Predicting Hypertension Subtypes with Machine Learning Using Targeted Metabolites and Their Ratios
Abstract
1. Introduction
2. Materials and Methods
2.1. Omic Dataset
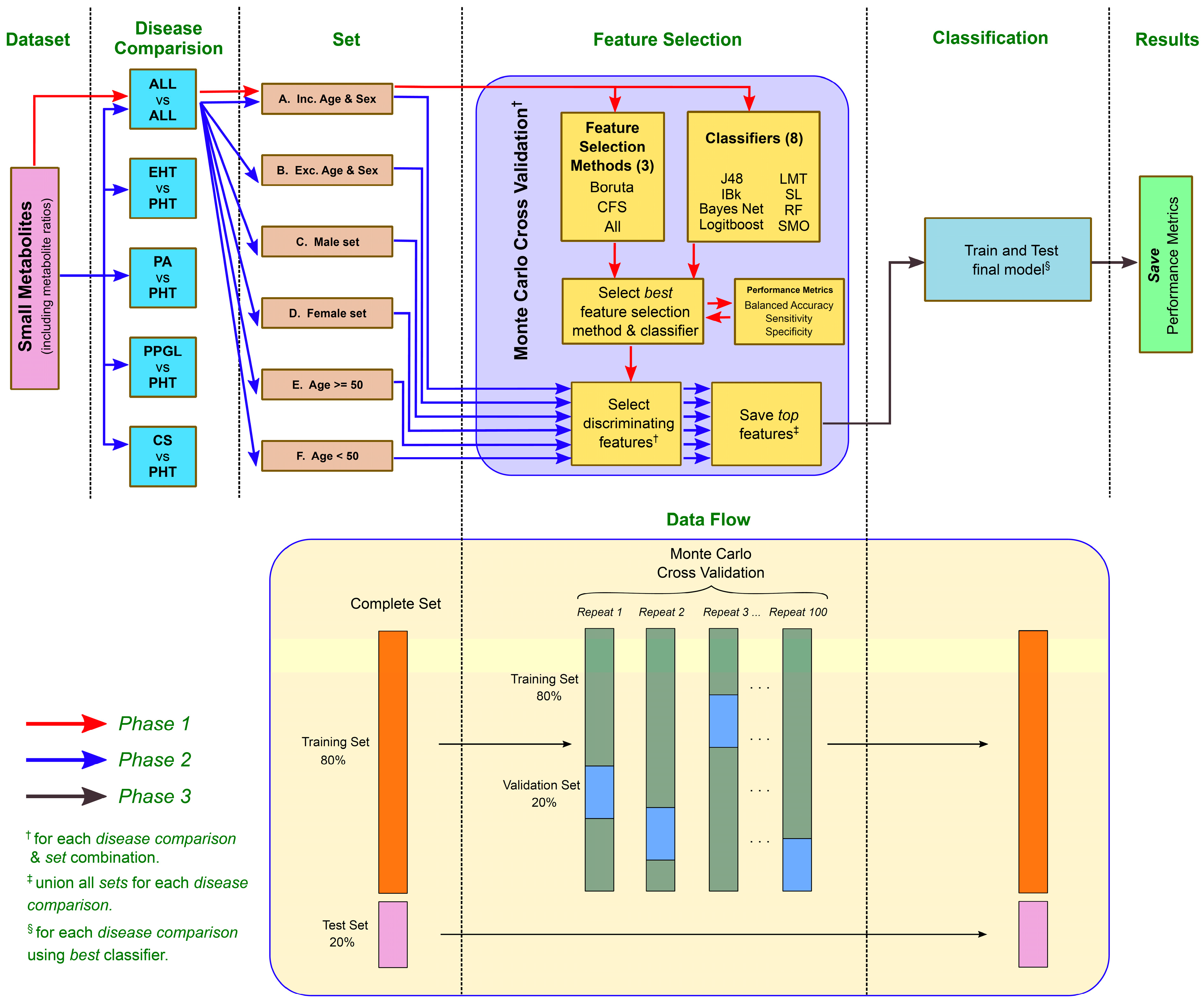
2.2. ML Analysis Pipeline
3. Results
3.1. Evaluation of Feature Selection Methods & Classifiers
3.2. Classification Performance and Discriminating Features
3.2.1. MCCV Classification Performance
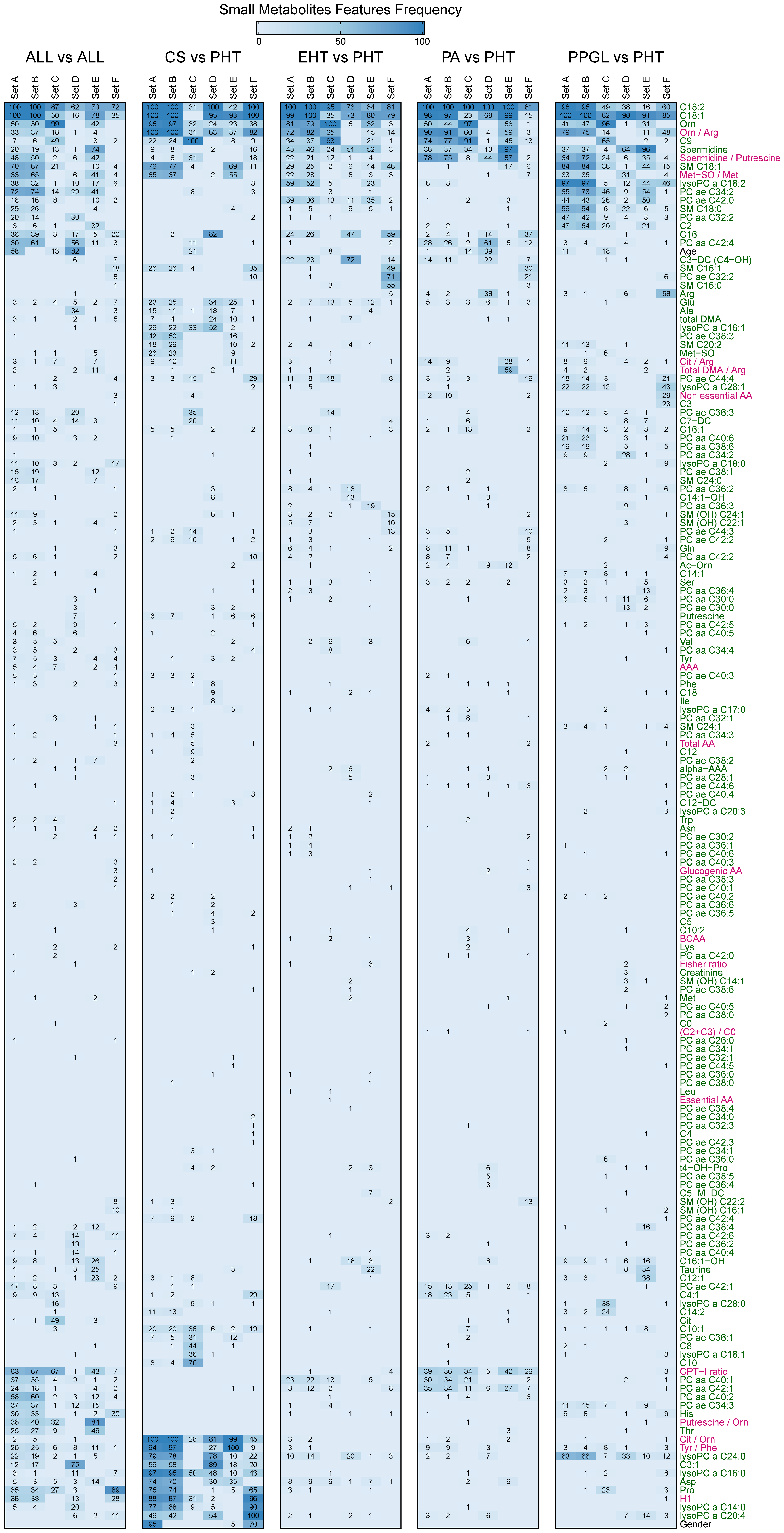
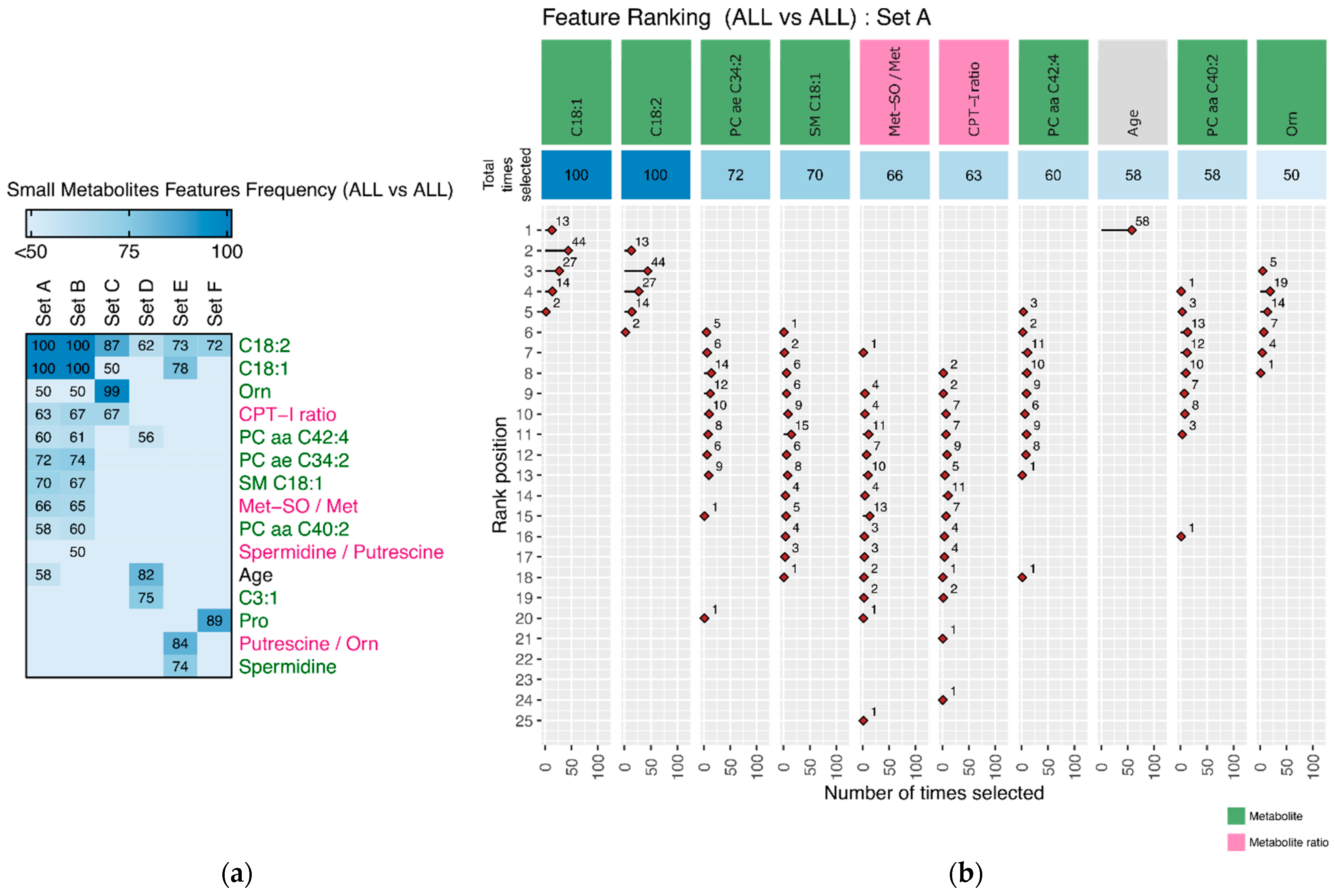
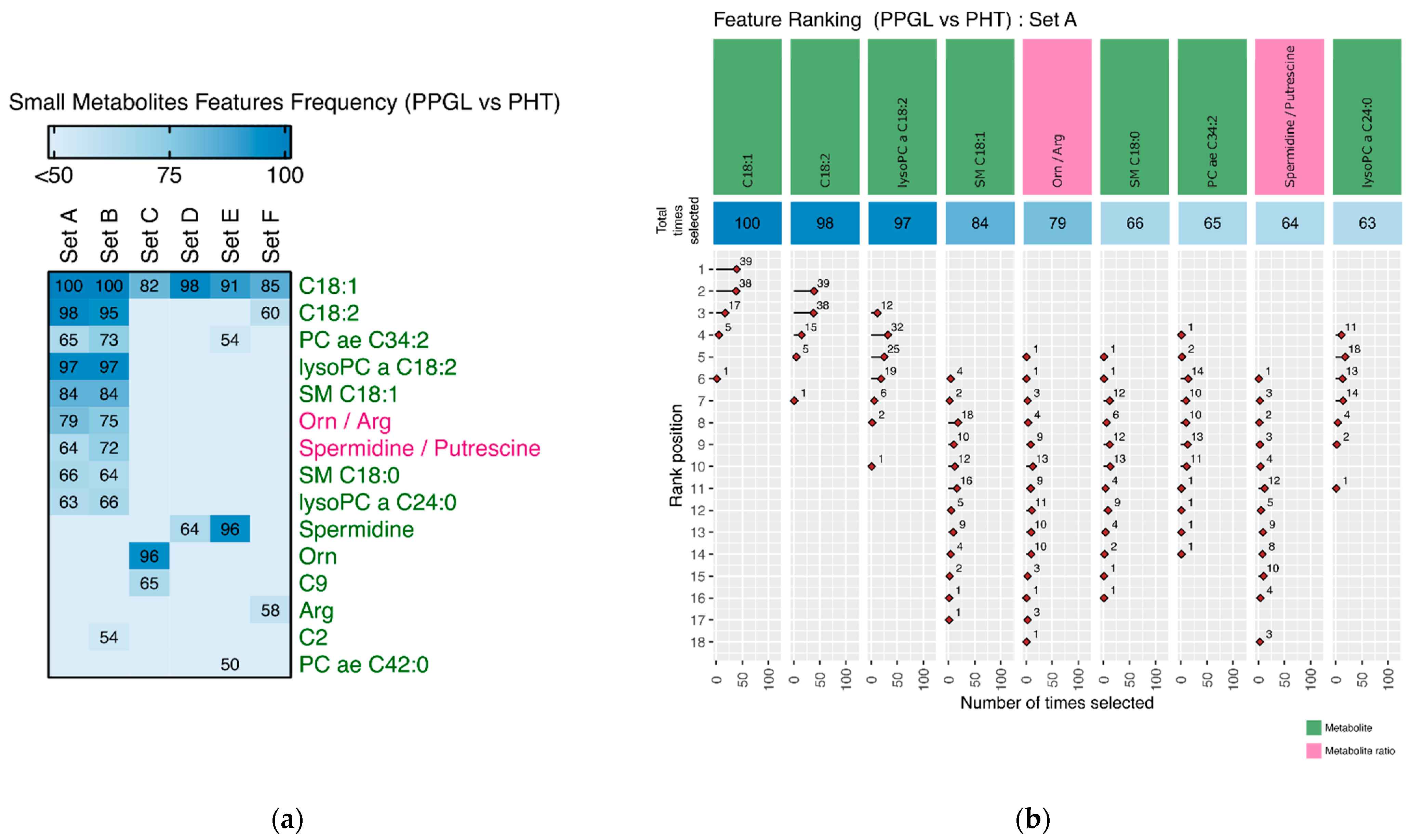
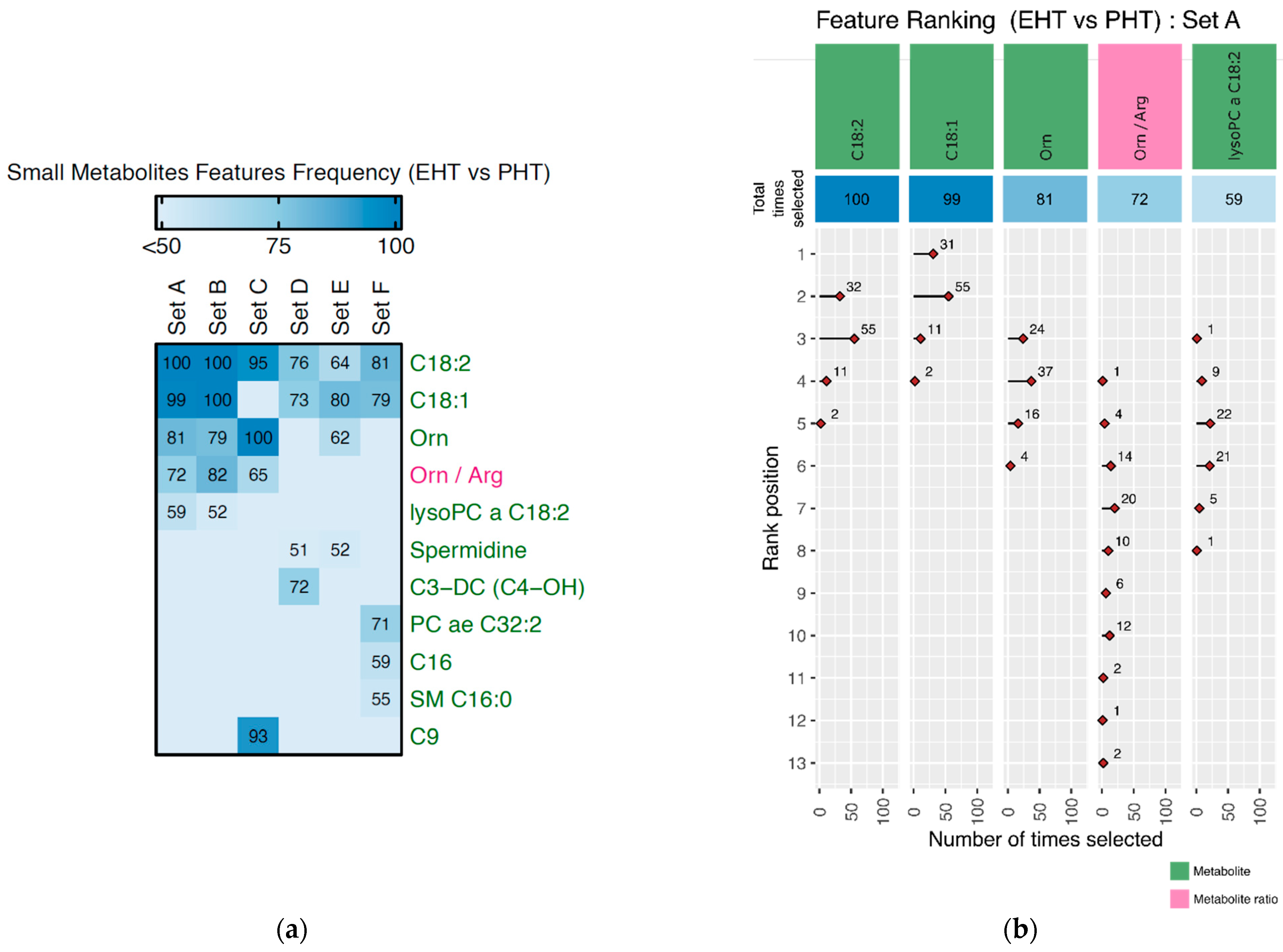
3.2.2. Discriminating Features
3.3. Final Model Training and Testing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Tables and Figures
| Acylcarnitines (40) | |||
| Abbreviation | Full-Name | Abbreviation | Full-Name |
| C0 | Carnitine | C10:1 | Decenoylcarnitine |
| C2 | Acetylcarnitine | C10:2 | Decadienylcarnitine |
| C3 | Propionylcarnitine | C12 | Dodecanoylcarnitine |
| C3:1 ** | Propenoylcarnitine | C12:1 | Dodecenoylcarnitine |
| C3-OH * | Hydroxypropionylcarnitine | C12-DC ** | Dodecanedioylcarnitine |
| C4 | Butyrylcarnitine | C14 | Tetradecanoylcarnitine |
| C4:1 | Butenoylcarnitine | C14:1 | Tetradecenoylcarnitine |
| C4-OH (C3-DC) | Hydroxybutyrylcarnitine | C14:1-OH | Hydroxytetradecenoylcarnitine |
| C5 | Valerylcarnitine | C14:2 | Tetradecadienylcarnitine |
| C5:1 * | Tiglylcarnitine | C14:2-OH * | Hydroxytetradecadienylcarnitine |
| C5:1-DC * | Glutaconylcarnitine | C16 | Hexadecanoylcarnitine |
| C5-DC (C6-OH) * | Glutarylcarnitine (Hydroxyhexanoylcarnitine) | C16:1 | Hexadecenoylcarnitine |
| C5-M-DC ** | Methylglutarylcarnitine | C16:1-OH | Hydroxyhexadecenoylcarnitine |
| C5-OH (C3-DC-M) * | Hydroxyvalerylcarnitine (Methylmalonylcarnitine) | C16:2 * | Hexadecadienylcarnitine |
| C6 (C4:1-DC) * | Hexanoylcarnitine (Fumarylcarnitine) | C16:2-OH * | Hydroxyhexadecadienylcarnitine |
| C6:1 * | Hexenoylcarnitine | C16-OH * | Hydroxyhexadecanoylcarnitine |
| C7-DC ** | Pimelylcarnitine | C18 | Octadecanoylcarnitine |
| C8 | Octanoylcarnitine | C18:1 | Octadecenoylcarnitine |
| C9 | Nonanoylcarnitine | C18:1-OH * | Hydroxyoctadecenoylcarnitine |
| C10 | Decanoylcarnitine | C18:2 | Octadecadienylcarnitine |
| Amino Acids (21) | |||
| Abbreviation | Full-Name | Abbreviation | Full-Name |
| Ala | Alanine | Lys | Lysine |
| Arg | Arginine | Met | Methionine |
| Asn | Asparagine | Orn | Ornithine |
| Asp | Aspartate | Phe | Phenylalanine |
| Cit | Citrulline | Pro | Proline |
| Gln | Glutamine | Ser | Serine |
| Glu | Glutamate | Thr | Threonine |
