Hepatic Transcriptome Analysis Provides New Insight into the Lipid-Reducing Effect of Dietary Taurine in High–Fat Fed Groupers (Epinephelus coioides)
Abstract
:1. Introduction
2. Results
2.1. Growth Performance and Tissue Lipid Contents
2.2. Illumina Sequencing and De Novo Assembly
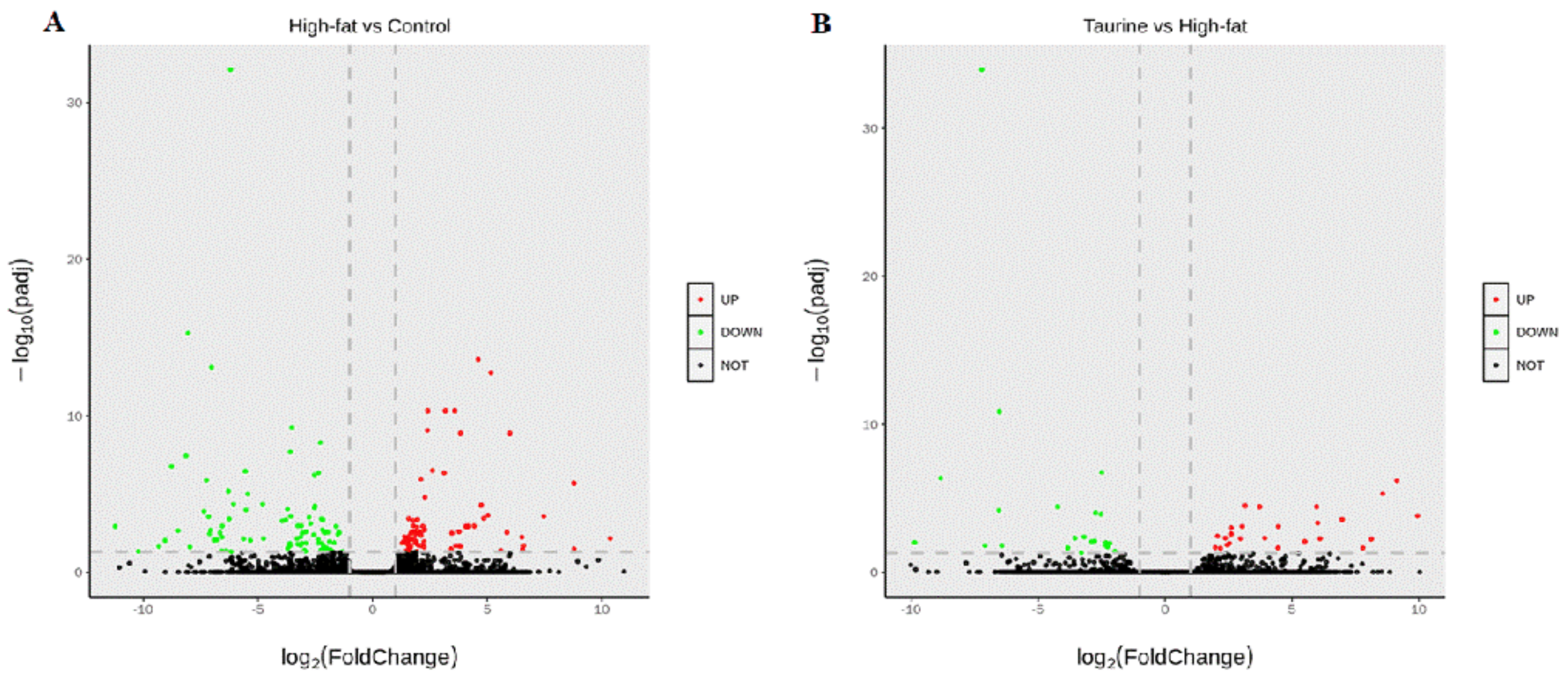
2.3. Identification of DEGs
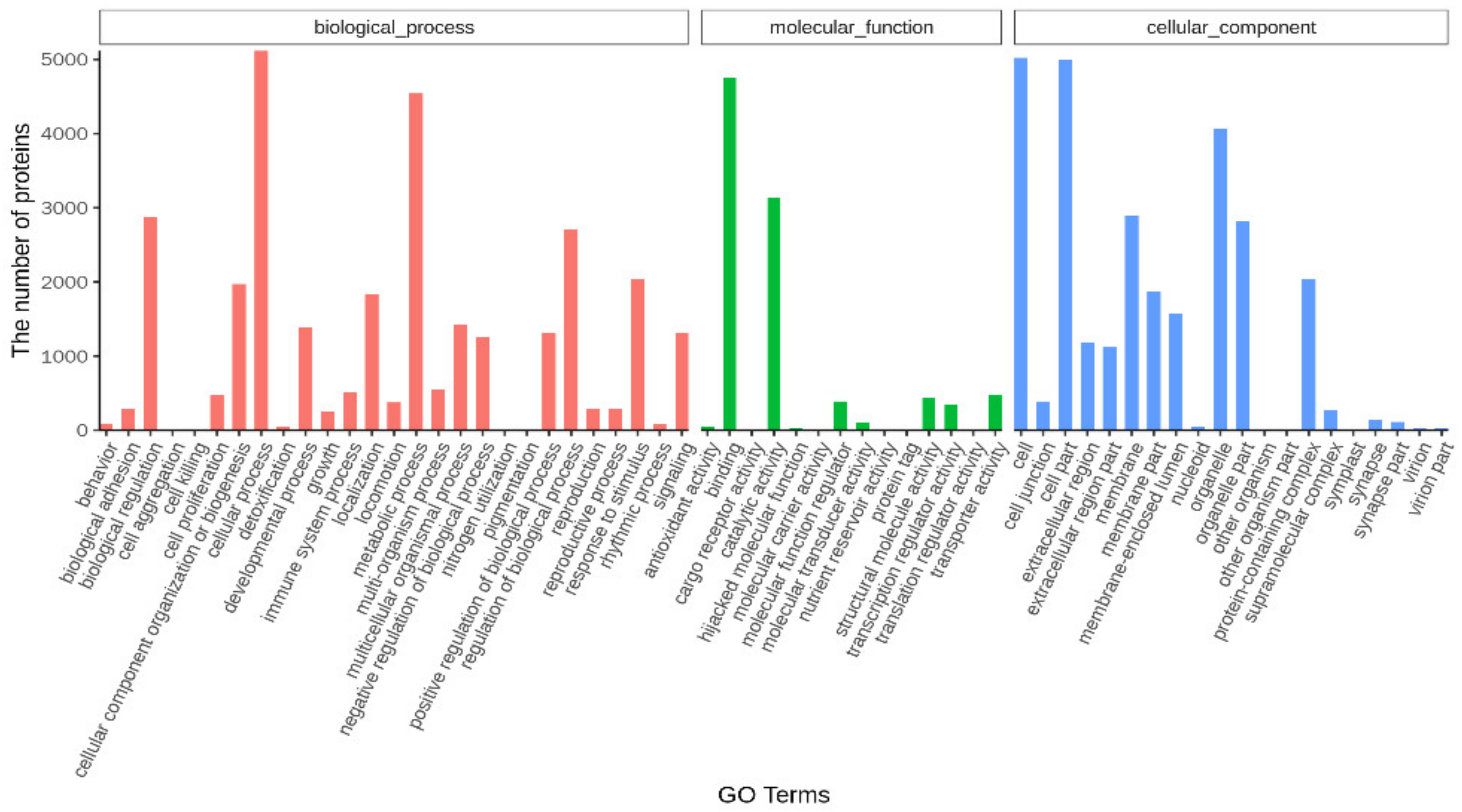
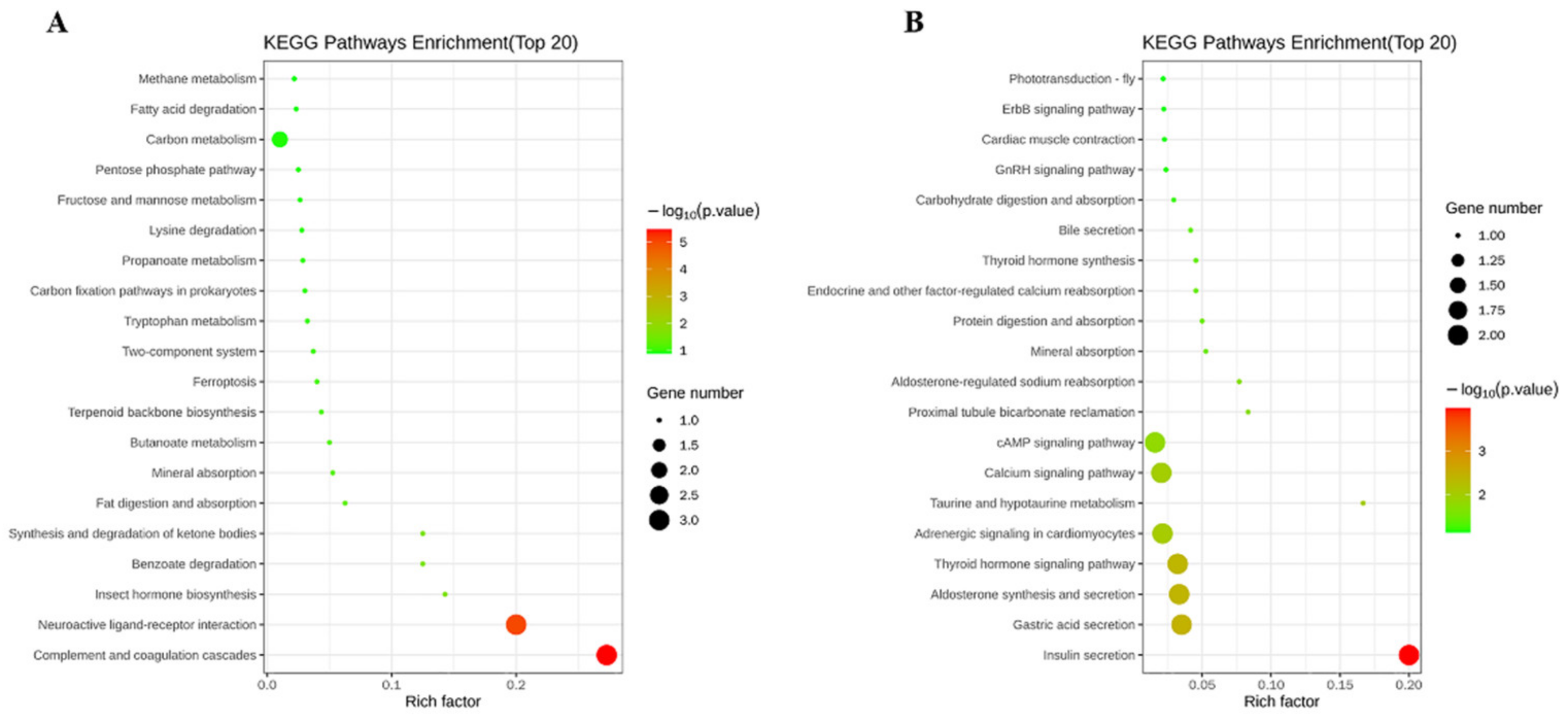
2.4. Function Annotation and Analysis for Unigenes
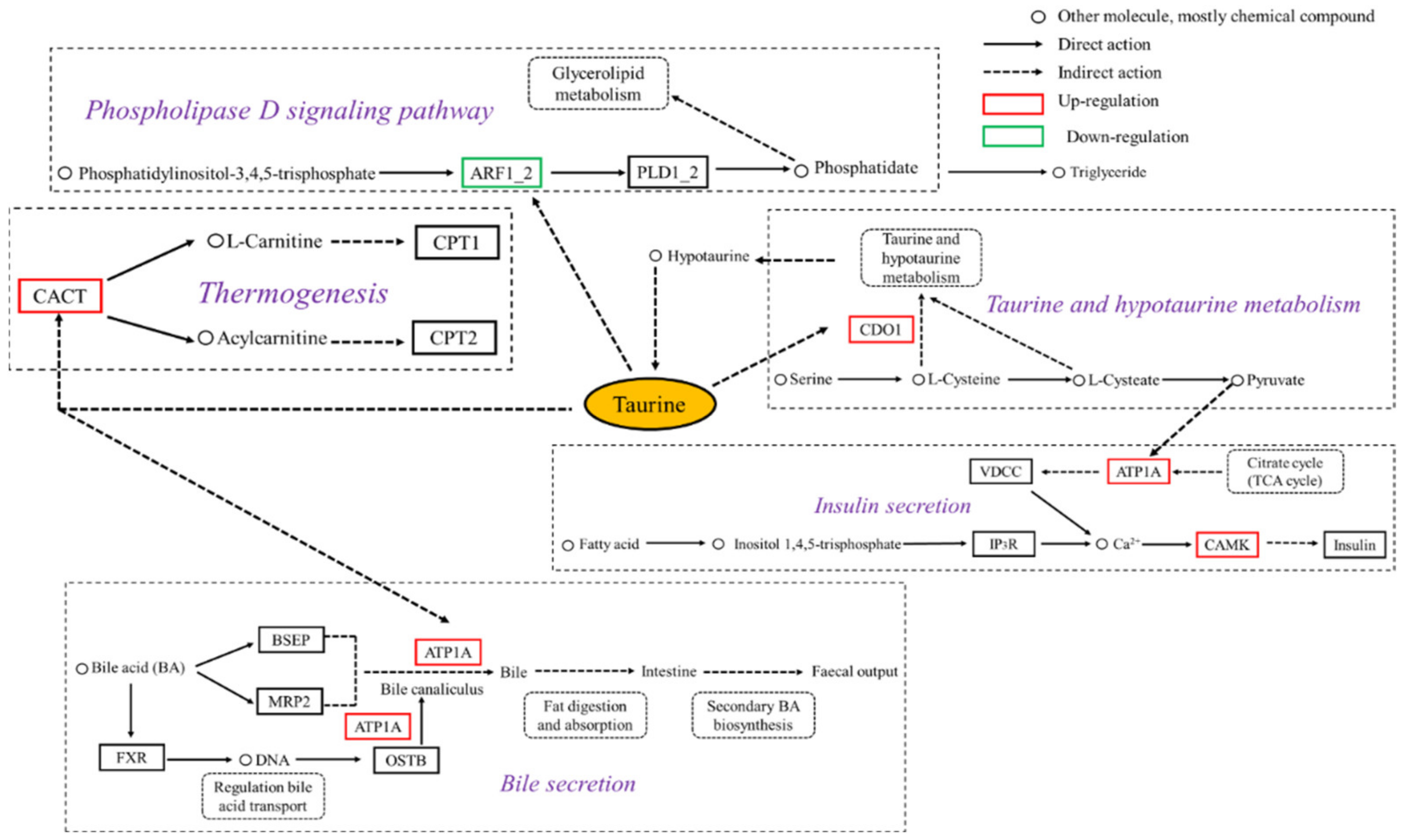
2.5. Signaling Pathway Network Related to Lipid Metabolism
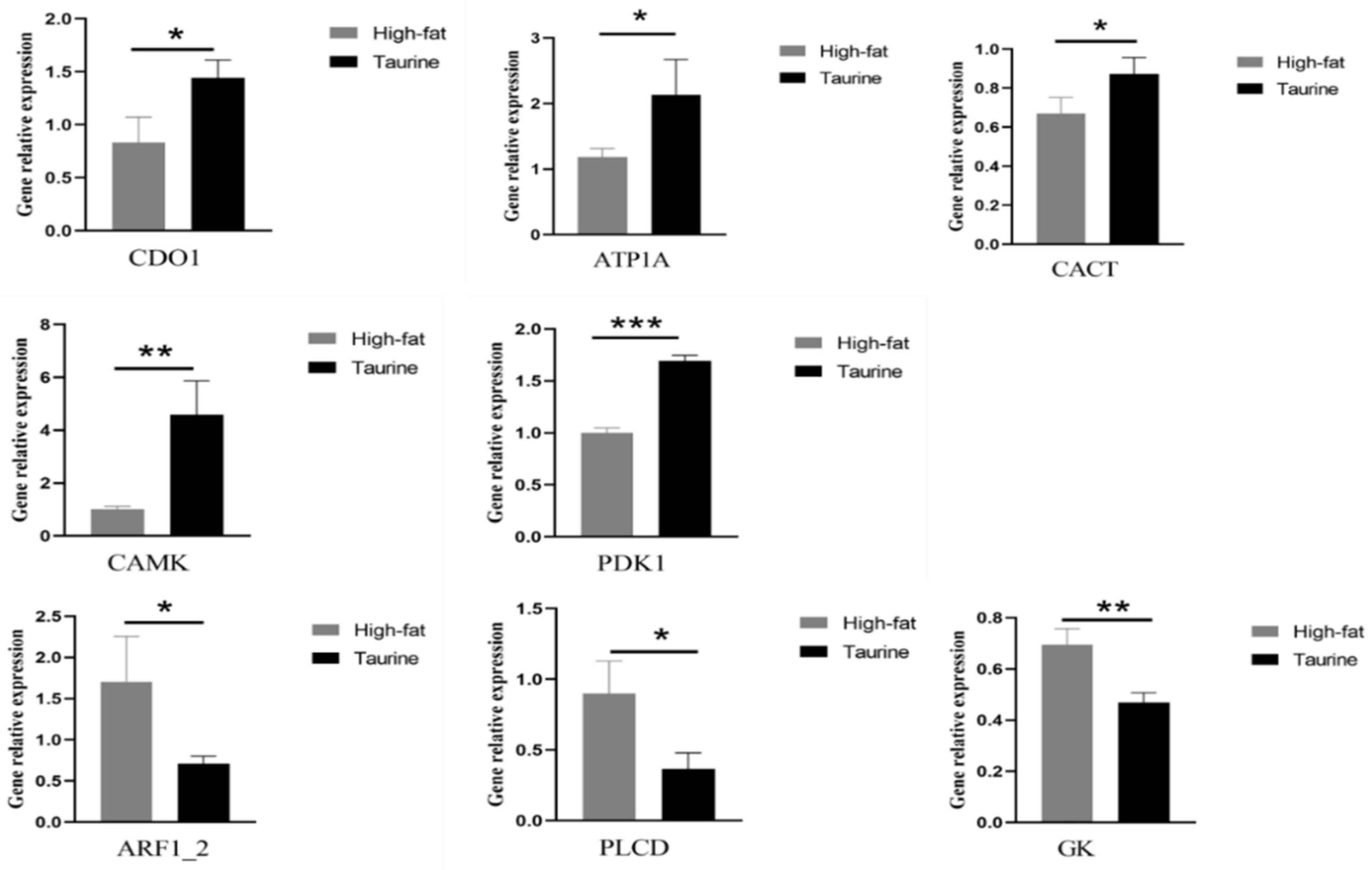
2.6. qRT–PCR Validation of DEGs
3. Discussion
4. Materials and Methods
4.1. Experimental Diets
4.2. Growth Trial
4.3. Sample Collection
4.4. Composition Analysis
4.5. RNA Extraction and cDNA Library Construction
4.6. Sequence Data Processing and Analysis
4.7. Identification and Enrichment Analysis of Differentially Expressed Genes
4.8. Quantitative Real–Time PCR Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambert, I.H.; Kristensen, D.M.; Holm, J.B.; Mortensen, O.H. Physiological role of taurine—From organism to organelle. Acta Physiol. 2015, 213, 191–212. [Google Scholar] [CrossRef]
- Chen, W.; Guo, J.; Chang, P. The effect of taurine on cholesterol metabolism. Mol. Nutr. Food Res. 2012, 56, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Yoshitama, A.; Sugita, S.; Fujita, M.; Murakami, S. Dietary taurine reduces hepatic secretion of cholesteryl ester and enhances fatty acid oxidation in rats fed a high-cholesterol diet. J. Nutr. Sci. Vitaminol. 2011, 57, 144–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brons, C.; Spohr, C.; Storgaard, H.; Dyerberg, J.; Vaag, A. Effect of taurine treatment on insulin secretion and action, and on serum lipid levels in overweight men with a genetic predisposition for type II diabetes mellitus. Eur. J. Clin. Nutr. 2004, 58, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, G.; Mai, K.; Xu, W.; Zhou, H. Differential regulation of taurine biosynthesis in rainbow trout and Japanese flounder. Sci. Rep. 2016, 6, 21231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salze, G.P.; Davis, D.A. Taurine: A critical nutrient for future fish feeds. Aquaculture 2015, 437, 215–229. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Q.; Kim, S.K.; Liao, Z.; Wei, Y.; Sun, B.; Jia, L.; Chi, S.; Liang, M. Dietary taurine stimulates the hepatic biosynthesis of both bile acids and cholesterol in the marine teleost, tiger puffer (Takifugu rubripes). Br. J. Nutr. 2020, 123, 1345–1356. [Google Scholar] [CrossRef] [Green Version]
- Martins, N.; Diógenes, A.F.; Magalhães, R.; Matas, I.; Oliva-Teles, A.; Peres, H. Dietary taurine supplementation affects lipid metabolism and improves the oxidative status of European seabass (Dicentrarchus labrax) juveniles. Aquaculture 2021, 531, 735820. [Google Scholar] [CrossRef]
- Li, M.; Lai, H.; Li, Q.; Gong, S.; Wang, R. Effects of dietary taurine on growth, immunity and hyperammonemia in juvenile yellow catfish Pelteobagrus fulvidraco fed all-plant protein diets. Aquaculture 2016, 450, 349–355. [Google Scholar] [CrossRef]
