Development of a Novel Targeted Metabolomic LC-QqQ-MS Method in Allergic Inflammation
Abstract
1. Introduction
2. Results
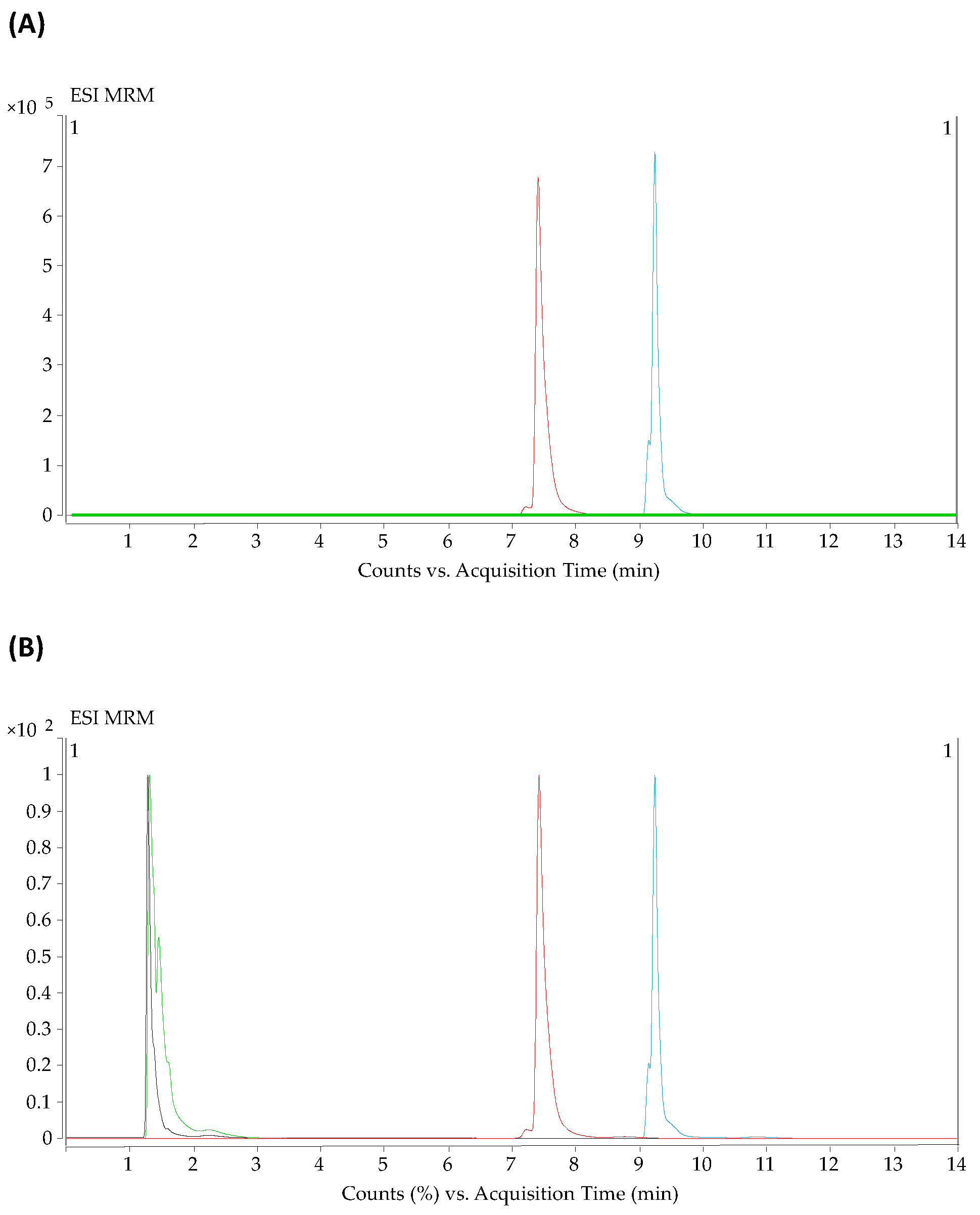
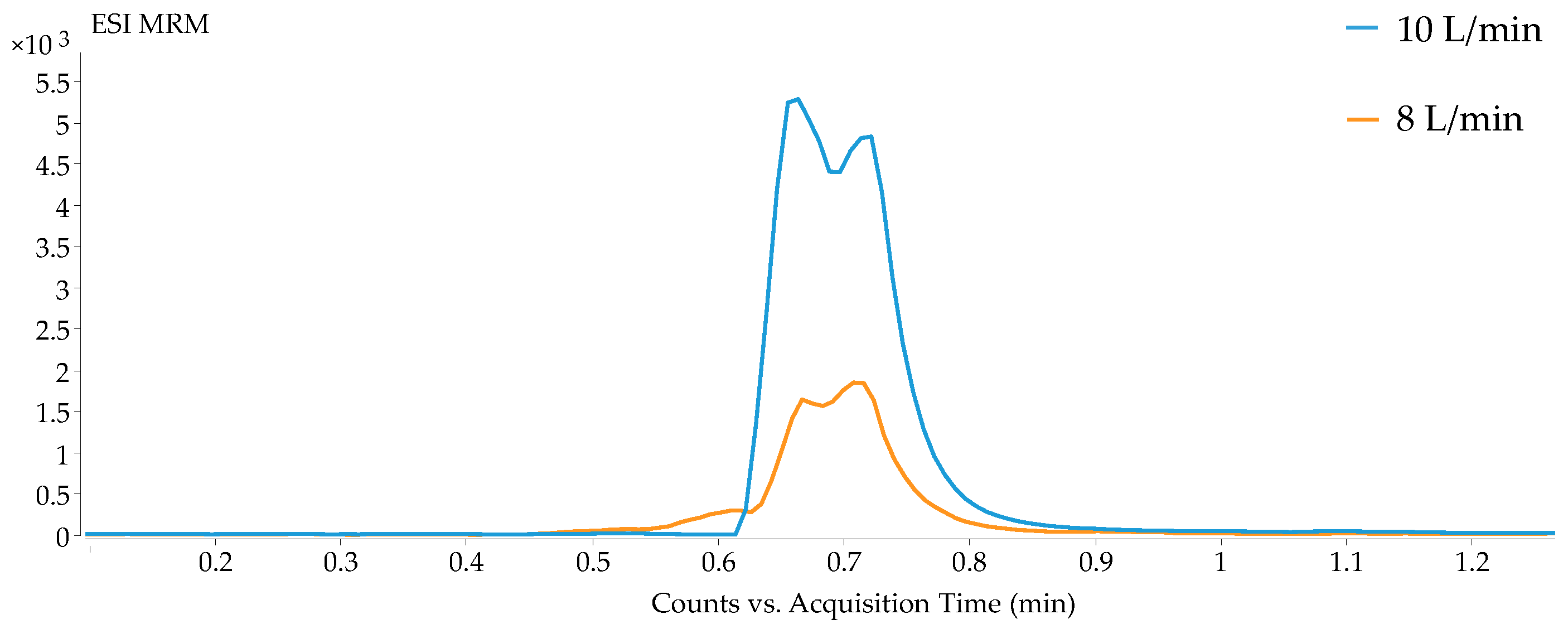
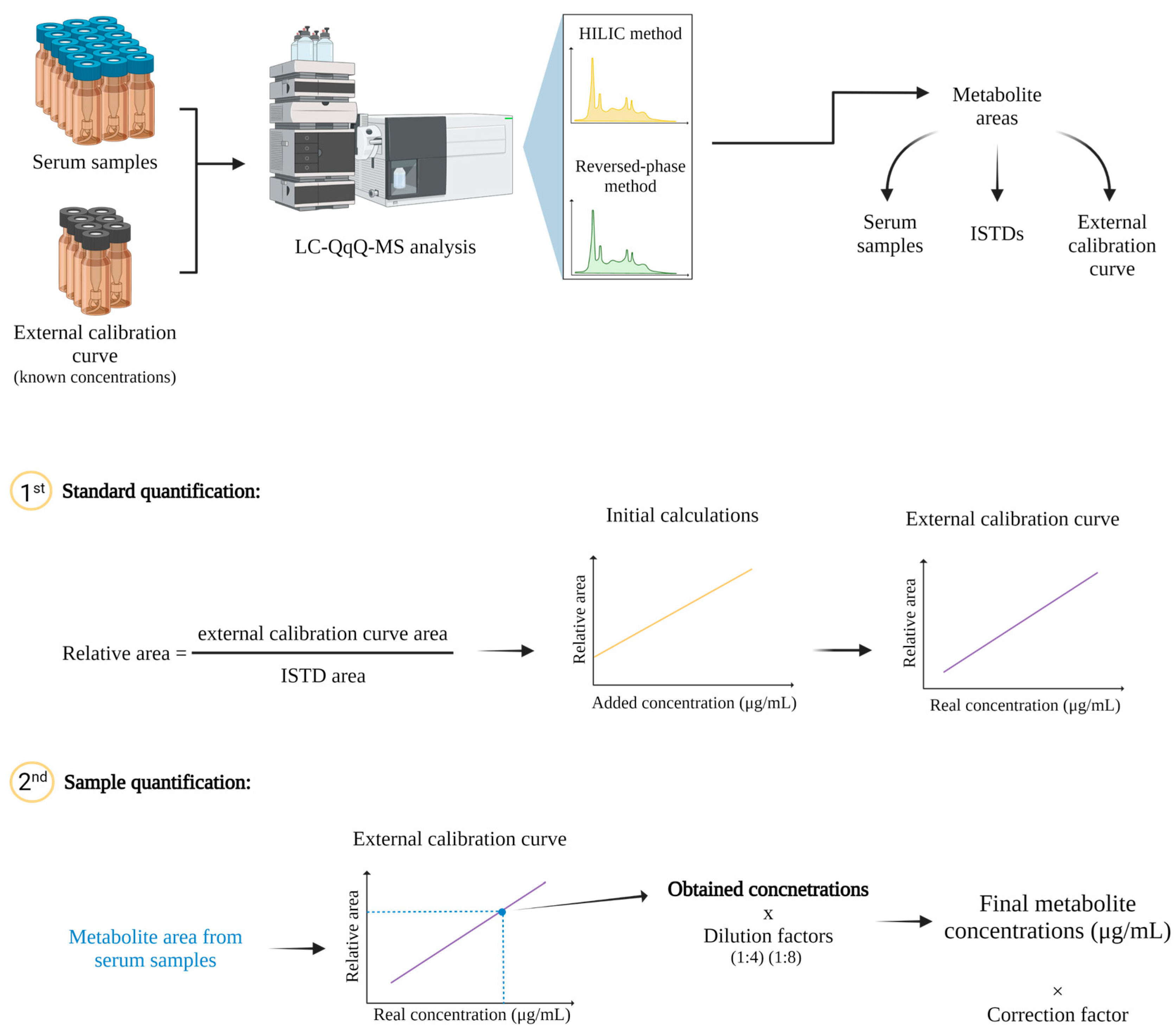
2.1. Optimization of the Parameters for HILIC and Reversed-Phase Methods in LC-QqQ-MS
2.2. Validation Study
2.3. Quantification of Metabolites in a Pooled Serum Sample
2.4. Clinical Models
2.4.1. Metabolite Quantification
2.4.2. Clinical Characteristics of Patients
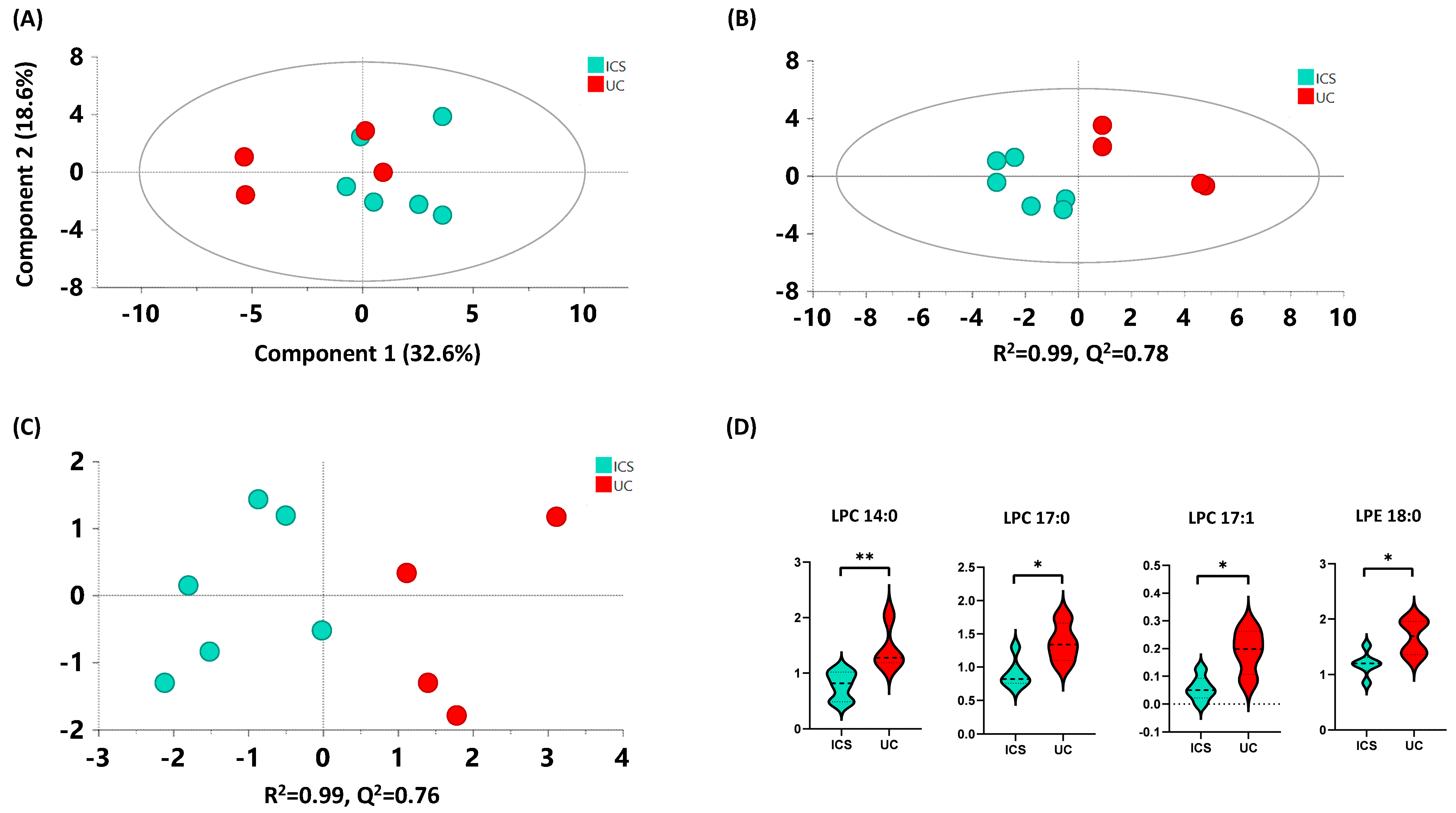
2.4.3. ICS versus UC
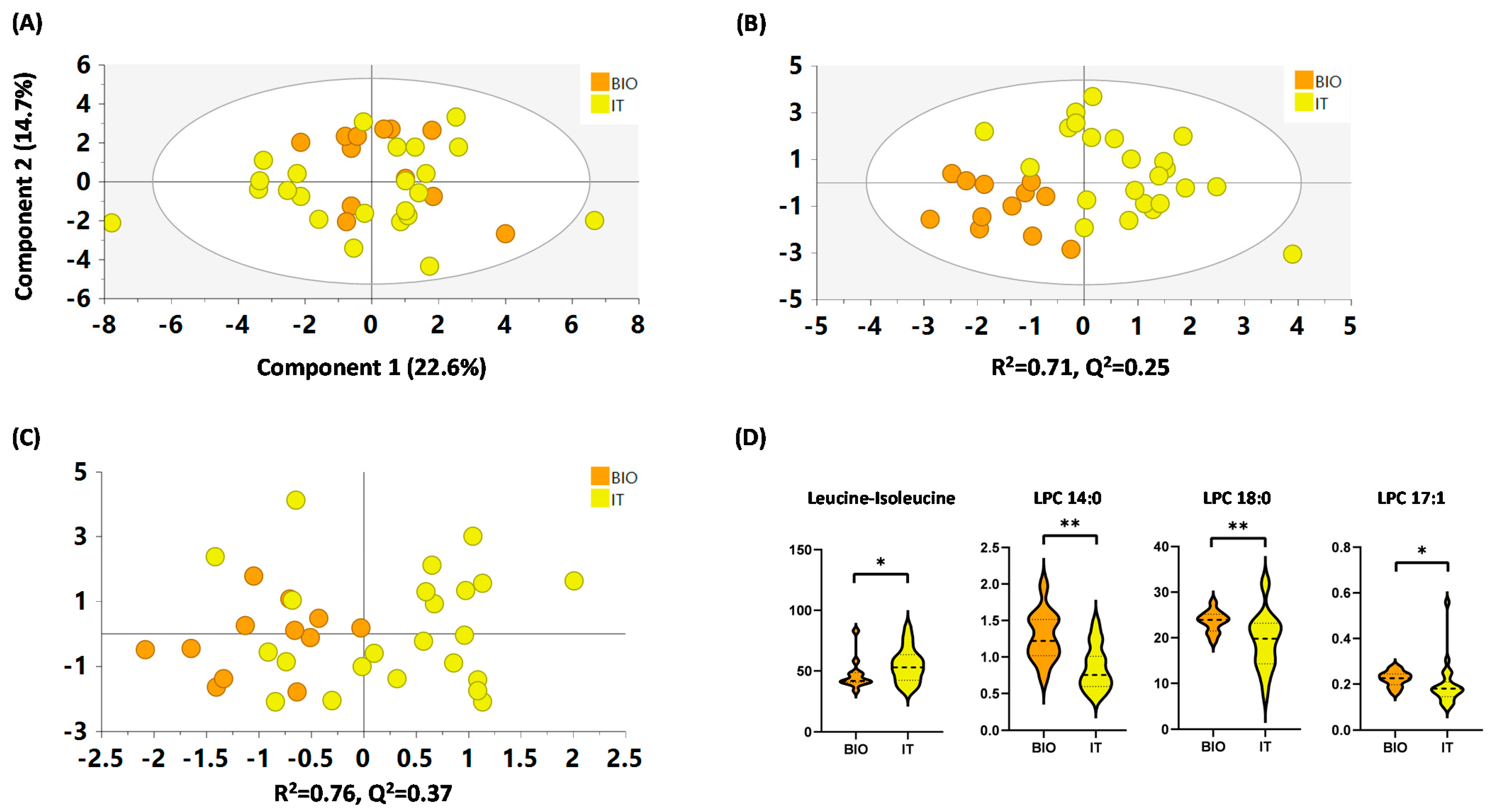
2.4.4. IT versus BIO
3. Discussion
4. Materials and Methods
4.1. Chemical and Reagents
4.2. HPLC-QqQ-MS Analytical Methods
4.3. Sample Preparation
4.4. Metabolite Quantification
4.5. Data Treatment
4.6. Method Development
4.7. Method Validation
4.8. Clinical Models and Sample Collection
4.9. Statistical Analysis of the Clinical Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J. Allergy Clin. Immunol. 2014, 133, 291–307.e5. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.; Johns, C.B. Food Allergy. Immunol. Allergy Clin. N. Am. 2015, 35, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Busse, W.W. Biological treatments for severe asthma: A major advance in asthma care. Allergol. Int. 2019, 68, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454. [Google Scholar] [CrossRef]
- Eguiluz-Gracia, I.; Tay, T.R.; Hew, M.; Escribese, M.M.; Barber, D.; O’Hehir, R.E.; Torres, M.J. Recent developments and highlights in biomarkers in allergic diseases and asthma. Allergy 2018, 73, 2290–2305. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Annesi-Maesano, I.; Bonertz, A.; Branca, F.; Cant, A.; Fras, Z.; Ingenrieth, F.; Namazova-Baranova, L.; Odemyr, M.; Spanevello, A.; et al. Prioritizing research challenges and funding for allergy and asthma and the need for translational research—The European Strategic Forum on Allergic Diseases. Allergy 2019, 74, 2064–2076. [Google Scholar] [CrossRef]
- Barber, D.; Villaseñor, A.; Escribese, M.M. Metabolomics strategies to discover new biomarkers associated to severe allergic phenotypes. Asia Pac. Allergy 2019, 9, e37. [Google Scholar] [CrossRef]
- Dierick, B.J.H.; van der Molen, T.; Flokstra-de Blok, B.M.J.; Muraro, A.; Postma, M.J.; Kocks, J.W.H.; van Boven, J.F.M. Burden and socioeconomics of asthma, allergic rhinitis, atopic dermatitis and food allergy. Expert Rev. Pharmacoecon. Outcomes Res. 2020, 20, 437–453. [Google Scholar] [CrossRef]
- Zhang, W.; Lin, C.; Sampath, V.; Nadeau, K. Impact of allergen immunotherapy in allergic asthma. Immunotherapy 2018, 10, 579–593. [Google Scholar] [CrossRef]
- Khalaf, K.; Paoletti, G.; Puggioni, F.; Racca, F.; De Luca, F.; Giorgis, V.; Canonica, G.W.; Heffler, E. Asthma from immune pathogenesis to precision medicine. Semin. Immunol. 2019, 46, 101294. [Google Scholar] [CrossRef]
- Wenzel, S.E. Asthma phenotypes: The evolution from clinical to molecular approaches. Nat. Med. 2012, 18, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.R.; Hew, M. Comorbid “treatable traits” in difficult asthma: Current evidence and clinical evaluation. Allergy 2018, 73, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Porsbjerg, C.; Menzies-Gow, A. Co-morbidities in severe asthma: Clinical impact and management: Co-morbidities in severe asthma. Respirology 2017, 22, 651–661. [Google Scholar] [CrossRef]