| Gly | Glycine | Trp | Tryptophan |
| His | Histidine | Tyr | Tyrosine |
| Ile | Isoleucine | Val | Valine |
| Leu | Leucine | ||
| Monosaccharides (1) | |||
| Abbreviation | Full-Name | ||
| H1 | Sum of Hexoses (including Glucose) | ||
| Glycerophospholipids (90) | |||
| Abbreviation | Full-Name | Abbreviation | Full-Name |
| lysoPC a C14:0 | PC aa C34:1 | PC aa C42:0 | PC ae C38:2 |
| lysoPC a C16:0 | PC aa C34:2 | PC aa C42:1 | PC ae C38:3 |
| lysoPC a C16:1 | PC aa C34:3 | PC aa C42:2 | PC ae C38:4 |
| lysoPC a C17:0 | PC aa C34:4 | PC aa C42:4 | PC ae C38:5 |
| lysoPC a C18:0 | PC aa C36:0 | PC aa C42:5 | PC ae C38:6 |
| lysoPC a C18:1 | PC aa C36:1 | PC aa C42:6 | PC ae C40:1 |
| lysoPC a C18:2 | PC aa C36:2 | PC ae C30:0 | PC ae C40:2 |
| lysoPC a C20:3 | PC aa C36:3 | PC ae C30:1* | PC ae C40:3 |
| lysoPC a C20:4 | PC aa C36:4 | PC ae C30:2 | PC ae C40:4 |
| lysoPC a C24:0 ** | PC aa C36:5 | PC ae C32:1 | PC ae C40:5 |
| lysoPC a C26:0 * | PC aa C36:6 | PC ae C32:2 | PC ae C40:6 |
| lysoPC a C26:1 * | PC aa C38:0 | PC ae C34:0 | PC ae C42:0 |
| lysoPC a C28:0 ** | PC aa C38:1 * | PC ae C34:1 | PC ae C42:1 |
| lysoPC a C28:1 ** | PC aa C38:3 | PC ae C34:2 | PC ae C42:2 |
| PC aa C24:0 * | PC aa C38:4 | PC ae C34:3 | PC ae C42:3 |
| PC aa C26:0 | PC aa C38:5 | PC ae C36:0 | PC ae C42:4 |
| PC aa C28:1 | PC aa C38:6 | PC ae C36:1 | PC ae C42:5 |
| PC aa C30:0 | PC aa C40:1 | PC ae C36:2 | PC ae C44:3 |
| PC aa C30:2 * | PC aa C40:2 | PC ae C36:3 | PC ae C44:4 |
| PC aa C32:0 | PC aa C40:3 | PC ae C36:4 | PC ae C44:5 |
| PC aa C32:1 | PC aa C40:4 | PC ae C36:5 | PC ae C44:6 |
| PC aa C32:2 ** | PC aa C40:5 | PC ae C38:0 | |
| PC aa C32:3 | PC aa C40:6 | PC ae C38:1 | |
| Sphingolipids (15) | |||
| Abbreviation | Full-Name | Abbreviation | Full-Name |
| SM (OH) C14:1 | SM C18:0 | SM (OH) C22:1 | SM (OH) C24:1 |
| SM C16:0 | SM C18:1 | SM (OH) C22:2 | SM C26:0 * |
| SM C16:1 | SM C20:2 | SM C24:0 | SM C26:1 * |
| SM (OH) C16:1 | SM C22:3 * | SM C24:1 | |
| Biogenic Amines (21) | |||
| Abbreviation | Full-Name | Abbreviation | Full-Name |
| Ac-Orn | Acetylornithine | PEA * | Phenylethylamine |
| ADMA * | Asymmetric dimethylarginine | cis-OH-Pro * | cis-4-Hydroxyproline |
| alpha-AAA | alpha-Aminoadipic acid | trans-OH-Pro | trans-4-Hydroxyproline |
| Carnosine * | Carnosine | Putrescine | Putrescine |
| Creatinine | Creatinine | SDMA * | Symmetric dimethylarginine |
| DOPA * | DOPA | Serotonin * | Serotonin |
| Dopamine * | Dopamine | Spermidine | Spermidine |
| Histamine * | Histamine | Spermine * | Spermine |
