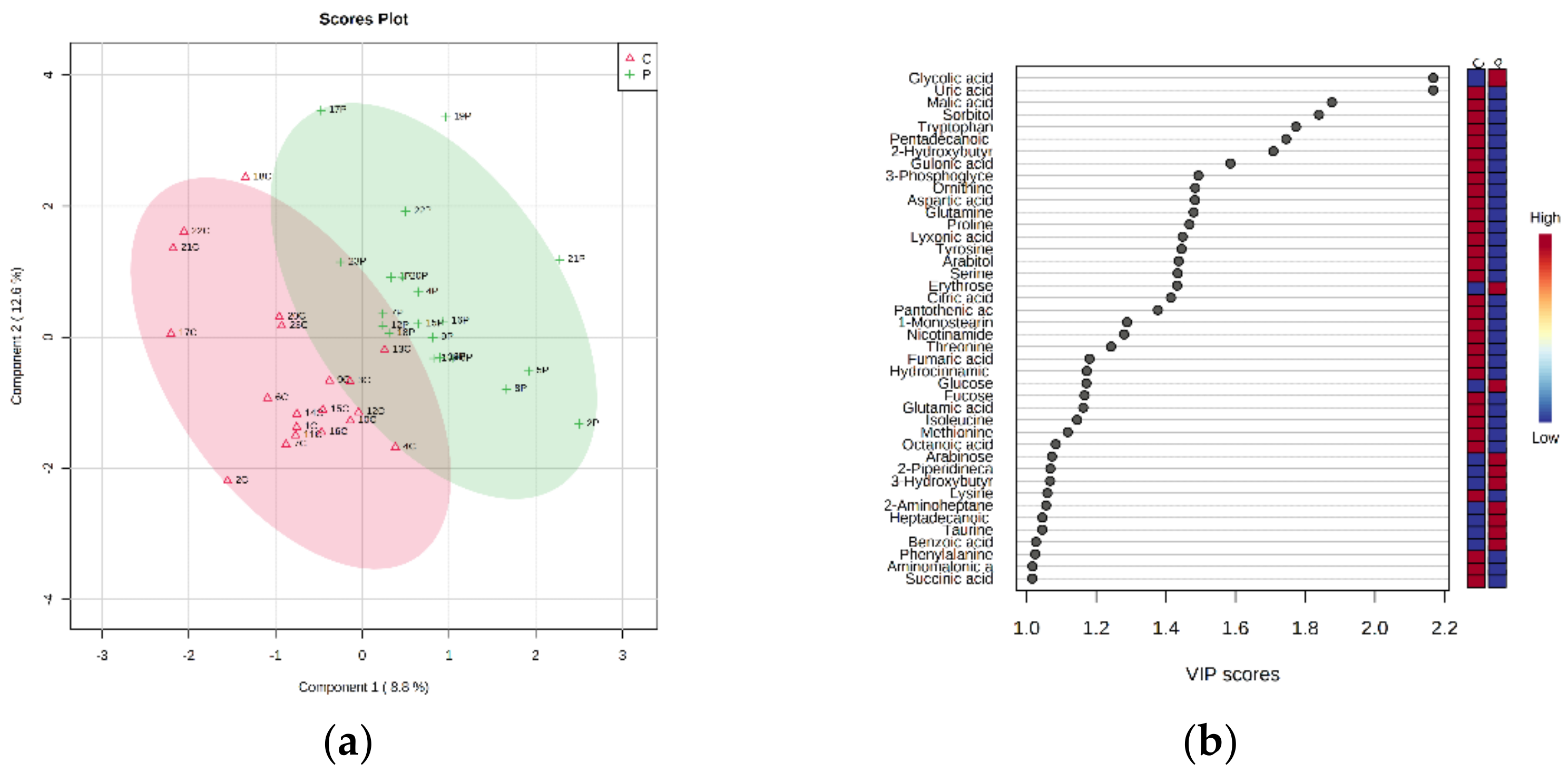
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Hoban, A.E.; Moloney, R.D.; Golubeva, A.V.; McVey Neufeld, K.A.; O’Sullivan, O.; Patterson, E.; Stanton, C.; Dinan, T.G.; Clarke, G.; Cryan, J.F. Behavioural and neurochemical consequences of chronic gut microbiota depletion during adulthood in the rat. Neuroscience 2016, 339, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.A.; Tillisch, K.; Gupta, A. Gut/brain axis and the microbiota. J. Clin. Investig. 2015, 125, 926–938. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Stilling, R.M.; Kennedy, P.J.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Minireview: Gut microbiota: The neglected endocrine organ. Mol. Endocrinol. 2014, 28, 1221–1238. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Lin, J.H.; Chiba, M.; Baillie, T.A. Is the role of the small intestine in first-pass metabolism overemphasized? Pharmacol. Rev. 1999, 51, 135–158. [Google Scholar] [PubMed]
- García-Cabrerizo, R.; Carbia, C.; O’Riordan, K.J.; Schellekens, H.; Cryan, J.F. Microbiota-gut-brain axis as a regulator of reward processes. J. Neurochem. 2021, 157, 1495–1524. [Google Scholar] [CrossRef]
- Distrutti, E.; Monaldi, L.; Ricci, P.; Fiorucci, S. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. WJG 2016, 22, 2219–2241. [Google Scholar] [CrossRef] [PubMed]
- Groen, R.N.; de Clercq, N.C.; Nieuwdorp, M.; Hoenders, H.J.R.; Groen, A.K. Gut microbiota, metabolism and psychopathology: A critical review and novel perspectives. Crit. Rev. Clin. Lab. Sci. 2018, 55, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Moya, A.; Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [CrossRef]
- Wallner-Liebmann, S.; Gralka, E.; Tenori, L.; Konrad, M.; Hofmann, P.; Dieber-Rotheneder, M.; Turano, P.; Luchinat, C.; Zatloukal, K. The impact of free or standardized lifestyle and urine sampling protocol on metabolome recognition accuracy. Genes Nutr. 2015, 10, 441. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, A.; Barbas, C. Chronic diseases and lifestyle biomarkers identification by metabolomics. In Metabolomics: From Fundamentals to Clinical Applications; Sussulini, A., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2017; Volume 965, pp. 235–263. ISBN 978-3-319-47655-1. [Google Scholar]
- Mejía-Granados, D.M.; Villasana-Salazar, B.; Coan, A.C.; Rizzi, L.; Balthazar, M.L.F.; de Godoi, A.B.; do Canto, A.M.; da Rosa, D.C.; Silva, L.S.; do Tacla, R.R.; et al. Gut microbiome in neuropsychiatric disorders. Arq. Neuro-Psiquiatr. 2022, 80, 192–207. [Google Scholar] [CrossRef]
- Nicholson, G.; Rantalainen, M.; Maher, A.D.; Li, J.V.; Malmodin, D.; Ahmadi, K.R.; Faber, J.H.; Hallgrímsdóttir, I.B.; Barrett, A.; Toft, H.; et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol. Syst. Biol. 2011, 7, 525. [Google Scholar] [CrossRef] [PubMed]
