Abstract
Genome-wide association studies (GWAS) of circulating metabolites have revealed the role of genetic regulation on the human metabolome. Most previous investigations focused on European ancestry, and few studies have been conducted among populations of African descent living in Africa, where the infectious disease burden is high (e.g., human immunodeficiency virus (HIV)). It is important to understand the genetic associations of the metabolome in diverse at-risk populations including people with HIV (PWH) living in Africa. After a thorough literature review, the reported significant gene–metabolite associations were tested among 490 PWH in South Africa. Linear regression was used to test associations between the candidate metabolites and genetic variants. GWAS of 154 plasma metabolites were performed to identify novel genetic associations. Among the 29 gene–metabolite associations identified in the literature, we replicated 10 in South Africans with HIV. The UGT1A cluster was associated with plasma levels of biliverdin and bilirubin; SLC16A9 and CPS1 were associated with carnitine and creatine, respectively. We also identified 22 genetic associations with metabolites using a genome-wide significance threshold (p-value < 5 × 10−8). In a GWAS of plasma metabolites in South African PWH, we replicated reported genetic associations across ancestries, and identified novel genetic associations using a metabolomics approach.
1. Introduction
Genome-wide associations studies (GWAS), which involve investigations on associations between genetic architecture and complex disease traits, have been widely conducted to understand how the genetic profile contributes to various diseases [1]. High-resolution metabolomics can measure large-scale profiles of small molecules in biological samples, which enables metabolome-wide association studies (MWAS) to understand a variety of diseases on a molecular basis [2]. The traditional GWAS has been hindered by the phenotyping of exposures and risk factors that are unmeasured or even unknown. By combining high-throughput phenotyping metabolomics data with GWAS (mGWAS), we have broadened our insight into many complex diseases and traits [3].
Early mGWAS research found that common single nucleotide polymorphisms (SNP) can account for as much as 12% of the variance in homeostasis of the human metabolic profile [4]. In recent years, a number of studies of blood and urine metabolites have been conducted, revealing many robust and reproducible associations between genetic variants and metabolites, with many involving enzyme catalysis and transporter proteins [5,6]. With a goal of building an integrated atlas of genomics and metabolomics, many investigations have focused on general populations of European descent [7,8], and only a few have focused on African Americans [9,10]. For example, the UGT1A1 gene encodes the enzyme in the bilirubin metabolism, and its association with bilirubin and biliverdin has been successfully established [9,11]. This particular locus is also involved in the metabolic pathways linking to several human immunodeficiency virus (HIV) medications; however, there have been no studies focusing on individuals of African ancestry who have HIV infection living in South Africa, which would permit an exploration of environmental and genetic influences on metabolic processes. In this study of people with HIV (PWH) living in South Africa who were antiretroviral therapy (ART) naive, we aimed to assess the generalizability of previously reported findings and explore the novel associations between genetic variants and metabolites in this unique cohort of genetic ancestry, geography, and health condition.
2. Results
2.1. Characteristics of Study Participants
All 490 participants were South Africans, with 305 from RK Khan Hospital and 185 from Bethesda Hospital. The mean age was 34.4 (standard deviation [SD] 10.0) years, and 312 (63.7%) were female (Table 1). The ancestry map of the South African study participants compared to the 1000 Genome Project [12] reference panel showed a well-defined South African genetic profile partially intersecting with African (AFR) ancestry but distinct from other ancestries (Figure 1).

Table 1.
Baseline characteristics of the study cohort.
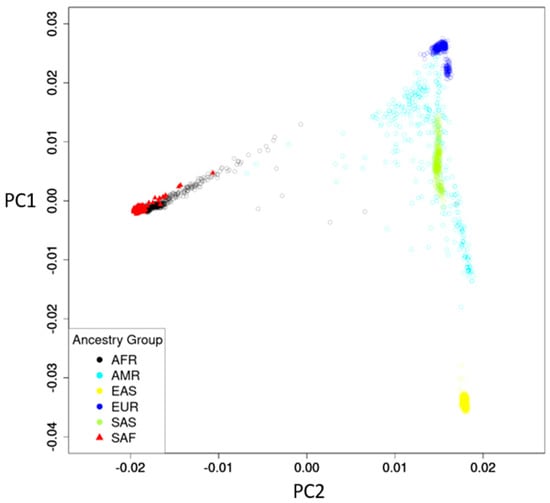
Figure 1.