- Gonzalez-Covarrubias, V.; Martínez-Martínez, E.; del Bosque-Plata, L. The Potential of Metabolomics in Biomedical Applications. Metabolites 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Gowda, S.G.B.; Gao, Z.-J.; Chen, Z.; Abe, T.; Hori, S.; Fukiya, S.; Ishizuka, S.; Yokota, A.; Chiba, H.; Hui, S.-P. Untargeted Lipidomic Analysis of Plasma from High-fat Diet-induced Obese Rats Using UHPLC-Linear Trap Quadrupole-Orbitrap MS. Anal. Sci. Int. J. Jpn. Soc. Anal. Chem. 2020, 36, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Obeso, D.; Mera-Berriatua, L.; Rodríguez-Coira, J.; Rosace, D.; Fernández, P.; Martín-Antoniano, I.A.; Santaolalla, M.; Marco Martín, G.; Chivato, T.; Fernández-Rivas, M.; et al. Multi-omics analysis points to altered platelet functions in severe food-associated respiratory allergy. Allergy 2018, 73, 2137–2149. [Google Scholar] [CrossRef]
- Perales-Chorda, C.; Obeso, D.; Twomey, L.; Rojas-Benedicto, A.; Puchades-Carrasco, L.; Roca, M.; Pineda-Lucena, A.; Laguna, J.J.; Barbas, C.; Esteban, V.; et al. Characterization of anaphylaxis reveals different metabolic changes depending on severity and triggers. Clin. Exp. Allergy 2021, 51, 1295–1309. [Google Scholar] [CrossRef]
- Delgado-Dolset, M.I.; Obeso, D.; Rodríguez-Coira, J.; Tarin, C.; Tan, G.; Cumplido, J.A.; Cabrera, A.; Angulo, S.; Barbas, C.; Sokolowska, M.; et al. Understanding uncontrolled severe allergic asthma by integration of omic and clinical data. Allergy 2022, 77, 1772–1785. [Google Scholar] [CrossRef]
- Barker-Tejeda, T.C.; Bazire, R.; Obeso, D.; Mera-Berriatua, L.; Rosace, D.; Vazquez-Cortes, S.; Ramos, T.; Rico, M.d.P.; Chivato, T.; Barbas, C.; et al. Exploring novel systemic biomarker approaches in grass-pollen sublingual immunotherapy using omics. Allergy 2021, 76, 1199–1212. [Google Scholar] [CrossRef]