- Garcia-Organista, A.A.; Mata-Sotres, J.A.; Viana, M.T.; Rombenso, A.N. The effects of high dietary methionine and taurine are not equal in terms of growth and lipid metabolism of juvenile California yellowtail (Seriola dorsalis). Aquaculture 2019, 512, 734304. [Google Scholar] [CrossRef]
- Koven, W.; Peduel, A.; Gada, M.; Nixon, O.; Ucko, M. Taurine improves the performance of white grouper juveniles (Epinephelus aeneus) fed a reduced fish meal diet. Aquaculture 2016, 460, 8–14. [Google Scholar] [CrossRef]
- Du, Z.Y.; Liu, Y.J.; Tian, L.X.; Wang, J.T.; Wang, Y.; Liang, G.Y. Effect of dietary lipid level on growth, feed utilization and body composition by juvenile grass carp (Ctenopharyngodon idella). Aquac. Nutr. 2005, 11, 139–146. [Google Scholar] [CrossRef]
- Meng, Y.; Tian, H.; Hu, X.; Han, B.; Li, X.; Cangzhong, L.; Ma, R.; Kubilay, A. Effects of dietary lipid levels on the lipid deposition and metabolism of subadult triploid rainbow trout (Oncorhynchus mykiss). Aquac. Nutr. 2022, 2022, 6924835. [Google Scholar] [CrossRef]
- Dai, Y.; Cao, X.; Zhang, D.; Li, X.; Liu, W.; Jiang, G. Chronic inflammation is a key to inducing liver injury in blunt snout bream (Megalobrama amblycephala) fed with high-fat diet. Dev. Comp. Immunol. 2019, 97, 28–37. [Google Scholar] [CrossRef]
- Fan, Z.; Li, J.; Zhang, Y.; Wu, D.; Zheng, X.; Wang, C.; Wang, L. Excessive dietary lipid affecting growth performance, feed utilization, lipid deposition, and hepatopancreas lipometabolism of large-sized common carp (Cyprinus carpio). Front. Nutr. 2021, 8, 694426. [Google Scholar] [CrossRef]
- Fei, S.; Xia, Y.; Chen, Z.; Liu, C.; Liu, H.; Han, D.; Jin, J.; Yang, Y.; Zhu, X.; Xie, S. A high-fat diet alters lipid accumulation and oxidative stress and reduces the disease resistance of overwintering hybrid yellow catfish (Pelteobagrus fulvidraco ♀ × P. vachelli ♂). Aquac. Rep. 2022, 23, 101043. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, Y.; Zhao, H.; Chen, W.; Chen, Y.; Lin, S. Effect of dietary lipid level on growth, lipid metabolism and oxidative status of largemouth bass, Micropterus salmoides. Aquaculture 2019, 506, 394–400. [Google Scholar] [CrossRef]
- Yin, P.; Xie, S.; Zhuang, Z.; He, X.; Tang, X.; Tian, L.; Liu, Y.; Niu, J. Dietary supplementation of bile acid attenuate adverse effects of high-fat diet on growth performance, antioxidant ability, lipid accumulation and intestinal health in juvenile largemouth bass (Micropterus salmoides). Aquaculture 2021, 531, 735864. [Google Scholar] [CrossRef]
- Jia, R.; Cao, L.P.; Du, J.L.; He, Q.; Gu, Z.Y.; Jeney, G.; Xu, P.; Yin, G.J. Effects of high-fat diet on antioxidative status, apoptosis and inflammation in liver of tilapia (Oreochromis niloticus) via Nrf2, TLRs and JNK pathways. Fish Shellfish Immun. 2020, 104, 391–401. [Google Scholar] [CrossRef]