| Kynurenine * | Kynurenine | Taurine | Taurine |
| Met-SO | Methionine sulfoxide | total DMA | Total dimethylarginine |
| Nitro-Tyr * | Nitrotyrosine | ||
| Data | Disease | Sex | Age Distribution | Total Count | ||
|---|---|---|---|---|---|---|
| Male | Female | Patient Age ≥ 50 | Patient Age < 50 | |||
| Training (80%) | CS | 3 | 29 | 17 | 15 | 32 |
| PA | 45 | 41 | 33 | 53 | 86 | |
| PPGL | 27 | 34 | 39 | 22 | 61 | |
| PHT | 29 | 18 | 22 | 25 | 47 | |
| Testing (20%) | CS | 1 | 7 | 5 | 3 | 8 |
| PA | 13 | 8 | 9 | 12 | 21 | |
| PPGL | 6 | 9 | 9 | 6 | 15 | |
| PHT | 11 | 1 | 1 | 11 | 12 | |
| EHT vs. PHT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classifier | All | CFS | Boruta | ||||||||||||
| B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | |
| IBk | 61 | 83 | 39 | 0.84 | 0.61 | 62 | 80 | 44 | 0.82 | 0.62 | 58 | 81 | 36 | 0.82 | 0.58 |
| J48 | 58 | 83 | 34 | 0.83 | 0.56 | 56 | 85 | 27 | 0.83 | 0.58 | 56 | 86 | 25 | 0.84 | 0.63 |
| LB | 61 | 89 | 33 | 0.87 | 0.74 | 59 | 89 | 30 | 0.86 | 0.74 | 59 | 88 | 29 | 0.86 | 0.75 |
| LMT | 62 | 91 | 33 | 0.87 | 0.76 | 56 | 93 | 18 | 0.87 | 0.70 | 55 | 92 | 19 | 0.86 | 0.69 |
| NB | 70 | 62 | 78 | 0.74 | 0.76 | 72 | 61 | 83 | 0.74 | 0.78 | 68 | 56 | 81 | 0.70 | 0.76 |
| RF | 53 | 99 | 7 | 0.89 | 0.77 | 58 | 94 | 22 | 0.88 | 0.75 | 57 | 90 | 24 | 0.86 | 0.74 |
| SL | 61 | 91 | 31 | 0.88 | 0.76 | 55 | 94 | 16 | 0.87 | 0.70 | 54 | 93 | 16 | 0.87 | 0.69 |
| SMO | 62 | 91 | 33 | 0.87 | 0.62 | 50 | 100 | 0 | 0.89 | 0.50 | 50 | 100 | 0 | 0.89 | 0.50 |
| CS vs. PHT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classifier | All | CFS | Boruta | ||||||||||||
| B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | |
| IBk | 82 | 73 | 91 | 0.77 | 0.82 | 83 | 74 | 91 | 0.78 | 82 | 0.87 | 80 | 94 | 0.84 | 0.87 |
| J48 | 76 | 73 | 78 | 0.71 | 0.75 | 74 | 70 | 78 | 0.68 | 74 | 0.74 | 71 | 78 | 0.69 | 0.74 |
| LB | 75 | 66 | 84 | 0.69 | 0.85 | 76 | 66 | 86 | 0.70 | 85 | 0.76 | 67 | 85 | 0.70 | 0.85 |
| LMT | 83 | 75 | 91 | 0.79 | 0.92 | 82 | 74 | 90 | 0.77 | 91 | 0.82 | 74 | 90 | 0.78 | 0.92 |
| NB | 81 | 74 | 88 | 0.76 | 0.87 | 81 | 67 | 95 | 0.75 | 91 | 0.83 | 70 | 96 | 0.78 | 0.94 |
| RF | 77 | 60 | 95 | 0.70 | 0.92 | 78 | 65 | 91 | 0.71 | 89 | 0.79 | 65 | 92 | 0.73 | 0.90 |
| SL | 83 | 75 | 91 | 0.79 | 0.92 | 82 | 74 | 90 | 0.77 | 91 | 0.82 | 74 | 90 | 0.78 | 0.91 |
| SMO | 87 | 82 | 93 | 0.84 | 0.87 | 81 | 69 | 93 | 0.76 | 81 | 0.83 | 70 | 95 | 0.78 | 0.83 |
| PA vs. PHT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classifier | All | CFS | Boruta | ||||||||||||