- Delgado-Dolset, M.I.; Obeso, D.; Sánchez-Solares, J.; Mera-Berriatua, L.; Fernández, P.; Barbas, C.; Fresnillo, M.; Chivato, T.; Barber, D.; Escribese, M.M.; et al. Understanding Systemic and Local Inflammation Induced by Nasal Polyposis: Role of the Allergic Phenotype. Front. Mol. Biosci. 2021, 8, 662792. [Google Scholar] [CrossRef]
- Díaz-Perales, A.; Escribese, M.M.; Garrido-Arandia, M.; Obeso, D.; Izquierdo-Alvarez, E.; Tome-Amat, J.; Barber, D. The Role of Sphingolipids in Allergic Disorders. Front. Allergy 2021, 2, 675557. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Casado, C.; Villaseñor, A.; Rodriguez-Nogales, A.; Bueno, J.; Barber, D.; Escribese, M. Understanding Platelets in Infectious and Allergic Lung Diseases. Int. J. Mol. Sci. 2019, 20, 1730. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Coira, J.; Villaseñor, A.; Izquierdo, E.; Huang, M.; Barker-Tejeda, T.C.; Radzikowska, U.; Sokolowska, M.; Barber, D. The Importance of Metabolism for Immune Homeostasis in Allergic Diseases. Front. Immunol. 2021, 12, 692004. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Gallart-Ayala, H.; Reinke, S.N.; Mathon, C.; Blankley, R.; Chaleckis, R.; Wheelock, C.E. Development of a Liquid Chromatography–High Resolution Mass Spectrometry Metabolomics Method with High Specificity for Metabolite Identification Using All Ion Fragmentation Acquisition. Anal. Chem. 2017, 89, 7933–7942. [Google Scholar] [CrossRef]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery: Mass spectrometry in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef]
- Dolores-Hernández, M.; Morales-Hipólito, E.A.; Villaseñor, A.; López-Arellano, R. Determination of zilpaterol in a residue depletion study using LC-MS/MS in cattle plasma, muscle, liver and kidney. Food Chem. 2022, 382, 132287. [Google Scholar] [CrossRef]