- Talani, G.; Biggio, F.; Mostallino, M.C.; Locci, V.; Porcedda, C.; Boi, L.; Saolini, E.; Piras, R.; Sanna, E.; Biggio, G. Treatment with gut bifidobacteria improves hippocampal plasticity and cognitive behavior in adult healthy rats. Neuropharmacology 2020, 165, 107909. [Google Scholar] [CrossRef]
- Ndagijimana, M.; Laghi, L.; Vitali, B.; Placucci, G.; Brigidi, P.; Guerzoni, M.E. Effect of a synbiotic food consumption on human gut metabolic profiles evaluated by 1H nuclear magnetic resonance spectroscopy. Int. J. Food Microbiol. 2009, 134, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.M.; Moeller, M.J.; Chey, W.D.; Schoenfeld, P.S. The utility of probiotics in the treatment of irritable bowel syndrome: A systematic review. Am. J. Gastroenterol. 2009, 104, 1033–1049. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; Suárez, A.; Mayo, B. Bifidobacterial diversity determined by culturing and by 16S RDNA sequence analysis in feces and mucosa from ten healthy Spanish adults. Dig. Dis. Sci. 2006, 51, 1878–1885. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; de’Angelis, G.L.; Shanahan, F.; et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Ferrario, C.; Lugli, G.A.; Mancabelli, L.; van Sinderen, D.; Ventura, M. Bifidobacteria and the infant gut: An example of co-evolution and natural selection. Cell. Mol. Life Sci. 2018, 75, 103–118. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 2016, 10, 1656–1668. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R. Prebiotics as gut microflora management tools. J. Clin. Gastroenterol. 2008, 42, S75–S79. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; O’Connell-Motherway, M.; Leahy, S.; Moreno-Munoz, J.A.; Fitzgerald, G.F.; van Sinderen, D. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 2007, 120, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.A.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Models Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Barberini, L.; Noto, A.; Fattuoni, C.; Satta, G.; Zucca, M.; Cabras, M.G.; Mura, M.; Cocco, P. The metabolomic profile of lymphoma subtypes: A pilot study. Molecules 2019, 24, 2367. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinf. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Fattuoni, C.; Palmas, F.; Noto, A.; Barberini, L.; Mussap, M.; Grapov, D.; Dessì, A.; Casu, M.; Casanova, A.; Furione, M.; et al. Primary HCMV infection in pregnancy from classic data towards metabolomics: An exploratory analysis. Clin. Chim. Acta 2016, 460, 23–32. [Google Scholar] [CrossRef]
- Chanpong, A.; Borrelli, O.; Thapar, N. Recent advances in understanding the roles of the enteric nervous system. Fac. Rev. 2022, 11, 7. [Google Scholar] [CrossRef]
- Manos, J. The human microbiome in disease and pathology. APMIS 2022, apm.13225. [Google Scholar] [CrossRef]