| B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | |
| IBk | 63 | 72 | 55 | 0.73 | 0.63 | 60 | 66 | 54 | 0.69 | 0.60 | 62 | 69 | 55 | 0.71 | 0.62 |
| J48 | 63 | 72 | 54 | 0.73 | 0.64 | 64 | 70 | 59 | 0.73 | 0.66 | 65 | 72 | 59 | 0.74 | 0.67 |
| LB | 65 | 76 | 53 | 0.76 | 0.74 | 65 | 78 | 52 | 0.76 | 0.75 | 65 | 76 | 54 | 0.76 | 0.75 |
| LMT | 67 | 77 | 56 | 0.77 | 0.78 | 66 | 75 | 57 | 0.75 | 0.77 | 66 | 76 | 57 | 0.76 | 0.77 |
| NB | 69 | 57 | 81 | 0.68 | 0.75 | 73 | 59 | 88 | 0.70 | 0.79 | 72 | 56 | 87 | 0.68 | 0.78 |
| RF | 62 | 88 | 37 | 0.79 | 0.78 | 65 | 78 | 52 | 0.77 | 0.76 | 64 | 77 | 51 | 0.76 | 0.75 |
| SL | 67 | 77 | 56 | 0.77 | 0.78 | 66 | 75 | 57 | 0.76 | 0.78 | 67 | 76 | 58 | 0.76 | 0.78 |
| SMO | 70 | 77 | 62 | 0.78 | 0.70 | 59 | 84 | 35 | 0.76 | 0.59 | 58 | 88 | 29 | 0.78 | 0.58 |
| PPGL vs. PHT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classifier | All | CFS | Boruta | ||||||||||||
| B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | |
| IBk | 62 | 54 | 71 | 0.61 | 0.62 | 66 | 63 | 70 | 0.67 | 0.66 | 65 | 64 | 66 | 0.67 | 0.65 |
| J48 | 66 | 71 | 62 | 0.71 | 0.66 | 66 | 72 | 60 | 0.71 | 0.67 | 68 | 73 | 63 | 0.72 | 0.69 |
| LB | 70 | 74 | 67 | 0.74 | 0.78 | 71 | 75 | 67 | 0.75 | 0.80 | 74 | 79 | 69 | 0.78 | 0.82 |
| LMT | 71 | 73 | 69 | 0.75 | 0.79 | 69 | 73 | 66 | 0.73 | 0.76 | 69 | 74 | 65 | 0.73 | 0.76 |
| NB | 73 | 67 | 79 | 0.73 | 0.81 | 73 | 64 | 82 | 0.72 | 0.81 | 70 | 59 | 80 | 0.68 | 0.79 |
| RF | 73 | 84 | 62 | 0.79 | 0.83 | 73 | 79 | 67 | 0.77 | 0.81 | 74 | 79 | 68 | 0.78 | 0.82 |
| SL | 72 | 74 | 70 | 0.75 | 0.79 | 70 | 73 | 67 | 0.73 | 0.76 | 70 | 74 | 65 | 0.73 | 0.77 |
| SMO | 74 | 79 | 68 | 0.78 | 0.74 | 71 | 74 | 68 | 0.75 | 0.71 | 70 | 73 | 66 | 0.74 | 0.70 |
| Reference | |||
|---|---|---|---|
| PA | PHT | ||
| Prediction | PA | 15 | 3 |
| PHT | 6 | 9 | |
| Reference | |||
|---|---|---|---|
| PPGL | PHT | ||
| Prediction | PPGL | 12 | 3 |
| PHT | 3 | 9 | |
| Reference | |||
|---|---|---|---|
| EHT | PHT | ||
| Prediction | EHT | 25 | 1 |
| PHT | 19 | 11 | |
| Reference | |||||
|---|---|---|---|---|---|
| CS | PA | PHT | PPGL | ||
| Prediction | CS | 2 | 2 | 0 | 5 |
| PA | 0 | 6 | 2 | 0 | |
| PHT | 2 | 10 | 8 | 3 | |
| PPGL | 4 | 3 | 2 | 7 | |


Appendix B. Patient Recruitment and Diagnostic Work-Up
Appendix C. Metabolite Quantification by AbsoluteIDQTM p180 Kit
References