Ancestry map of study participants and reference panel. Reference panel of 1000 Genome project: AFR, African; AMR, American; EAS, East Asian; EUR, European; SAS, South Asian. Study participants: SAF, South African.
2.2. Candidate Gene–Metabolite Associations
From the 24 reported GWAS of blood-based metabolic profiles (Figure 2, Table 2), we searched for the metabolites analyzed and annotated in the present study. We identified a total of 29 candidate SNP-metabolite associations, including 18 independent genetic loci and 14 metabolites from previous studies, and tested these associations in the South African cohort (Table 3). Five metabolites including citrulline, glutamine, histidine, serine, and urate, were identified from two different liquid chromatography platforms and were both analyzed, resulting in 36 replications of SNP-metabolite associations. Among the 36, a total of 24 were consistent in direction regardless of statistical significance, 7 were inconsistent, and 5 could not be compared due to lack of beta coefficients in the publications. A total of seven were both consistent in direction and statistically significant. The UGT1A1/UGT1A cluster locus showed robust association, with the T allele of the lead SNP rs887829 associated with higher bilirubin (0.31 ± 0.07, p = 3.38 × 10−6) and biliverdin (0.39 ± 0.07, p = 1.04 × 10−8) levels. The G allele of rs1171617 in SLC16A9 and A allele of rs7422339 in CPS1 were associated with lower carnitine (−0.23 ± 0.07, p = 0.0014) and higher creatine (0.19 ± 0.07, p = 0.0045), respectively (Figure 3, Table 3).
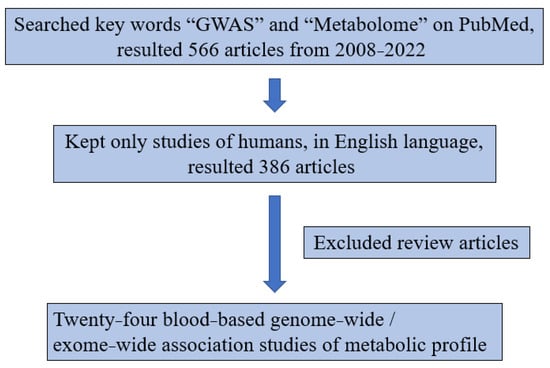
Figure 2.
Diagram of literature review.

Table 2.
Articles selected for the candidate gene–metabolite associations.

Table 3.
Previously reported genetic associations with metabolites.

Figure 3.
Diagram of the candidate gene–metabolite associations.
2.3. GWAS
The 154 inflation factors (lambda) of GWAS had a range of 0.98–1.02 (Supplementary Table S1). The Q–Q plots and Manhattan plots are presented in Supplementary Figures S1 and S2. A total of 22 genetic associations with metabolites passed the genome-wide significance level of p < 5 × 10−8 (Table 4). The rs887829 variant of UGT1A1/UGT1A cluster locus was one of the top associations in the GWAS of biliverdin. Several novel loci were identified, such as the G allele of rs1401798 associated with higher hypoxanthine (0.37 ± 0.06, p = 8.80 × 10−10), and the G allele of rs6874865 associated with lower 3-methyl-2-oxindole (−0.48 ± 0.08, p = 8.66 × 10−9).

Table 4.
Genetic associations with metabolites identified in GWAS (p < 5 × 10−8).