- Zhou, Y.L.; Guo, J.L.; Tang, R.J.; Ma, H.J.; Chen, Y.J.; Lin, S.M. High dietary lipid level alters the growth, hepatic metabolism enzyme, and anti-oxidative capacity in juvenile largemouth bass Micropterus salmoides. Fish Physiol. Biochem. 2020, 46, 125–134. [Google Scholar] [CrossRef]
- Cao, X.; Dai, Y.; Liu, M.; Yuan, X.; Wang, C.; Huang, Y.; Liu, W.; Jiang, G. High-fat diet induces aberrant hepatic lipid secretion in blunt snout bream by activating endoplasmic reticulum stress-associated IRE1/XBP1 pathway. BBA Mol. Cell Biol. Lipids 2019, 1864, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Yi, K.; Qian, X.; Niu, X.; Sun, Y.; Ye, J. Growth and metabolic responses of juvenile grouper (Epinephelus coioides) to dietary methionine/cystine ratio at constant sulfur amino acid levels. Aquaculture 2020, 518, 734869. [Google Scholar] [CrossRef]
- Shapawi, R.; Abdullah, F.C.; Senoo, S.; Mustafa, S. Nutrition, growth and resilience of tiger grouper (Epinephelus fuscoguttatus) × giant grouper (Epinephelus lanceolatus) hybrid—A review. Rev. Aquac. 2019, 11, 1285–1296. [Google Scholar] [CrossRef]
- Bai, F.; Niu, X.; Wang, X.; Ye, J. Growth performance, biochemical composition and expression of lipid metabolism related genes in groupers (Epinephelus coioides) are altered by dietary taurine. Aquac. Nutr. 2021, 27, 2690–2702. [Google Scholar] [CrossRef]
- Bai, F.; Wang, X.; Niu, X.; Shen, G.; Ye, J. Lipidomic profiling reveals the reducing lipid accumulation effect of dietary taurine in groupers (Epinephelus coioides). Front. Mol. Biosci. 2021, 8, 814318. [Google Scholar] [CrossRef]
- Wang, X.; Bai, F.; Niu, X.; Sun, Y.; Ye, J. The lipid-lowering effect of dietary taurine in orange-spotted groupers (Epinephelus coioides) involves both bile acids and lipid metabolism. Front. Mar. Sci. 2022, 9, 859428. [Google Scholar] [CrossRef]
- Jin, M.; Zhu, T.; Tocher, D.R.; Luo, J.; Shen, Y.; Li, X.; Pan, T.; Yuan, Y.; Betancor, M.B.; Jiao, L.; et al. Dietary fenofibrate attenuated high-fat-diet-induced lipid accumulation and inflammation response partly through regulation of pparalpha and sirt1 in juvenile black seabream (Acanthopagrus schlegelii). Dev. Comp. Immunol. 2020, 109, 103691. [Google Scholar] [CrossRef]
- Ding, T.; Xu, N.; Liu, Y.; Du, J.; Xiang, X.; Xu, D.; Liu, Q.; Yin, Z.; Li, J.; Mai, K.; et al. Effect of dietary bile acid (BA) on the growth performance, body composition, antioxidant responses and expression of lipid metabolism-related genes of juvenile large yellow croaker (Larimichthys crocea) fed high-lipid diets. Aquaculture 2020, 518, 734768. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Tian, L.; Mai, K.; Du, Z.; Wang, Y.; Yang, H. Effect of dietary lipid level on growth performance, lipid deposition, hepatic lipogenesis in juvenile cobia (Rachycentron canadum). Aquaculture 2005, 249, 439–447. [Google Scholar] [CrossRef]
- Han, T.; Li, X.; Wang, J.; Hu, S.; Jiang, Y.; Zhong, X. Effect of dietary lipid level on growth, feed utilization and body composition of juvenile giant croaker Nibea japonica. Aquaculture 2014, 434, 145–150. [Google Scholar] [CrossRef]