- Buszewski, B.; Noga, S. Hydrophilic interaction liquid chromatography (HILIC)—A powerful separation technique. Anal. Bioanal. Chem. 2012, 402, 231–247. [Google Scholar] [CrossRef]
- Panuwet, P.; Hunter, R.E.; D’Souza, P.E.; Chen, X.; Radford, S.A.; Cohen, J.R.; Marder, M.E.; Kartavenka, K.; Ryan, P.B.; Barr, D.B. Biological Matrix Effects in Quantitative Tandem Mass Spectrometry-Based Analytical Methods: Advancing Biomonitoring. Crit. Rev. Anal. Chem. 2016, 46, 93–105. [Google Scholar] [CrossRef]
- Monnard, I.; Bénet, T.; Jenni, R.; Austin, S.; Silva-Zolezzi, I.; Godin, J.-P. Plasma and urinary inositol isomer profiles measured by UHPLC-MS/MS reveal differences in scyllo-inositol levels between non-pregnant and pregnant women. Anal. Bioanal. Chem. 2020, 412, 7871–7880. [Google Scholar] [CrossRef]
- Barker-Tejeda, T.C.; Villaseñor, A.; Gonzalez-Riano, C.; López-López, Á.; Gradillas, A.; Barbas, C. In vitro generation of oxidized standards for lipidomics. Application to major membrane lipid components. J. Chromatogr. A 2021, 1651, 462254. [Google Scholar] [CrossRef]
- Kim, S.; Moon, H.-G.; Park, G.Y. The roles of autotaxin/lysophosphatidic acid in immune regulation and asthma. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2020, 1865, 158641. [Google Scholar] [CrossRef] [PubMed]
- Yoder, M.; Zhuge, Y.; Yuan, Y.; Holian, O.; Kuo, S.; van Breemen, R.; Thomas, L.L.; Lum, H. Bioactive Lysophosphatidylcholine 16:0 and 18:0 Are Elevated in Lungs of Asthmatic Subjects. Allergy Asthma Immunol. Res. 2014, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Knuplez, E.; Curcic, S.; Theiler, A.; Bärnthaler, T.; Trakaki, A.; Trieb, M.; Holzer, M.; Heinemann, A.; Zimmermann, R.; Sturm, E.M.; et al. Lysophosphatidylcholines inhibit human eosinophil activation and suppress eosinophil migration in vivo. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2020, 1865, 158686. [Google Scholar] [CrossRef]