- Mills, K.T.; Stefanescu, A.; He, J. The Global Epidemiology of Hypertension. Nat. Rev. Nephrol. 2020, 16, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Mancia, G.; Spiering, W.; Rosei, E.A.; Azizi, M.; Burnier, M.; Clement, D.L.; Coca, A.; de Simone, G.; Dominiczak, A.; et al. 2018 ESC/ESH Guidelines for the Management of Arterial Hypertension. Eur. Heart J. 2018, 39, 3021–3104. [Google Scholar] [CrossRef] [PubMed]
- Di Dalmazi, G.; Quinkler, M.; Deutschbein, T.; Prehn, C.; Rayes, N.; Kroiss, M.; Berr, C.M.; Stalla, G.; Fassnacht, M.; Adamski, J.; et al. Cortisol-Related Metabolic Alterations Assessed by Mass Spectrometry Assay in Patients with Cushing’s Syndrome. Eur. J. Endocrinol. 2017, 177, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Rhayem, Y.; Kunzke, T.; Sun, N.; Feuchtinger, A.; Ludwig, P.; Strom, T.M.; Gomez-Sanchez, C.; Knösel, T.; Kirchner, T.; et al. In Situ Metabolomics of Aldosterone-Producing Adenomas. JCI Insight 2019, 4, e130356. [Google Scholar] [CrossRef] [PubMed]
- Erlic, Z.; Kurlbaum, M.; Deutschbein, T.; Nölting, S.; Prejbisz, A.; Timmers, H.; Richter, S.; Prehn, C.; Weismann, D.; Adamski, J.; et al. Metabolic Impact of Pheochromocytoma/Paraganglioma: Targeted Metabolomics in Patients before and after Tumor Removal. Eur. J. Endocrinol. 2019, 181, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Erlic, Z.; Reel, P.; Reel, S.; Amar, L.; Pecori, A.; Larsen, C.K.; Tetti, M.; Pamporaki, C.; Prehn, C.; Adamski, J.; et al. Targeted Metabolomics as a Tool in Discriminating Endocrine from Primary Hypertension. J. Clin. Endocrinol. Metab. 2020, 106, e1111–e1128. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.2.1–30.2.24. [Google Scholar] [CrossRef]
- Ramasubbu, R.; Brown, M.R.G.; Cortese, F.; Gaxiola, I.; Goodyear, B.; Greenshaw, A.J.; Dursun, S.M.; Greiner, R. Accuracy of Automated Classification of Major Depressive Disorder as a Function of Symptom Severity. NeuroImage Clin. 2016, 12, 320–331. [Google Scholar] [CrossRef]
- Nouretdinov, I.; Costafreda, S.G.; Gammerman, A.; Chervonenkis, A.; Vovk, V.; Vapnik, V.; Fu, C.H.Y. Machine Learning Classification with Confidence: Application of Transductive Conformal Predictors to MRI-Based Diagnostic and Prognostic Markers in Depression. Neuroimage 2011, 56, 809–813. [Google Scholar] [CrossRef]
- Leclercq, M.; Vittrant, B.; Martin-Magniette, M.L.; Boyer, M.P.S.; Perin, O.; Bergeron, A.; Fradet, Y.; Droit, A. Large-Scale Automatic Feature Selection for Biomarker Discovery in High-Dimensional OMICs Data. Front. Genet. 2019, 10, 452. [Google Scholar] [CrossRef]