3. Discussion
Low- and middle-income countries, such as South Africa, are facing unique public health challenges with high prevalence of infectious disease, and the growing burden of noncommunicable diseases [29]. However, most genomic and metabolic studies have been conducted on individuals of European ancestry from high-income countries. Due to different genetic ancestries and environmental exposures, the genomic and metabolomic profiles and associations with European ancestry may not apply to people with non-European ancestry from low- and middle-income countries. The present mGWAS in South Africans provides one of the first catalogs to start filling the knowledge gap in diverse global populations. Understanding the genomic and metabolic characteristics from underrepresented populations is critically needed to identify molecular mechanisms driving risks for chronic infections and noncommunicable diseases. The -omics findings will also shed light on novel precision medicine applications for at-risk populations from low- and middle-income countries to reduce health disparities.
In this mGWAS conducted among 490 PWH from Africa, we replicated previously reported genetic associations primarily from European individuals living in high-income countries. The UGT1A1/UGT1A cluster locus was associated with biliverdin and bilirubin, which are both bile pigments formed during the breakdown of hemoglobin in red blood cells. Biliverdin can be converted as unconjugated bilirubin by biliverdin reductase before being further metabolized. The UGT1A1 gene encodes the bilirubin uridine diphosphate-glucuronosyltransferase, which is the enzyme responsible for conjugation, detoxification, and clearance of bilirubin, and it has been found among European, African, and Asian populations; mutations of the gene may cause unconjugated hyperbilirubinemia [30,31,32,33,34]. Further, elevated bilirubin levels were found associated with lower risk in cardiovascular events among PWH [35], and the UGT1A1 gene is particularly of interest in future studies of HIV, since it was involved in the metabolic pathways linking to several HIV medications, such as Bictegravir, Cabotegravir, Dolutegravir, Elvitegravir, and Raltegravir (https://clinicalinfo.hiv.gov/en/guidelines/hiv-clinical-guidelines-adult-and-adolescent-arv/characteristics-integrase-inhibitors, accessed on 15 June 2022). The CPS1 locus encodes a mitochondrial enzyme responsible for carbamoyl phosphate synthesis from ammonia and bicarbonate with the use of adenosine triphosphate. This is the first step of the urea cycle, which can generate arginine, a critical precursor of creatine synthesis [17]. Creatine can be both naturally synthesized or ingested in a usual diet to supply energy to the muscles, and then is converted to creatinine, filtered from the blood by the glomerulus [36]. The association between CPS1 and creatine was previously reported among participants of European descent in the Framingham Heart Study [17], and another study of European participants, which demonstrated that the locus was also associated with creatinine [37]. As another critical compound in energy metabolism, carnitine transports long-chain fatty acids into the mitochondria to be oxidized and produce energy [38,39]. The product of SLC16A9 is a carnitine efflux transporter [8]; its association with carnitine was previously established among several studies in European populations but has not been generalized to other ancestries [8,17,22]. In addition, we found a borderline statistically significant association of serine and the PHGDH gene, which encodes the enzyme involved in the process of serine synthesis [7,8].
To the best of our knowledge, this is the first population study of genome-metabolome cross-talk among individuals of African ancestry living in Africa, which represents a population facing a unique public health burden of both non-communicable diseases and chronic infectious diseases. Although the relatively moderate sample size limited us from identifying more genome-wide significant association with metabolites, our results demonstrated the value of such a mGWAS in this under-represented population. Beyond several gene–metabolite associations that are generalizable across ancestries, several novel discoveries provide candidates for replication and may benefit future research endeavors in African countries. The identified gene–metabolite associations can be used as genetic instrumental variables in the Mendelian Randomization framework to investigative potential causal relationship between metabolites and disease outcomes. There are a few limitations associated with the present study. For example, the plasma samples were not collected during fasting, and information on diet was not surveyed, which may influence the abundance of certainly diet-related metabolites. A causal pathway may not be directly inferred from these observed associations. Future studies are warranted to provide guidance on translation and clinical implementations of identified genes and metabolites among the PWH population. In addition, future studies with improved metabolomic coverage and annotation will help establish a more complete atlas of genome-metabolome relationship in this unique population. With more and larger mGWAS in African populations, improved statistical power will facilitate robust findings of gene–metabolite associations to better understand molecular mechanisms, and to investigate the functional role of less frequent genetic variants on disease risk.