- Matsunari, H.; Takeuchi, T.; Takahashi, M.; Mushiake, K. Effect of dietary taurine supplementation on growth performance of yellowtail juveniles Seriola quinqueradiata. Fish. Sci. 2005, 71, 1131–1135. [Google Scholar] [CrossRef]
- Matsunari, H.; Furuita, H.; Yamamoto, T.; Kim, S.; Sakakura, Y.; Takeuchi, T. Effect of dietary taurine and cystine on growth performance of juvenile red sea bream Pagrus major. Aquaculture 2008, 274, 142–147. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Oh, D.; Khosravi, S.; Cha, J.; Park, S.; Kim, K.; Lee, K. Taurine is an essential nutrient for juvenile parrot fish Oplegnathus fasciatus. Aquaculture 2013, 414–415, 274–279. [Google Scholar] [CrossRef]
- Lunger, A.N.; Mclean, E.; Gaylord, T.G.; Kuhn, D.; Craig, S.R. Taurine supplementation to alternative dietary proteins used in fish meal replacement enhances growth of juvenile cobia (Rachycentron canadum). Aquaculture 2007, 271, 401–410. [Google Scholar] [CrossRef]
- Martins, N.; Estevão-Rodrigues, T.; Diógenes, A.F.; Diaz-Rosales, P.; Oliva-Teles, A.; Peres, H. Taurine requirement for growth and nitrogen accretion of European sea bass (Dicentrarchus labrax, L.) juveniles. Aquaculture 2018, 494, 19–25. [Google Scholar] [CrossRef]
- Yun, B.; Ai, Q.; Mai, K.; Xu, W.; Qi, G.; Luo, Y. Synergistic effects of dietary cholesterol and taurine on growth performance and cholesterol metabolism in juvenile turbot (Scophthalmus maximus L.) fed high plant protein diets. Aquaculture 2012, 324, 85–91. [Google Scholar] [CrossRef]
- Candebat, C.L.; Booth, M.; Codabaccus, M.B.; Pirozzi, I. Dietary methionine spares the requirement for taurine in juvenile yellowtail kingfish (Seriola lalandi). Aquaculture 2020, 522, 735090. [Google Scholar] [CrossRef]
- De Moura, L.B.; Diógenes, A.F.; Campelo, D.A.V.; Almeida, F.L.A.D.; Pousão-Ferreira, P.M.; Furuya, W.M.; Oliva-Teles, A.; Peres, H. Taurine and methionine supplementation as a nutritional strategy for growth promotion of meagre (Argyrosomus regius) fed high plant protein diets. Aquaculture 2018, 497, 389–395. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, G.; Li, Z.; Hu, Y.; Zhong, L.; Zhou, Q.; Peng, M. Effect of dietary taurine supplementation on growth, digestive enzyme, immunity and resistant to dry stress of rice field eel (Monopterus albus) fed low fish meal diets. Aquac. Res. 2018, 49, 2108–2118. [Google Scholar] [CrossRef]
- Dehghani, R.; Oujifard, A.; Mozanzadeh, M.T.; Morshedi, V.; Bagheri, D. Effects of dietary taurine on growth performance, antioxidant status, digestive enzymes activities and skin mucosal immune responses in yellowfin seabream, Acanthopagrus latus. Aquaculture 2020, 517, 734795. [Google Scholar] [CrossRef]