- Bonnechère, B.; Amin, N.; van Duijn, C. The role of gut microbiota in neuropsychiatric diseases—Creation of an atlas-based on quantified evidence. Front. Cell. Infect. Microbiol. 2022, 12, 831666. [Google Scholar] [CrossRef] [PubMed]
- Noto, A.; Fanos, V.; Dessì, A. Metabolomics in newborns. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2016; Volume 74, pp. 35–61. ISBN 978-0-12-804689-0. [Google Scholar]
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and their health-promoting effects. Microbiol. Spectr. 2017, 5, 73–98. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Vaughan, E.E. Probiotic and gut lactobacilli and bifidobacteria: Molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 2009, 63, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 3160–3165. [Google Scholar] [CrossRef]
- Schell, M.A.; Karmirantzou, M.; Snel, B.; Vilanova, D.; Berger, B.; Pessi, G.; Zwahlen, M.-C.; Desiere, F.; Bork, P.; Delley, M.; et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 2002, 99, 14422–14427. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; O’Flaherty, S.; Claesson, M.J.; Turroni, F.; Klaenhammer, T.R.; van Sinderen, D.; O’Toole, P.W. Genome-scale analyses of health-promoting bacteria: Probiogenomics. Nat. Rev. Microbiol. 2009, 7, 61–71. [Google Scholar] [CrossRef]
- Jäger, R.; Purpura, M.; Farmer, S.; Cash, H.A.; Keller, D. Probiotic Bacillus coagulans GBI-30, 6086 improves protein absorption and utilization. Probiotics Antimicro. Prot. 2018, 10, 611–615. [Google Scholar] [CrossRef]
- Jäger, R.; Mohr, A.E.; Carpenter, K.C.; Kerksick, C.M.; Purpura, M.; Moussa, A.; Townsend, J.R.; Lamprecht, M.; West, N.P.; Black, K.; et al. International society of sports nutrition position stand: Probiotics. J. Int. Soc. Sports Nutr. 2019, 16, 62. [Google Scholar] [CrossRef]
- Lamprecht, M.; Bogner, S.; Schippinger, G.; Steinbauer, K.; Fankhauser, F.; Hallstroem, S.; Schuetz, B.; Greilberger, J.F. Probiotic supplementation affects markers of intestinal barrier, oxidation, and inflammation in trained men; a randomized, double-blinded, placebo-controlled trial. J. Int. Soc. Sports Nutr. 2012, 9, 45. [Google Scholar] [CrossRef]
- Jäger, R.; Purpura, M.; Stone, J.; Turner, S.; Anzalone, A.; Eimerbrink, M.; Pane, M.; Amoruso, A.; Rowlands, D.; Oliver, J. Probiotic streptococcus thermophilus FP4 and Bifidobacterium breve BR03 supplementation attenuates performance and range-of-motion decrements following muscle damaging exercise. Nutrients 2016, 8, 642. [Google Scholar] [CrossRef]
- Del Olmo, A.; Calzada, J.; Nuñez, M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 2017, 57, 3084–3103. [Google Scholar] [CrossRef]
- Knarreborg, A.; Miquel, N.; Granli, T.; Jensen, B.B. Establishment and application of an in vitro methodology to study the effects of organic acids on coliform and lactic acid bacteria in the proximal part of the gastrointestinal tract of piglets. Anim. Feed Sci. Technol. 2002, 99, 131–140. [Google Scholar] [CrossRef]