- Hwang, J.-S.; Im, C.-R.; Im, S.-H. Immune Disorders and Its Correlation with Gut Microbiome. Immune Netw. 2012, 12, 129. [Google Scholar] [CrossRef]
- Jenkins, B.; West, J.; Koulman, A. A Review of Odd-Chain Fatty Acid Metabolism and the Role of Pentadecanoic Acid (C15:0) and Heptadecanoic Acid (C17:0) in Health and Disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef]
- Stråvik, M.; Barman, M.; Hesselmar, B.; Sandin, A.; Wold, A.E.; Sandberg, A.-S. Maternal Intake of Cow’s Milk during Lactation Is Associated with Lower Prevalence of Food Allergy in Offspring. Nutrients 2020, 12, 3680. [Google Scholar] [CrossRef] [PubMed]
- Gai, X.; Guo, C.; Zhang, L.; Zhang, L.; Abulikemu, M.; Wang, J.; Zhou, Q.; Chen, Y.; Sun, Y.; Chang, C. Serum Glycerophospholipid Profile in Acute Exacerbation of Chronic Obstructive Pulmonary Disease. Front. Physiol. 2021, 12, 646010. [Google Scholar] [CrossRef]
- Bansal, P.; Gaur, S.N.; Arora, N. Lysophosphatidylcholine plays critical role in allergic airway disease manifestation. Sci. Rep. 2016, 6, 27430. [Google Scholar] [CrossRef]
- Choi, Y.H.; Jin, G.Y.; Li, L.C.; Yan, G.H. Inhibition of Protein Kinase C Delta Attenuates Allergic Airway Inflammation through Suppression of PI3K/Akt/mTOR/HIF-1 Alpha/VEGF Pathway. PLoS ONE 2013, 8, e81773. [Google Scholar] [CrossRef]
- Available online: https://www.ema.europa.eu/en/ich-q2r2-validation-analytical-procedures#current-effective-version---currently-under-revision,-see-below-section (accessed on 31 May 2022).