- Jurkowska, H.; Roman, H.B.; Hirschberger, L.L.; Sasakura, K.; Nagano, T.; Hanaoka, K.; Krijt, J.; Stipanuk, M.H. Primary hepatocytes from mice lacking cysteine dioxygenase show increased cysteine concentrations and higher rates of metabolism of cysteine to hydrogen sulfide and thiosulfate. Amino Acids 2014, 46, 1353–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betancor, M.B.; Laurent, G.R.; Ortega, A.; de la Gándara, F.; Tocher, D.R.; Mourente, G. Taurine metabolism and effects of inclusion levels in rotifer (Brachionus rotundiformis, Tschugunoff, 1921) on Atlantic bluefin tuna (Thunnus thynnus, L.) larvae. Aquaculture 2019, 510, 353–363. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, Q.; Xu, H.; Liang, M. Taurine requirement and metabolism response of tiger puffer Takifugu rubripes to graded taurine supplementation. Aquaculture 2020, 524, 735237. [Google Scholar] [CrossRef]
- Wang, L.; Su, H.; Wang, C.L.; Mu, C.H. Structure and functional mechanism of the ADP-ribosylation factor. Chin. J. Cell Biol. 2007, 29, 675–681. (In Chinese) [Google Scholar]
- Mcdermott, M.; Wakelam, M.J.; Morris, A.J. Phospholipase D. Biochem. Cell Biol. 2004, 82, 225–253. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Sena, T.; Soluyanova, P.; Guzman, C.; Valdivielso, J.M.; Castell, J.V.; Jover, R. The vitamin D receptor regulates glycerolipid and phospholipid metabolism in human hepatocytes. Biomolecules 2020, 10, 493. [Google Scholar] [CrossRef] [Green Version]
- Kullak-Ublick, G.A.; Beuers, U.; Paumgartner, G. Hepatobiliary transport. J. Hepatol. 2000, 32, 3–18. [Google Scholar] [CrossRef]
- Deuschle, U.; Birkel, M.; Hambruch, E.; Hornberger, M.; Kinzel, O.; Perovic-Ottstadt, S.; Schulz, A.; Hahn, U.; Burnet, M.; Kremoser, C. The nuclear bile acid receptor FXR controls the liver derived tumor suppressor histidine-rich glycoprotein. Int. J. Cancer 2015, 136, 2693–2704. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Song, X.; Valanejad, L.; Vasilenko, A.; More, V.; Qiu, X.; Chen, W.; Lai, Y.; Slitt, A.; Stoner, M.; et al. Bile salt export pump is dysregulated with altered farnesoid X receptor isoform expression in patients with hepatocellular carcinoma. Hepatology 2013, 57, 1530–1541. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.L.; Ding, Y.; Chen, Z.L.; Wang, Y.; Liu, P.; Qin, H.; Zhou, L.S.; Zhang, L.L.; Huang, J.; Zhao, L. Emodin rescues intrahepatic cholestasis via stimulating FXR/BSEP pathway in promoting the canalicular export of accumulated bile. Front. Pharmacol. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Kubitz, R.; Dröge, C.; Stindt, J.; Weissenberger, K.; Häussinger, D. The bile salt export pump (BSEP) in health and disease. Clin. Res. Hepatol. Gas. 2012, 36, 536–553. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Sun, B.; Zhang, Q.; Jia, L.; Wei, Y.; Liang, M.; Xu, H. Dietary bile acids regulate the hepatic lipid homeostasis in tiger puffer fed normal or high-lipid diets. Aquaculture 2020, 519, 734935. [Google Scholar] [CrossRef]
- Di Leo, M.A.; Santini, S.A.; Cercone, S.; Lepore, D.; Gentiloni, S.N.; Caputo, S.; Greco, A.V.; Giardina, B.; Franconi, F.; Ghirlanda, G. Chronic taurine supplementation ameliorates oxidative stress and Na+ K+ ATPase impairment in the retina of diabetic rats. Amino Acids 2002, 23, 401–406. [Google Scholar] [CrossRef]