- Kluge, H.; Broz, J.; Eder, K. Effect of Benzoic acid on growth performance, nutrient digestibility, nitrogen balance, gastrointestinal microflora and parameters of microbial metabolism in piglets. J. Anim. Physiol. Anim. Nutr. 2006, 90, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Halas, D.; Hansen, C.F.; Hampson, D.J.; Mullan, B.P.; Wilson, R.H.; Pluske, J.R. Effect of dietary supplementation with inulin and/or benzoic acid on the incidence and severity of post-weaning diarrhoea in weaner pigs after experimental challenge with enterotoxigenic Escherichia coli. Arch. Anim. Nutr. 2009, 63, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Lugli, G.A.; Duranti, S.; Turroni, F.; Mancabelli, L.; Ferrario, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Salazar, N.; Solís, G.; Fernández, N.; Hernández-Barranco, A.M.; Cuesta, I.; Gueimonde, M.; de los Reyes-Gavilán, C.G. Assessment of intestinal microbiota modulation ability of bifidobacterium strains in in vitro fecal batch cultures from preterm neonates. Anaerobe 2013, 19, 9–16. [Google Scholar] [CrossRef]
- Sugahara, H.; Odamaki, T.; Fukuda, S.; Kato, T.; Xiao, J.; Abe, F.; Kikuchi, J.; Ohno, H. Probiotic bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Sci. Rep. 2015, 5, 13548. [Google Scholar] [CrossRef]
- Heinken, A.; Sahoo, S.; Fleming, R.M.T.; Thiele, I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 2013, 4, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Heinken, A.; Ravcheev, D.A.; Baldini, F.; Heirendt, L.; Fleming, R.M.T.; Thiele, I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome 2019, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Heinken, A.; Fleming, R.M. A systems biology approach to studying the role of microbes in human health. Curr. Opin. Biotechnol. 2013, 24, 4–12. [Google Scholar] [CrossRef]
- Mierziak, J.; Burgberger, M.; Wojtasik, W. 3-Hydroxybutyrate as a metabolite and a signal molecule regulating processes of living organisms. Biomolecules 2021, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Yudkoff, M.; Daikhin, Y.; Nissim, I.; Lazarow, A.; Nissim, I. Brain amino acid metabolism and ketosis. J. Neurosci. Res. 2001, 66, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Yudkoff, M.; Daikhin, Y.; Horyn, O.; Nissim, I.; Nissim, I. Ketosis and brain handling of glutamate, glutamine, and GABA. Epilepsia 2008, 49, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Erecińska, M.; Nelson, D.; Daikhin, Y.; Yudkoff, M. Regulation of GABA level in rat brain synaptosomes: Fluxes through enzymes of the GABA shunt and effects of glutamate, calcium, and ketone bodies. J. Neurochem. 2002, 67, 2325–2334. [Google Scholar] [CrossRef]