- Ko, J.; Baldassano, S.N.; Loh, P.-L.; Kording, K.; Litt, B.; Issadore, D. Machine Learning to Detect Signatures of Disease in Liquid Biopsies—A User’s Guide. Lab Chip 2018, 18, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Casanova, R.; Varma, S.; Simpson, B.; Kim, M.; An, Y.; Saldana, S.; Riveros, C.; Moscato, P.; Griswold, M.; Sonntag, D.; et al. Blood Metabolite Markers of Preclinical Alzheimer’s Disease in Two Longitudinally Followed Cohorts of Older Individuals. Alzheimer’s Dement. 2016, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Ottas, A.; Fishman, D.; Okas, T.-L.; Kingo, K.; Soomets, U. The Metabolic Analysis of Psoriasis Identifies the Associated Metabolites While Providing Computational Models for the Monitoring of the Disease. Arch. Dermatol. Res. 2017, 309, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Bahado-Singh, R.O.; Yilmaz, A.; Bisgin, H.; Turkoglu, O.; Kumar, P.; Sherman, E.; Mrazik, A.; Odibo, A.; Graham, S.F. Artificial Intelligence and the Analysis of Multi-Platform Metabolomics Data for the Detection of Intrauterine Growth Restriction. PLoS ONE 2019, 14, e0214121. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.; Böhm, C.; Baumgartner, D.; Marini, G.; Weinberger, K.; Olgemöller, B.; Liebl, B.; Roscher, A.A. Supervised Machine Learning Techniques for the Classification of Metabolic Disorders in Newborns. Bioinformatics 2004, 20, 2985–2996. [Google Scholar] [CrossRef]
- Takahashi, Y.; Ueki, M.; Yamada, M.; Tamiya, G.; Motoike, I.N.; Saigusa, D.; Sakurai, M.; Nagami, F.; Ogishima, S.; Koshiba, S.; et al. Improved Metabolomic Data-Based Prediction of Depressive Symptoms Using Nonlinear Machine Learning with Feature Selection. Transl. Psychiatry 2020, 10, 157. [Google Scholar] [CrossRef]
- Braun, L.T.; Vogel, F.; Reincke, M. Long-Term Morbidity and Mortality in Patients with Cushing’s Syndrome. J. Neuroendocrinol. 2022, e13113. [Google Scholar] [CrossRef]
- Bothou, C.; Beuschlein, F.; Spyroglou, A. Links between Aldosterone Excess and Metabolic Complications: A Comprehensive Review. Diabetes Metab. 2020, 46, 1–7. [Google Scholar] [CrossRef]
- Erlic, Z.; Beuschlein, F. Metabolic Alterations in Patients with Pheochromocytoma. Exp. Clin. Endocrinol. Diabetes 2019, 127, 129–136. [Google Scholar] [CrossRef]
- Römisch-Margl, W.; Prehn, C.; Bogumil, R.; Röhring, C.; Suhre, K.; Adamski, J. Procedure for Tissue Sample Preparation and Metabolite Extraction for High-Throughput Targeted Metabolomics. Metabolomics 2012, 8, 133–142. [Google Scholar] [CrossRef]