4. Materials and Methods
4.1. Study Participants
Ethics approval from the Biomedical Research Ethics Committee of the University of KwaZulu-Natal, the Emory University, and Mass General Brigham institutional review boards was obtained prior to the start of the study. After signed informed consent, people with HIV (PWH) who were at least 18 years of age and qualified for anti-retroviral therapy, were enrolled into the HIV AIDS Drug Resistance Surveillance Study (ADReSS). This study was based on a sub-cohort from the ADReSS participants recruited from KwaZulu-Natal, South Africa. The recruitment was conducted in 2014–2016 at two participating clinical sites, the RK Khan Hospital and Bethesda Hospital in KwaZulu-Natal, South Africa [40]. A total of 490 participants from the study with both genomics and metabolomics data available were included in the analysis. Full summary statistics of the results presented in the study are available upon request. Individual level dataset underlying this study can be requested and shared in compliance with the informed consent and IRB approval from participating institutes.
4.2. Genotyping and Imputation
Genotyping was performed on DNA extracted from blood samples of 998 participants, using the Illumina Global Screening Array. Genotype imputation for the 998 samples was performed based on the TOPMed reference panel in the genome build GRCh38 [41], using 590,511 genotyped SNPs. A total of 8,568,848 genetic variants with imputation quality R2 > 0.5, genotype call rate >95%, Hardy–Weinberg equilibrium p > 10−6, and minor allele frequency >0.05 entered analysis. Total of 490 participants with African ancestry also had metabolomics data from plasma samples after quality control procedures.
4.3. Metabolomic Profiling
The high-resolution metabolic profiling was performed using liquid chromatography with high-resolution mass spectrometry (Thermo Scientific Fusion, Waltham, MA, US) based on an established protocol [42,43,44]. Plasma samples collected from participants prior to antiretroviral therapy initiation were stored at −80 Celsius before thawing for analysis by liquid-chromatography mass spectrometry (Thermo Scientific Fusion, Waltham, MA, USA). Thawed plasma was treated with acetonitrile containing an internal isotopic standard mix [44,45], then centrifuged for 10 min at 4 Celsius to remove protein. The remaining supernatant was then maintained in a Dionex Ultimate 3000 autosampler at 4 Celsius until analysis. Two liquid chromatography strategies were used, including Supelco Ascentis Express HILIC 100 × 2.1 mm (53939-U) columns and Higgins C18100 × 2.1 mm (TS-1021-C185) columns with positive and negative ionization mode, respectively. All samples were analyzed in triplicate, with the standard reference samples of National Institute of Standards and Technology 1950 analyzed at the beginning and end of the analysis, and pooled plasma reference samples inserted at the beginning and end of each batch of 20 study samples [46]. Metabolic feature extraction was performed using apLCMS and xMSanalyzer [47,48]. Batch effect was corrected using Combat [49]. A metabolic feature was defined by the mass-to-charge ratio (m/z) ranging from 85 to 1275, and retention time up to 5 min. Pearson correlation coefficient ≥0.7 within triplicates of the metabolic feature intensities were retained, and the triplicates of intensities were median summarized based on the non-zero readings. Then, the zero readings of each metabolic feature were imputed as random numbers below the minimum intensity across all study samples. Annotations of metabolic features were matched to an in-house library of confirmed metabolites, which were of Level 1 identification confidence per established criteria [50,51]. The matching of annotations allowed m/z differences of 10 parts-per-million and retention time differences of 30 s. After removing metabolic features with multiple matches of annotations, a total of 154 metabolites were successfully and uniquely matched, as shown in Supplementary Table S1.
4.4. Literature Review
The diagram of the literature review is shown in Figure 2. We searched PubMed for the key words “GWAS” and “Metabolome” and found 566 articles from 2008–2022. Among those, a total of 386 articles were human studies in the English language. We reviewed these articles and found 24 blood-based GWAS of metabolic profile in Table 2. We then identified statistically significant gene–metabolite associations to pursue replication in this present study of South Africans.