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. PCA as a Practical Indicator of OPLS-DA Model Reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef] [PubMed]

| dMRM | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Precursor Ion (m/z) | Product Ion (m/z) | Transition Type | Ret Time (min) | Delta Ret Time (min) | Fragmentor (eV) | CE (eV) | CAV (eV) | ESI Mode |
| HILIC Method | |||||||||
| Adenosine | 268.11 | 135.90 | Quantifier | 3.02 | 5 | 100 | 20 | 7 | Positive |
| Adenosine | 268.11 | 118.90 | Qualifier | 3.02 | 5 | 100 | 56 | 7 | Positive |
| Arginine | 175.12 | 70.10 | Quantifier | 12.41 | 5 | 89 | 29 | 7 | Positive |
| Arginine | 175.12 | 60.10 | Qualifier | 12.41 | 5 | 89 | 13 | 7 | Positive |
| Betaine | 118.09 | 58.00 | Quantifier | 9.90 | 5 | 100 | 36 | 7 | Positive |
| Betaine | 118.09 | 42.00 | Qualifier | 9.90 | 5 | 100 | 72 | 7 | Positive |
| Carnitine | 162.11 | 85.00 | Qualifier | 11.80 | 5 | 80 | 19 | 7 | Positive |
| Carnitine | 162.11 | 60.10 | Quantifier | 11.80 | 5 | 80 | 15 | 7 | Positive |
| Carnitine d3 | 165.13 | 63.00 | Qualifier | 11.79 | 5 | 100 | 20 | 7 | Positive |
| Carnitine d3 | 165.13 | 43.00 | Quantifier | 11.79 | 5 | 100 | 36 | 7 | Positive |
| Cortisol | 363.22 | 121.00 | Qualifier | 0.91 | 5 | 121 | 25 | 7 | Positive |
| Cortisol | 363.22 | 91.00 | Quantifier | 0.91 | 5 | 121 | 77 | 7 | Positive |
| Creatine | 132.08 | 44.20 | Quantifier | 9.81 | 5 | 99 | 25 | 7 | Positive |
| Creatine | 132.08 | 43.10 | Qualifier | 9.81 | 5 | 99 | 57 | 7 | Positive |
| Creatinine | 114.07 | 44.20 | Quantifier | 6.45 | 5 | 104 | 17 | 7 | Positive |
| Creatinine | 114.07 | 43.10 | Qualifier | 6.45 | 5 | 104 | 57 | 7 | Positive |
| Hexanoylcarnitine | 260.19 | 84.90 | Quantifier | 9.99 | 5 | 100 | 24 | 7 | Positive |
| Hexanoylcarnitine | 260.19 | 29.10 | Qualifier | 9.99 | 5 | 100 | 64 | 7 | Positive |
| Hippuric acid | 180.07 | 105.00 | Quantifier | 6.54 | 5 | 72 | 13 | 7 | Positive |
| Hippuric acid | 180.07 | 77.00 | Qualifier | 6.54 | 5 | 72 | 37 | 7 | Positive |
| Hypoxanthine | 137.05 | 118.90 | Qualifier | 3.01 | 5 | 100 | 24 | 7 | Positive |
| Hypoxanthine | 137.05 | 55.00 | Quantifier | 3.01 | 5 | 100 | 36 | 7 | Positive |
| Isoleucine d7 | 139.16 | 92.00 | Quantifier | 8.86 | 5 | 100 | 8 | 7 | Positive |
| Isoleucine d7 | 139.16 | 47.00 | Qualifier | 8.86 | 5 | 100 | 28 | 7 | Positive |
| Leucine/isoleucine | 132.10 | 86.00 | Quantifier | 8.64 | 5 | 100 | 8 | 7 | Positive |
| Leucine/isoleucine | 132.10 | 30.10 | Qualifier | 8.64 | 5 | 100 | 20 | 7 | Positive |
| Phenylalanine | 166.09 | 119.90 | Quantifier | 8.33 | 5 | 100 | 16 | 7 | Positive |
| Phenylalanine | 166.09 | 76.90 | Qualifier | 8.33 | 5 | 100 | 48 | 7 | Positive |
| Phenylalanine d5 | 171.12 | 124.90 | Quantifier | 8.4 | 5 | 131 | 16 | 7 | Positive |
| Proline | 116.07 | 70.00 | Quantifier | 9.47 | 5 | 100 | 20 | 7 | Positive |