- Stilling, R.M.; van de Wouw, M.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. The neuropharmacology of butyrate: The bread and butter of the microbiota-gut-brain axis? Neurochem. Int. 2016, 99, 110–132. [Google Scholar] [CrossRef]
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial genes, brain & behaviour—Epigenetic regulation of the gut-brain axis: Microbial genes, brain & behaviour. Genes Brain Behav. 2014, 13, 69–86. [Google Scholar] [CrossRef]
- Clarke, K.; Tchabanenko, K.; Pawlosky, R.; Carter, E.; Todd King, M.; Musa-Veloso, K.; Ho, M.; Roberts, A.; Robertson, J.; VanItallie, T.B.; et al. Kinetics, safety and tolerability of (R)-3-hydroxybutyl (R)-3-hydroxybutyrate in healthy adult subjects. Regulat. Toxicol. Pharmacol. 2012, 63, 401–408. [Google Scholar] [CrossRef]
- Soto-Mota, A.; Norwitz, N.G.; Clarke, K. Why a d-β-hydroxybutyrate monoester? Biochem. Soc. Transact. 2020, 48, 51–59. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, Y.; Luan, Y.; Li, Y.; Liu, J.; Yue, Z.; Yuan, B.; Sun, J.; Xie, C.; Li, L.; et al. BHBA treatment improves cognitive function by targeting pleiotropic mechanisms in transgenic mouse model of alzheimer’s disease. FASEB J. 2020, 34, 1412–1429. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-P.; Li, S.-N.; Wang, J.-F.; Li, Y.; Xie, S.-S.; Xue, W.-J.; Liu, H.-M.; Huang, B.-X.; Lv, Q.-K.; Lei, L.-C.; et al. BHBA suppresses LPS-induced inflammation in BV-2 cells by inhibiting NF-κ B activation. Med. Inflamm. 2014, 2014, 983401. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-P.; Wang, J.-F.; Xue, W.-J.; Liu, H.-M.; Liu, B.; Zeng, Y.-L.; Li, S.-N.; Huang, B.-X.; Lv, Q.-K.; Wang, W.; et al. Anti-inflammatory effects of BHBA in both in vivo and in vitro Parkinson’s disease models are mediated by GPR109A-dependent mechanisms. J. Neuroinflamm. 2015, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Ruskin, D.N.; Ross, J.L.; Kawamura, M.; Ruiz, T.L.; Geiger, J.D.; Masino, S.A. A ketogenic diet delays weight loss and does not impair working memory or motor function in the R6/2 1J mouse model of Huntington’s disease. Physiol. Behav. 2011, 103, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Chesser, A.S.; Grima, J.C.; Rappold, P.M.; Blum, D.; Przedborski, S.; Tieu, K. D-β-hydroxybutyrate is protective in mouse models of Huntington’s disease. PLoS ONE 2011, 6, e24620. [Google Scholar] [CrossRef] [PubMed]
- Kajitani, N.; Iwata, M.; Miura, A.; Tsunetomi, K.; Yamanashi, T.; Matsuo, R.; Nishiguchi, T.; Fukuda, S.; Nagata, M.; Shibushita, M.; et al. Prefrontal cortex infusion of beta-hydroxybutyrate, an endogenous NLRP3 inflammasome inhibitor, produces antidepressant-like effects in a rodent model of depression. Neuropsychopharmacol. Rep. 2020, 40, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Mao, X.; Li, R.W.; Hou, E.; Wang, Y.; Xue, C.; Tang, Q. Neoagarotetraose protects mice against intense exercise-induced fatigue damage by modulating gut microbial composition and function. Mol. Nutr. Food Res. 2017, 61, 1600585. [Google Scholar] [CrossRef]