4.5. Statistical Analysis
Genetic background of the 490 study participants were analyzed coupled with the 1000 Genome Project [12] reference panel, which included African (AFR), American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS). Principal components (PCs) were computed, and the ancestry map was generated based on the top two PCs that explained the most variance. For the analysis of gene–metabolite associations, the intensities of metabolites were rank-based inverse normal transformed to achieve normality. Linear regression and additive genetic models were used to test the associations between the candidate metabolites and SNPs, controlling for age, gender, study sites, and the top ten PCs, which were derived from PC analysis of the study participants. We also performed GWAS for each of the 154 metabolites identified to explore novel genetic loci, using the same statistical model. For candidate gene–metabolite association analysis, we considered p < 0.05 as statistically significant, and the genome-wide significance threshold p < 5 × 10−8 was used for GWAS. All analyses were conducted in R version 4.0.2 (Vienna, Austria. URL Available online: https://www.R-project.org/ (accessed on 15 June 2022) and the PLINK software [52].
5. Conclusions
Among a cohort of PWH living in South Africa prior to ART initiation, we identified genetic associations with metabolites across ancestries as well as unique to ancestry group, which supports the importance of genomic association studies in diverse ancestries. Findings in gene–metabolite associations from diverse ancestries and geographic regions can provide insights to the disease risk at the molecular level and reduce health disparities for underrepresented populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo12070624/s1, Supplementary Table S1: The 154 metabolites analyzed in GWAS; Supplementary Figure S1: Q-Q plots for the GWAS of 154 metabolites; Supplementary Figure S2: Manhattan plots for the GWAS of 154 metabolites.
Author Contributions
Conceptualization, C.L. and Y.V.S.; methodology, C.L. and Y.V.S.; software, C.L.; validation, Z.W.; formal analysis, C.L.; investigation, C.L. and Y.V.S.; resources, Y.V.S.; data curation, C.L., Z.W., and Q.H.; writing—original draft preparation, C.L.; writing—review and editing, Z.W., Q.H., Y.C., J.C., J.B., J.A.E., C.E.O., M.R.D., H.S., S.P., P.M., D.R.K., M.Y.S.M., D.P.J., V.C.M. and Y.V.S.; visualization, Q.H.; supervision, Y.V.S.; project administration, S.P., Y.C., V.C.M. and Y.V.S.; funding acquisition, V.C.M. and Y.V.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly funded by the National Institute of Health grant number R01DK125187, and Emory CFAR grant number P30AI050409.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Emory University (protocol code IRB00063791, last approved on 10 December 2021).” for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
V.C.M. has received investigator-initiated research grants (to the institution) and consultation fees (both unrelated to the current work) from Eli Lilly, Bayer, Gilead Sciences, and ViiV. The remaining authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Manolio, T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010, 363, 166–176. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Elliott, P. The metabolome-wide association study: A new look at human disease risk factors. J. Proteome Res. 2008, 7, 3637–3638. [Google Scholar] [CrossRef] [PubMed]