- Huang, M.; Yang, X.; Zhou, Y.; Ge, J.; Davis, D.A.; Dong, Y.; Gao, Q.; Dong, S. Growth, serum biochemical parameters, salinity tolerance and antioxidant enzyme activity of rainbow trout (Oncorhynchus mykiss) in response to dietary taurine levels. Mar. Life Sci. Technol. 2021, 3, 449–462. [Google Scholar] [CrossRef]
- Haeusler, R.A.; Camastra, S.; Astiarraga, B.; Nannipieri, M.; Anselmino, M.; Ferrannini, E. Decreased expression of hepatic glucokinase in type 2 diabetes. Mol. Metab. 2015, 4, 222–226. [Google Scholar] [CrossRef]
- Caseras, A.; Meton, I.; Vives, C.; Egea, M.; Fernandez, F.; Baanante, I.V. Nutritional regulation of glucose-6-phosphatase gene expression in liver of the gilthead sea bream (Sparus aurata). Br. J. Nutr. 2002, 88, 607–614. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yang, J.; Liu, B.; Zou, Y.; Sun, M.; Li, Z.; Yang, R.; Xu, X.; Zou, L.; Li, G.; et al. P2Y12 shRNA normalizes inflammatory dysfunctional hepatic glucokinase activity in type 2 diabetic rats. Biomed. Pharmacother. 2020, 132, 110803. [Google Scholar] [CrossRef]
- Satsu, H.; Manabe, M.; Shimizu, M. Activation of Ca2+/calmodulin-dependent protein kinase II is involved in hyperosmotic induction of the human taurine transporter. FEBS Lett. 2004, 569, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Kim, H.E.; Choi, S.E.; Shin, H.C.; Kwag, W.J.; Lee, B.K.; Cho, K.W.; Kang, Y. Involvement of Ca2+/calmodulin kinase II (CaMK II) in genistein-induced potentiation of leucine/glutamine-stimulated insulin secretion. Mol. Cells 2009, 28, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Vettorazzi, J.F.; Ribeiro, R.A.; Santos-Silva, J.C.; Borck, P.C.; Batista, T.M.; Nardelli, T.R.; Boschero, A.C.; Carneiro, E.M. Taurine supplementation increases K (ATP) channel protein content, improving Ca2+ handling and insulin secretion in islets from malnourished mice fed on a high-fat diet. Amino Acids 2014, 46, 2123–2136. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.A.; Bonfleur, M.L.; Amaral, A.G.; Vanzela, E.C.; Rocco, S.A.; Boschero, A.C.; Carneiro, E.M. Taurine supplementation enhances nutrient-induced insulin secretion in pancreatic mice islets. Diabetes Metab. Res. Rev. 2009, 25, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.A.; Sheridan, M.A. New insights into the signaling system and function of insulin in fish. Gen. Comp. Endocr. 2011, 173, 227–247. [Google Scholar] [CrossRef] [PubMed]
- Polakof, S.; Médale, F.; Skiba-Cassy, S.; Corraze, G.; Panserat, S. Molecular regulation of lipid metabolism in liver and muscle of rainbow trout subjected to acute and chronic insulin treatments. Domest. Anim. Endocrinol. 2010, 39, 26–33. [Google Scholar] [CrossRef]
- Pan, Y.; Luo, Z.; Wu, K.; Zhang, L.; Xu, Y.; Chen, Q. Cloning, mRNA expression and transcriptional regulation of five retinoid X receptor subtypes in yellow catfish Pelteobagrus fulvidraco by insulin. Gen. Comp. Endocr. 2016, 225, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.Y.; Zhai, Z.Z.; Li, Z.F.; Wang, L. High fat diet-triggered non-alcoholic fatty liver disease: A review of proposed mechanisms. Chem. Biol. Interact. 2020, 330, 109199. [Google Scholar] [CrossRef]