- Torres-Gonzalez, U. Kraft dairy fat consumption and the risk of metabolic syndrome: An examination of the saturated fatty acids in dairy. Nutrients 2019, 11, 2200. [Google Scholar] [CrossRef]
- Warensjö, E.; Jansson, J.-H.; Cederholm, T.; Boman, K.; Eliasson, M.; Hallmans, G.; Johansson, I.; Sjögren, P. Biomarkers of milk fat and the risk of myocardial infarction in men and women: A prospective, matched case-control study. Am. J. Clin. Nutr. 2010, 92, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Weitkunat, K.; Stuhlmann, C.; Postel, A.; Rumberger, S.; Fankhänel, M.; Woting, A.; Petzke, K.J.; Gohlke, S.; Schulz, T.J.; Blaut, M.; et al. Short-chain fatty acids and inulin, but not guar gum, prevent diet-induced obesity and insulin resistance through differential mechanisms in mice. Sci. Rep. 2017, 7, 6109. [Google Scholar] [CrossRef]
- Iggman, D.; Ärnlöv, J.; Vessby, B.; Cederholm, T.; Sjögren, P.; Risérus, U. Adipose tissue fatty acids and insulin sensitivity in elderly men. Diabetologia 2010, 53, 850–857. [Google Scholar] [CrossRef]
- Forouhi, N.G.; Koulman, A.; Sharp, S.J.; Imamura, F.; Kröger, J.; Schulze, M.B.; Crowe, F.L.; Huerta, J.M.; Guevara, M.; Beulens, J.W.; et al. Differences in the prospective association between individual plasma phospholipid saturated fatty acids and incident type 2 diabetes: The EPIC-InterAct case-cohort study. Lancet Diabetes Endocrinol. 2014, 2, 810–818. [Google Scholar] [CrossRef]
- Kröger, J.; Zietemann, V.; Enzenbach, C.; Weikert, C.; Jansen, E.H.; Döring, F.; Joost, H.-G.; Boeing, H.; Schulze, M.B. Erythrocyte membrane phospholipid fatty acids, desaturase activity, and dietary fatty acids in relation to risk of type 2 diabetes in the European prospective investigation into cancer and nutrition (EPIC)—Potsdam study. Am. J. Clin. Nutr. 2011, 93, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Lin, J.; Aris, I.M.; Yang, G.; Chen, W.-Q.; Li, L.-J. Circulating saturated fatty acids and incident type 2 diabetes: A systematic review and meta-analysis. Nutrients 2019, 11, 998. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, F.; de Vos, W.M. Mouse models for human intestinal microbiota research: A critical evaluation. Cell. Mol. Life Sci. 2018, 75, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Blacher, E.; Elinav, E.; Pettersson, S. Our gut microbiome: The evolving inner self. Cell 2017, 171, 1481–1493. [Google Scholar] [CrossRef]
- Simon, J.-C.; Marchesi, J.R.; Mougel, C.; Selosse, M.-A. Host-microbiota interactions: From Holobiont theory to analysis. Microbiome 2019, 7, 5. [Google Scholar] [CrossRef] [PubMed]
| Bifidobacterium Strain | CFU/kg/Day | Proportion |
|---|---|---|