- Adamski, J. Genome-wide association studies with metabolomics. Genome Med. 2012, 4, 34. [Google Scholar] [CrossRef]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; de Angelis, M.H.; Kronenberg, F.; Meitinger, T.; Mewes, H.W.; Wichmann, H.E.; Weinberger, K.M.; Adamski, J.; et al. Genetics meets metabolomics: A genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef]
- Draisma, H.H.M.; Pool, R.; Kobl, M.; Jansen, R.; Petersen, A.K.; Vaarhorst, A.A.M.; Yet, I.; Haller, T.; Demirkan, A.; Esko, T.; et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 2015, 6, 7208. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sekula, P.; Wuttke, M.; Wahrheit, J.; Hausknecht, B.; Schultheiss, U.T.; Gronwald, W.; Schlosser, P.; Tucci, S.; Ekici, A.B.; et al. Genome-Wide Association Studies of Metabolites in Patients with CKD Identify Multiple Loci and Illuminate Tubular Transport Mechanisms. J. Am. Soc. Nephrol. 2018, 29, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Fauman, E.B.; Petersen, A.K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef]
- Suhre, K.; Shin, S.Y.; Petersen, A.K.; Mohney, R.P.; Meredith, D.; Wagele, B.; Altmaier, E.; CardioGram; Deloukas, P.; Erdmann, J.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef]
- Yu, B.; Zheng, Y.; Alexander, D.; Morrison, A.C.; Coresh, J.; Boerwinkle, E. Genetic determinants influencing human serum metabolome among African Americans. PLoS Genet. 2014, 10, e1004212. [Google Scholar] [CrossRef]
- Luo, S.; Feofanova, E.V.; Tin, A.; Tung, S.; Rhee, E.P.; Coresh, J.; Arking, D.E.; Surapaneni, A.; Schlosser, P.; Li, Y.; et al. Genome-wide association study of serum metabolites in the African American Study of Kidney Disease and Hypertension. Kidney Int. 2021, 100, 430–439. [Google Scholar] [CrossRef]
- Rhee, E.P.; Surapaneni, A.; Zheng, Z.; Zhou, L.; Dutta, D.; Arking, D.E.; Zhang, J.; Duong, T.; Chatterjee, N.; Luo, S.; et al. Trans-ethnic genome-wide association study of blood metabolites in the Chronic Renal Insufficiency Cohort (CRIC) study. Kidney Int. 2022, 101, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Illig, T.; Gieger, C.; Zhai, G.; Romisch-Margl, W.; Wang-Sattler, R.; Prehn, C.; Altmaier, E.; Kastenmuller, G.; Kato, B.S.; Mewes, H.W.; et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010, 42, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Kettunen, J.; Tukiainen, T.; Sarin, A.P.; Ortega-Alonso, A.; Tikkanen, E.; Lyytikainen, L.P.; Kangas, A.J.; Soininen, P.; Wurtz, P.; Silander, K.; et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012, 44, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Ripatti, S.; Kettunen, J.; Lyytikainen, L.P.; Oksala, N.; Laurila, P.P.; Kangas, A.J.; Soininen, P.; Savolainen, M.J.; Viikari, J.; et al. Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012, 8, e1002907. [Google Scholar] [CrossRef]
- Krumsiek, J.; Suhre, K.; Evans, A.M.; Mitchell, M.W.; Mohney, R.P.; Milburn, M.V.; Wagele, B.; Romisch-Margl, W.; Illig, T.; Adamski, J.; et al. Mining the unknown: A systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012, 8, e1003005. [Google Scholar] [CrossRef]
- Rhee, E.P.; Ho, J.E.; Chen, M.H.; Shen, D.; Cheng, S.; Larson, M.G.; Ghorbani, A.; Shi, X.; Helenius, I.T.; O’Donnell, C.J.; et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013, 18, 130–143. [Google Scholar] [CrossRef]
- Ried, J.S.; Shin, S.Y.; Krumsiek, J.; Illig, T.; Theis, F.J.; Spector, T.D.; Adamski, J.; Wichmann, H.E.; Strauch, K.; Soranzo, N.; et al. Novel genetic associations with serum level metabolites identified by phenotype set enrichment analyses. Hum. Mol. Genet. 2014, 23, 5847–5857. [Google Scholar] [CrossRef][Green Version]