- Zukunft, S.; Sorgenfrei, M.; Prehn, C.; Möller, G.; Adamski, J. Targeted Metabolomics of Dried Blood Spot Extracts. Chromatographia 2013, 76, 1295–1305. [Google Scholar] [CrossRef]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing Value Estimation Methods for DNA Microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Nind, T.; Galloway, J.; McAllister, G.; Scobbie, D.; Bonney, W.; Hall, C.; Tramma, L.; Reel, P.; Groves, M.; Appleby, P.; et al. The Research Data Management Platform (RDMP): A Novel, Process Driven, Open-Source Tool for the Management of Longitudinal Cohorts of Clinical Data. GigaScience 2018, 7, giy060. [Google Scholar] [CrossRef]
- Hall, M.A. Correlation-Based Feature Selection for Machine Learning. Ph.D. Thesis, University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Chapman & Hall: Boca Raton, FL, USA, 1998; ISBN 978-0-412-04841-8. [Google Scholar]
- Bentley, J.L. Multidimensional Binary Search Trees Used for Associative Searching. Commun. ACM 1975, 18, 509–517. [Google Scholar] [CrossRef]
- Zheng, Z.; Webb, G.I. Lazy Learning of Bayesian Rules. Mach. Learn. 2000, 41, 53–84. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Additive Logistic Regression: A Statistical View of Boosting. Ann. Stat. 1998, 28, 337–407. [Google Scholar] [CrossRef]
- Landwehr, N.; Hall, M.; Frank, E. Logistic Model Trees. Mach. Learn. 2005, 59, 161–205. [Google Scholar] [CrossRef]
- Sumner, M.; Frank, E.; Hall, M. Speeding up Logistic Model Tree Induction. In Proceedings of the 9th European Conference on European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases, Porto, Portugal, 3 October 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 675–683. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Platt, J. Fast Training of Support Vector Machines Using Sequential Minimal Optimization; Technical Report MSR-TR-98-14; Microsoft Reserch: Redmond, WA, USA, 1998. [Google Scholar]
- Simon, R. Resampling Strategies for Model Assessment and Selection. In Fundamentals of Data Mining in Genomics and Proteomics; Dubitzky, W., Granzow, M., Berrar, D., Eds.; Springer: Boston, MA, USA, 2007; pp. 173–186. ISBN 978-0-387-47509-7. [Google Scholar]
- Velez, D.R.; White, B.C.; Motsinger, A.A.; Bush, W.S.; Ritchie, M.D.; Williams, S.M.; Moore, J.H. A Balanced Accuracy Function for Epistasis Modeling in Imbalanced Datasets Using Multifactor Dimensionality Reduction. Genet. Epidemiol. 2007, 31, 306–315. [Google Scholar] [CrossRef] [PubMed]
- ConfusionMatrix: Create a Confusion Matrix in Caret: Classification and Regression Training. Available online: https://rdrr.io/cran/caret/man/confusionMatrix.html (accessed on 24 July 2022).