- Sakurai, M.; Takamura, T.; Ota, T.; Ando, H.; Akahori, H.; Kaji, K.; Sasaki, M.; Nakanuma, Y.; Miura, K.; Kaneko, S. Liver steatosis, but not fibrosis, is associated with insulin resistance in nonalcoholic fatty liver disease. J. Gastroenterol. 2007, 42, 312–317. [Google Scholar] [CrossRef] [Green Version]
- Batista, T.M.; Ribeiro, R.A.; Da, S.P.; Camargo, R.L.; Lollo, P.C.; Boschero, A.C.; Carneiro, E.M. Taurine supplementation improves liver glucose control in normal protein and malnourished mice fed a high-fat diet. Mol. Nutr. Food Res. 2013, 57, 423–434. [Google Scholar] [CrossRef]
- Nakaya, Y.; Minami, A.; Harada, N.; Sakamoto, S.; Niwa, Y.; Ohnaka, M. Taurine improves insulin sensitivity in the Otsuka Long-Evans Tokushima Fatty rat, a model of spontaneous type 2 diabetes. Am. J. Clin. Nutr. 2000, 71, 54–58. [Google Scholar] [CrossRef] [Green Version]
- Musso, G.; Gambino, R.; Cassader, M. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog. Lipid Res. 2009, 48, 1–26. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentile, C.L.; Nivala, A.M.; Gonzales, J.C.; Pfaffenbach, K.T.; Wang, D.; Wei, Y.; Jiang, H.; Orlicky, D.J.; Petersen, D.R.; Pagliassotti, M.J.; et al. Experimental evidence for therapeutic potential of taurine in the treatment of nonalcoholic fatty liver disease. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R1710–R1722. [Google Scholar] [CrossRef] [PubMed]
- Giudetti, A.M.; Stanca, E.; Siculella, L.; Gnoni, G.V.; Damiano, F. Nutritional and hormonal regulation of citrate and carnitine/acylcarnitine transporters: Two mitochondrial carriers involved in fatty acid metabolism. Int. J. Mol. Sci. 2016, 17, 817. [Google Scholar] [CrossRef]
- Tian, J.; Lu, R.; Ji, H.; Sun, J.; Li, C.; Liu, P.; Lei, C.; Chen, L.; Du, Z. Comparative analysis of the hepatopancreas transcriptome of grass carp (Ctenopharyngodon idellus) fed with lard oil and fish oil diets. Gene 2015, 565, 192–200. [Google Scholar] [CrossRef]
- Priore, P.; Stanca, E.; Gnoni, G.V.; Siculella, L. Dietary fat types differently modulate the activity and expression of mitochondrial carnitine/acylcarnitine translocase in rat liver. Biochim. Biophys. Acta 2012, 1821, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Liu, Y.; Mai, K.; Tian, L.; Liu, D.; Tan, X.; Lin, H. Effect of dietary lipid level on growth performance, feed utilization and body composition of grouper Epinephelus coioides juveniles fed isonitrogenous diets in floating netcages. Aquac. Int. 2005, 13, 257–269. [Google Scholar] [CrossRef]
- Zhou, M.W.; Wang, H.W.; Ye, J.D. Responses to growth performance, body composition and tissue free amino acid contents of grouper (Epinephelus coioides) to dietary taurine content. Chin. J. Anim. Nutr. 2015, 27, 785–794. (In Chinese) [Google Scholar]
- Horwitz, W. Official Methods of Analysis of AOAC International, 16th ed.; AOAC International: Rockville, MD,USA, 1995. [Google Scholar]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Aragão, C.; Colen, R.; Ferreira, S.; Pinto, W.; Conceição, L.E.C.; Dias, J. Microencapsulation of taurine in Senegalese sole diets improves its metabolic availability. Aquaculture 2014, 431, 53–58. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Parameters | Diets (Fat Level/Taurine Level) | ||