- Demirkan, A.; Henneman, P.; Verhoeven, A.; Dharuri, H.; Amin, N.; van Klinken, J.B.; Karssen, L.C.; de Vries, B.; Meissner, A.; Goraler, S.; et al. Insight in genome-wide association of metabolite quantitative traits by exome sequence analyses. PLoS Genet. 2015, 11, e1004835. [Google Scholar] [CrossRef]
- Rhee, E.P.; Yang, Q.; Yu, B.; Liu, X.; Cheng, S.; Deik, A.; Pierce, K.A.; Bullock, K.; Ho, J.E.; Levy, D.; et al. An exome array study of the plasma metabolome. Nat. Commun. 2016, 7, 12360. [Google Scholar] [CrossRef]
- Kettunen, J.; Demirkan, A.; Wurtz, P.; Draisma, H.H.; Haller, T.; Rawal, R.; Vaarhorst, A.; Kangas, A.J.; Lyytikainen, L.P.; Pirinen, M.; et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 2016, 7, 11122. [Google Scholar] [CrossRef]
- Yet, I.; Menni, C.; Shin, S.Y.; Mangino, M.; Soranzo, N.; Adamski, J.; Suhre, K.; Spector, T.D.; Kastenmuller, G.; Bell, J.T. Genetic Influences on Metabolite Levels: A Comparison across Metabolomic Platforms. PLoS ONE 2016, 11, e0153672. [Google Scholar] [CrossRef] [PubMed]
- Long, T.; Hicks, M.; Yu, H.C.; Biggs, W.H.; Kirkness, E.F.; Menni, C.; Zierer, J.; Small, K.S.; Mangino, M.; Messier, H.; et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017, 49, 568–578. [Google Scholar] [CrossRef]
- Yousri, N.A.; Fakhro, K.A.; Robay, A.; Rodriguez-Flores, J.L.; Mohney, R.P.; Zeriri, H.; Odeh, T.; Kader, S.A.; Aldous, E.K.; Thareja, G.; et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat. Commun. 2018, 9, 333. [Google Scholar] [CrossRef] [PubMed]
- Teslovich, T.M.; Kim, D.S.; Yin, X.; Stancakova, A.; Jackson, A.U.; Wielscher, M.; Naj, A.; Perry, J.R.B.; Huyghe, J.R.; Stringham, H.M.; et al. Identification of seven novel loci associated with amino acid levels using single-variant and gene-based tests in 8545 Finnish men from the METSIM study. Hum. Mol. Genet. 2018, 27, 1664–1674. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, R.; Ramo, J.T.; Ripatti, P.; Koskela, J.T.; Kurki, M.; Karjalainen, J.; Palta, P.; Hassan, S.; Nunez-Fontarnau, J.; Kiiskinen, T.T.J.; et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat. Commun. 2019, 10, 4329. [Google Scholar] [CrossRef]
- Feofanova, E.V.; Chen, H.; Dai, Y.; Jia, P.; Grove, M.L.; Morrison, A.C.; Qi, Q.; Daviglus, M.; Cai, J.; North, K.E.; et al. A Genome-wide Association Study Discovers 46 Loci of the Human Metabolome in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2020, 107, 849–863. [Google Scholar] [CrossRef]
- Harshfield, E.L.; Fauman, E.B.; Stacey, D.; Paul, D.S.; Ziemek, D.; Ong, R.M.Y.; Danesh, J.; Butterworth, A.S.; Rasheed, A.; Sattar, T.; et al. Genome-wide analysis of blood lipid metabolites in over 5000 South Asians reveals biological insights at cardiometabolic disease loci. BMC Med. 2021, 19, 232. [Google Scholar] [CrossRef]
- Agan, B.K.; Marconi, V.C. Noncommunicable Diseases: Yet Another Challenge for Human Immunodeficiency Virus Treatment and Care in Sub-Saharan Africa. Clin. Infect. Dis. 2020, 71, 1874–1876. [Google Scholar] [CrossRef]
- Shi, X.; Aronson, S.; Khan, A.S.; Bosma, P.J. A novel UGT1A1 gene mutation causing severe unconjugated hyperbilirubinemia: A case report. BMC Pediatr. 2019, 19, 173. [Google Scholar] [CrossRef]
- Chen, G.; Adeyemo, A.; Zhou, J.; Doumatey, A.P.; Bentley, A.R.; Ekoru, K.; Shriner, D.; Rotimi, C.N. A UGT1A1 variant is associated with serum total bilirubin levels, which are causal for hypertension in African-ancestry individuals. NPJ Genom. Med. 2021, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ramos, E.; Adeyemo, A.; Shriner, D.; Zhou, J.; Doumatey, A.P.; Huang, H.; Erdos, M.R.; Gerry, N.P.; Herbert, A.; et al. UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur. J. Hum. Genet. 2012, 20, 463–468. [Google Scholar] [CrossRef]