- Kuhn, M.; Johnson, K. Over-Fitting and Model Tuning. In Applied Predictive Modeling; Kuhn, M., Johnson, K., Eds.; Springer: New York, NY, USA, 2013; pp. 61–92. ISBN 978-1-4614-6849-3. [Google Scholar]
- Hornik, K.; Buchta, C.; Zeileis, A. Open-Source Machine Learning: R Meets Weka. Comput. Stat. 2009, 24, 225–232. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Leong, L.K.; Abdullah, A.A. Prediction of Alzheimer’s Disease (AD) Using Machine Learning Techniques with Boruta Algorithm as Feature Selection Method. J. Phys. Conf. Ser. 2019, 1372, 012065. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-Sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Duchnowski, P.; Hryniewiecki, T.; Zatorska, K.; Żebrowska, A.; Kuśmierczyk, M.; Szymański, P. High-sensitivity Troponin T as a Prognostic Marker in Patients Undergoing Aortic Valve Replacement. Pol. Arch. Intern. Med. 2017, 127, 628–630. [Google Scholar] [CrossRef][Green Version]
- Mulatero, P.; Monticone, S.; Deinum, J.; Amar, L.; Prejbisz, A.; Zennaro, M.-C.; Beuschlein, F.; Rossi, G.P.; Nishikawa, T.; Morganti, A.; et al. Genetics, Prevalence, Screening and Confirmation of Primary Aldosteronism: A Position Statement and Consensus of the Working Group on Endocrine Hypertension of The European Society of Hypertension∗. J. Hypertens. 2020, 38, 1919–1928. [Google Scholar] [CrossRef]
- Lenders, J.W.M.; Kerstens, M.N.; Amar, L.; Prejbisz, A.; Robledo, M.; Taieb, D.; Pacak, K.; Crona, J.; Zelinka, T.; Mannelli, M.; et al. Genetics, Diagnosis, Management and Future Directions of Research of Phaeochromocytoma and Paraganglioma: A Position Statement and Consensus of the Working Group on Endocrine Hypertension of the European Society of Hypertension. J. Hypertens. 2020, 38, 1443–1456. [Google Scholar] [CrossRef]
- European Medicines Agency. Guideline on Bioanalytical Method Validation; Committee for Medicinal Products for Human Use (CHMP): London, UK, 2011. [Google Scholar]

| Disease | Patient Count (n=) | Sex | Age Distribution | ||
|---|---|---|---|---|---|
| Male (n=) | Female (n=) | Patient Age ≥ 50 | Patient Age < 50 | ||
| Cushing’s Syndrome (CS) | 40 | 4 | 36 | 22 | 18 |
| Primary Aldosteronism (PA) | 107 | 58 | 49 | 42 | 65 |
| Pheochromocytoma or Paraganglioma (PPGL) | 76 | 33 | 43 | 48 | 28 |
| Primary Hypertension (PHT) | 59 | 40 | 19 | 23 | 36 |
| ALL vs. ALL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classifier | All | CFS | Boruta | ||||||||||||
| B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | B. Acc (%) | Sen (%) | Spec (%) | F1 | AUC | |
| IBk | 60 | 41 | 79 | 0.39 | 0.60 | 57 | 35 | 78 | 0.29 | 0.57 | 58 | 37 | 79 | 0.35 | 0.58 |
| J48 | 56 | 35 | 78 | 0.30 | 0.58 | 57 | 36 | 78 | 0.31 | 0.60 | 56 | 34 | 78 | 0.27 | 0.57 |
| LB | 61 | 42 | 80 | 0.41 | 0.71 | 60 | 40 | 80 | 0.31 | 0.68 | 60 | 40 | 80 | 0.32 | 0.68 |
| LMT | 69 | 54 | 84 | 0.53 | 0.81 | 58 | 38 | 79 | 0.32 | 0.69 | 60 | 41 | 80 | 0.36 | 0.69 |
| NB | 64 | 48 | 81 | 0.44 | 0.73 | 59 | 40 | 79 | 0.26 | 0.68 | 60 | 41 | 80 | 0.29 | 0.68 |
| RF | 60 | 40 | 80 | 0.24 | 0.76 | 59 | 38 | 79 | 0.29 | 0.68 | 59 | 38 | 79 | 0.28 | 0.70 |
| SL | 69 | 54 | 84 | 0.54 | 0.82 | 58 | 38 | 79 | 0.31 | 0.69 | 60 | 41 | 80 | 0.35 | 0.70 |
| SMO | 71 | 56 | 85 | 0.57 | 0.78 | 51 | 27 | 76 | 0.2 | 0.63 | 54 | 31 | 77 | 0.06 | 0.64 |
| Disease Comparisons | Classifier | Features Used | B. Accuracy (%) | Sensitivity (%) | Specificity (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age Included? | Sex Included? | No of Metabolites | No of Metabolite Ratios | Total | F1 | AUC | |||||
| PA vs. PHT | SL | ✕ | ✕ | 6 | 3 | 9 | 73 | 71 | 75 | 0.8 | 0.7 |
| CS vs. PHT | LMT | ✕ | ✔ | 16 | 5 | 22 | 83 | 75 | 92 | 0.8 | 0.8 |
| PPGL vs. PHT | LB | ✕ | ✕ | 13 | 2 | 15 | 78 | 80 | 75 | 0.8 | 0.8 |
| EHT vs. PHT | RF | ✕ | ✕ | 10 | 1 | 11 | 74 | 57 | 92 | 0.7 | 0.8 |
| ALL vs. ALL | LMT | ✔ | ✕ | 10 | 4 | 15 | 61 | 42 | 81 | 0.4 | 0.7 |
| Reference | |||
|---|---|---|---|
| CS | PHT | ||
| Prediction | CS | 6 | 1 |
| PHT | 2 | 11 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reel, S.; Reel, P.S.; Erlic, Z.; Amar, L.; Pecori, A.; Larsen, C.K.; Tetti, M.; Pamporaki, C.; Prehn, C.; Adamski, J.; et al. Predicting Hypertension Subtypes with Machine Learning Using Targeted Metabolites and Their Ratios. Metabolites 2022, 12, 755. https://doi.org/10.3390/metabo12080755
Reel S, Reel PS, Erlic Z, Amar L, Pecori A, Larsen CK, Tetti M, Pamporaki C, Prehn C, Adamski J, et al. Predicting Hypertension Subtypes with Machine Learning Using Targeted Metabolites and Their Ratios. Metabolites. 2022; 12(8):755. https://doi.org/10.3390/metabo12080755
Chicago/Turabian StyleReel, Smarti, Parminder S. Reel, Zoran Erlic, Laurence Amar, Alessio Pecori, Casper K. Larsen, Martina Tetti, Christina Pamporaki, Cornelia Prehn, Jerzy Adamski, and et al. 2022. "Predicting Hypertension Subtypes with Machine Learning Using Targeted Metabolites and Their Ratios" Metabolites 12, no. 8: 755. https://doi.org/10.3390/metabo12080755
APA StyleReel, S., Reel, P. S., Erlic, Z., Amar, L., Pecori, A., Larsen, C. K., Tetti, M., Pamporaki, C., Prehn, C., Adamski, J., Prejbisz, A., Ceccato, F., Scaroni, C., Kroiss, M., Dennedy, M. C., Deinum, J., Eisenhofer, G., Langton, K., Mulatero, P., ... Jefferson, E. R. (2022). Predicting Hypertension Subtypes with Machine Learning Using Targeted Metabolites and Their Ratios. Metabolites, 12(8), 755. https://doi.org/10.3390/metabo12080755