| Bifidobacterium longum BB536 | (3 × 109 CFU) | 3/5 of total |
| Bifidobacterium breve M-16V | (1 × 109 CFU) | 1/5 of total |
| Bifidobacterium infantis M-63 | (1 × 109 CFU) | 1/5 of total |
| Metabolite | Class b | HMDB ID |
|---|---|---|
| Glycolic acid | HA | HMDB0000115 |
| Uric acid | P | HMDB0000289 |
| Malic acid | HA | HMDB0000156 |
| Sorbitol | PO | HMDB0000247 |
| Tryptophan | AA | HMDB0000929 |
| Pentadecanoic acid a | FA | HMDB0000826 |
| 2-Hydroxybutyric acid | HA | HMDB0000008 |
| Gulonic acid | HA | HMDB0003290 |
| 3-Phosphoglyceric acid | HA | HMDB0000807 |
| Ornithine | AA | HMDB0000214 |
| Aspartic acid | AA | HMDB0000191 |
| Glutamine | AA | HMDB0000641 |
| Proline | AA | HMDB0000162 |
| Lyxonic acid a | HA | HMDB0060255 |
| Tyrosine | AA | HMDB0000158 |
| Arabitol | PO | HMDB0000568 |
| Serine | AA | HMDB0000187 |
| Erythrose | S | HMDB0002649 |
| Citric acid | HA | HMDB0000094 |
| Pantothenic acid a | HA | HMDB0000210 |
| 1-Monostearin | MG | HMDB0011131 |
| Nicotinamide | Am | HMDB0001406 |
| Threonine | AA | HMDB0000167 |
| Fumaric acid | A | HMDB0000134 |
| Hydrocinnamic acid a | A | HMDB0000764 |
| Glucose | S | HMDB0000122 |
| Fucose | S | HMDB0000174 |
| Glutamic acid | AA | HMDB0000148 |
| Isoleucine | AA | HMDB0000172 |
| Methionine | AA | HMDB0000696 |
| Octanoic acid | FA | HMDB0000482 |
| Arabinose | S | HMDB0029942 |
| 2-Piperidinecarboxylic acid | A | HMDB0000070 |
| 3-Hydroxybutyric acid a | HA | HMDB0000011 |
| Lysine | AA | HMDB0000182 |
| 2-Aminoheptanedioic acid a | AA | HMDB0034252 |
| Heptadecanoic acid | FA | HMDB0002259 |
| Taurine | AA | HMDB0000251 |
| Benzoic acid | A | HMDB0001870 |
| Phenylalanine | AA | HMDB0000159 |
| Aminomalonic acid | AA | HMDB0001147 |
| Succinic acid | A | HMDB0000254 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biggio, F.; Fattuoni, C.; Mostallino, M.C.; Follesa, P. Effects of Chronic Bifidobacteria Administration in Adult Male Rats on Plasma Metabolites: A Preliminary Metabolomic Study. Metabolites 2022, 12, 762. https://doi.org/10.3390/metabo12080762
Biggio F, Fattuoni C, Mostallino MC, Follesa P. Effects of Chronic Bifidobacteria Administration in Adult Male Rats on Plasma Metabolites: A Preliminary Metabolomic Study. Metabolites. 2022; 12(8):762. https://doi.org/10.3390/metabo12080762
Chicago/Turabian StyleBiggio, Francesca, Claudia Fattuoni, Maria Cristina Mostallino, and Paolo Follesa. 2022. "Effects of Chronic Bifidobacteria Administration in Adult Male Rats on Plasma Metabolites: A Preliminary Metabolomic Study" Metabolites 12, no. 8: 762. https://doi.org/10.3390/metabo12080762
APA StyleBiggio, F., Fattuoni, C., Mostallino, M. C., & Follesa, P. (2022). Effects of Chronic Bifidobacteria Administration in Adult Male Rats on Plasma Metabolites: A Preliminary Metabolomic Study. Metabolites, 12(8), 762. https://doi.org/10.3390/metabo12080762