- Yang, H.; Lin, F.; Chen, Z.K.; Zhang, L.; Xu, J.X.; Wu, Y.H.; Gu, J.Y.; Ma, Y.B.; Li, J.D.; Yang, L.Y. UGT1A1 mutation association with increased bilirubin levels and severity of unconjugated hyperbilirubinemia in ABO incompatible newborns of China. BMC Pediatr. 2021, 21, 259. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.P.; O’Donnell, C.J.; Schwaiger, J.P.; Cupples, L.A.; Lingenhel, A.; Hunt, S.C.; Yang, S.; Kronenberg, F. Association between the UGT1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation 2006, 114, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Marconi, V.C.; Duncan, M.S.; So-Armah, K.; Re, V.L., 3rd; Lim, J.K.; Butt, A.A.; Goetz, M.B.; Rodriguez-Barradas, M.C.; Alcorn, C.W.; Lennox, J.; et al. Bilirubin Is Inversely Associated With Cardiovascular Disease Among HIV-Positive and HIV-Negative Individuals in VACS (Veterans Aging Cohort Study). J. Am. Heart Assoc. 2018, 7, e007792. [Google Scholar] [CrossRef]
- Wyss, M.; Kaddurah-Daouk, R. Creatine and creatinine metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef]
- Kottgen, A.; Pattaro, C.; Boger, C.A.; Fuchsberger, C.; Olden, M.; Glazer, N.L.; Parsa, A.; Gao, X.; Yang, Q.; Smith, A.V.; et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010, 42, 376–384. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Sawicka, A.K.; Renzi, G.; Olek, R.A. The bright and the dark sides of L-carnitine supplementation: A systematic review. J. Int. Soc. Sports Nutr. 2020, 17, 49. [Google Scholar] [CrossRef]
- Brijkumar, J.; Johnson, B.A.; Zhao, Y.; Edwards, J.; Moodley, P.; Pathan, K.; Pillay, S.; Castro, K.G.; Sunpath, H.; Kuritzkes, D.R.; et al. A packaged intervention to improve viral load monitoring within a deeply rural health district of South Africa. BMC Infect. Dis. 2020, 20, 836. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Yu, T.; Strobel, F.H.; Jones, D.P. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst 2010, 135, 2864–2870. [Google Scholar] [CrossRef]
- Go, Y.M.; Walker, D.I.; Liang, Y.; Uppal, K.; Soltow, Q.A.; Tran, V.; Strobel, F.; Quyyumi, A.A.; Ziegler, T.R.; Pennell, K.D.; et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol. Sci. 2015, 148, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Soltow, Q.A.; Strobel, F.H.; Mansfield, K.G.; Wachtman, L.; Park, Y.; Jones, D.P. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013, 9, S132–S143. [Google Scholar] [CrossRef] [PubMed]
- Frediani, J.K.; Jones, D.P.; Tukvadze, N.; Uppal, K.; Sanikidze, E.; Kipiani, M.; Tran, V.T.; Hebbar, G.; Walker, D.I.; Kempker, R.R.; et al. Plasma metabolomics in human pulmonary tuberculosis disease: A pilot study. PLoS ONE 2014, 9, e108854. [Google Scholar] [CrossRef]
- Simon-Manso, Y.; Lowenthal, M.S.; Kilpatrick, L.E.; Sampson, M.L.; Telu, K.H.; Rudnick, P.A.; Mallard, W.G.; Bearden, D.W.; Schock, T.B.; Tchekhovskoi, D.V.; et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem. 2013, 85, 11725–11731. [Google Scholar] [CrossRef]
- Yu, T.; Park, Y.; Johnson, J.M.; Jones, D.P. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009, 25, 1930–1936. [Google Scholar] [CrossRef]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Liu, K.H.; Nellis, M.; Uppal, K.; Ma, C.; Tran, V.; Liang, Y.; Walker, D.I.; Jones, D.P. Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem. 2020, 92, 8836–8844. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).