|---|---|---|---|
| Control (10/0) | High–Fat (15/0) | Taurine (15/1) | |
| Weight gain (%) | 151.44 ± 9.86 a | 157.38 ± 4.77 a | 180.07 ± 3.45 b |
| Feed conversion ratio | 0.99 ± 0.01 b | 1.00 ± 0.01 b | 0.91 ± 0.02 a |
| Hepatosomatic index (%) | 2.79 ± 0.14 b | 2.89 ± 0.06 b | 1.56 ± 0.24 a |
| Liver lipid content (%) | 9.57 ± 0.56 a | 13.38 ± 0.47 b | 9.05 ± 0.31 a |
| Muscle lipid content (%) | 1.31 ± 0.72 a | 1.95 ± 0.11 b | 1.79 ± 0.01 b |
| Items | Min Length | Max Length | Mean Length | N50 | N90 |
|---|---|---|---|---|---|
| Unigene number | 200 | 16,130 | 532 | 653 | 243 |
| Type of Database Annotation | Number of Unigenes | Percentage (%) |
|---|---|---|
| NR | 72,125 | 20.65 |
| SwissProt | 13,104 | 3.75 |
| PFAM | 22,814 | 6.53 |
| GO | 6988 | 2.00 |
| KO | 5124 | 1.47 |
| Annotated in all databases | 487 | 0.14 |
| Annotated in at least one database | 72,679 | 20.81 |
| Total Unigenes | 349,211 | 100 |
| Ingredients | Diets (Fat Level/Taurine Level) | ||
|---|---|---|---|
| Control (10/0) | High-Fat (15/0) | Taurine (15/1) | |
| Animal by–product (casein:gelatin = 4:1) | 50 | 50 | 50 |
| Shrimp meal | 4 | 4 | 4 |
| Corn starch | 25 | 25 | 25 |
| Oil (fish:soy oil = 1:1) | 6 | 10 | 10 |
| Soy lecithin | 4 | 4 | 4 |
| Premix | 0.8 | 0.8 | 0.8 |
| Ca(H2PO4)2 | 2 | 2 | 2 |
| Microcrystalline cellulose | 7.2 | 3.2 | 2.2 |
| Sodium alginate | 1 | 1 | 1 |
| Taurine | 0 | 0 | 1 |
| Nutrient level (analyzed values) | |||
| Dry matter | 91.18 | 90.24 | 90.38 |
| Crude protein | 46.55 | 46.87 | 46.56 |
| Crude lipid | 10.44 | 14.79 | 14.89 |
| Taurine | 0.04 | 0.04 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Bai, F.; Song, T.; Niu, X.; Wang, X.; Wang, K.; Ye, J. Hepatic Transcriptome Analysis Provides New Insight into the Lipid-Reducing Effect of Dietary Taurine in High–Fat Fed Groupers (Epinephelus coioides). Metabolites 2022, 12, 670. https://doi.org/10.3390/metabo12070670
Chen M, Bai F, Song T, Niu X, Wang X, Wang K, Ye J. Hepatic Transcriptome Analysis Provides New Insight into the Lipid-Reducing Effect of Dietary Taurine in High–Fat Fed Groupers (Epinephelus coioides). Metabolites. 2022; 12(7):670. https://doi.org/10.3390/metabo12070670
Chicago/Turabian StyleChen, Mingfan, Fakai Bai, Tao Song, Xingjian Niu, Xuexi Wang, Kun Wang, and Jidan Ye. 2022. "Hepatic Transcriptome Analysis Provides New Insight into the Lipid-Reducing Effect of Dietary Taurine in High–Fat Fed Groupers (Epinephelus coioides)" Metabolites 12, no. 7: 670. https://doi.org/10.3390/metabo12070670
APA StyleChen, M., Bai, F., Song, T., Niu, X., Wang, X., Wang, K., & Ye, J. (2022). Hepatic Transcriptome Analysis Provides New Insight into the Lipid-Reducing Effect of Dietary Taurine in High–Fat Fed Groupers (Epinephelus coioides). Metabolites, 12(7), 670. https://doi.org/10.3390/metabo12070670