| Proline | 116.07 | 28.10 | Qualifier | 9.47 | 5 | 100 | 48 | 7 | Positive |
| Propionylcarnitine | 218.14 | 84.90 | Quantifier | 11.19 | 5 | 100 | 20 | 7 | Positive |
| Propionylcarnitine | 218.14 | 29.10 | Qualifier | 11.19 | 5 | 100 | 52 | 7 | Positive |
| Urea | 61.04 | 44.00 | Quantifier | 1.90 | 5 | 100 | 24 | 7 | Positive |
| Urea | 61.04 | 29.10 | Qualifier | 1.90 | 5 | 100 | 90 | 7 | Positive |
| Valine d8 | 126.14 | 80.10 | Quantifier | 9.64 | 5 | 67 | 13 | 7 | Positive |
| Reversed-phase method | |||||||||
| Arachidonic acid | 303.20 | 303.20 | Quantifier | 9.86 | 6 | 131 | 0 | 7 | Negative |
| Arachidonic acid | 303.20 | 259.20 | Qualifier | 9.86 | 6 | 131 | 13 | 7 | Negative |
| Bilirubin | 585.27 | 299.10 | Quantifier | 10.11 | 6 | 131 | 25 | 7 | Positive |
| Bilirubin | 585.27 | 271.10 | Qualifier | 10.11 | 6 | 131 | 50 | 7 | Positive |
| Lactic acid | 89.00 | 89.00 | Quantifier | 1.00 | 6 | 45 | 0 | 7 | Negative |
| Lactic acid | 89.00 | 43.10 | Qualifier | 1.00 | 6 | 45 | 9 | 7 | Negative |
| Lauric acid | 199.20 | 199.20 | Quantifier | 8.74 | 6 | 45 | 0 | 7 | Negative |
| LPC 14:0 | 468.31 | 183.90 | Quantifier | 7.83 | 6 | 100 | 28 | 7 | Positive |
| LPC 14:0 | 468.31 | 103.90 | Qualifier | 7.83 | 6 | 100 | 56 | 7 | Positive |
| LPC 16:0 | 496.34 | 183.80 | Quantifier | 8.63 | 6 | 100 | 28 | 7 | Positive |
| LPC 16:0 | 496.34 | 103.90 | Qualifier | 8.63 | 6 | 100 | 28 | 7 | Positive |
| LPC 17:0 | 510.36 | 184.00 | Quantifier | 9.06 | 6 | 180 | 29 | 7 | Positive |
| LPC 17:0 | 510.36 | 104.00 | Qualifier | 9.06 | 6 | 180 | 29 | 7 | Positive |
| LPC 17:1 | 508.34 | 183.90 | Quantifier | 8.45 | 6 | 185 | 29 | 7 | Positive |
| LPC 17:1 | 508.34 | 104.00 | Qualifier | 8.45 | 6 | 185 | 29 | 7 | Positive |
| LPC 18:0 | 524.37 | 183.80 | Quantifier | 9.55 | 6 | 100 | 28 | 7 | Positive |
| LPC 18:0 | 524.37 | 103.90 | Qualifier | 9.55 | 6 | 100 | 28 | 7 | Positive |
| LPC 18:1 | 522.36 | 183.80 | Quantifier | 8.85 | 6 | 100 | 28 | 7 | Positive |
| LPC 18:1 | 522.36 | 103.90 | Qualifier | 8.85 | 6 | 100 | 28 | 7 | Positive |
| LPC 18:1 d7 | 529.40 | 183.80 | Quantifier | 9.76 | 6 | 100 | 32 | 7 | Positive |
| LPC 18:1 d7 | 529.40 | 103.90 | Qualifier | 9.76 | 6 | 100 | 28 | 7 | Positive |
| LPC 19:0 | 539.39 | 183.80 | Quantifier | 10.48 | 6 | 100 | 32 | 7 | Positive |
| LPC 19:0 | 538.39 | 183.90 | Qualifier | 10.48 | 6 | 100 | 28 | 7 | Positive |
| LPE 18:0 | 482.33 | 341.10 | Qualifier | 8.90 | 6 | 100 | 20 | 7 | Positive |
| LPE 18:0 | 482.33 | 44.10 | Quantifier | 8.90 | 6 | 100 | 20 | 7 | Positive |
| LPI 16:0 | 573.30 | 555.20 | Qualifier | 9.83 | 6 | 104 | 5 | 7 | Positive |
| LPI 16:0 | 573.30 | 313.20 | Quantifier | 9.83 | 6 | 104 | 25 | 7 | Positive |
| LPI 20:4 | 621.31 | 603.30 | Qualifier | 9.77 | 6 | 89 | 5 | 7 | Positive |
| LPI 20:4 | 621.31 | 361.30 | Quantifier | 9.77 | 6 | 89 | 13 | 7 | Positive |
| Oleamide | 282.28 | 55.10 | Qualifier | 9.84 | 6 | 104 | 45 | 7 | Positive |
| Oleamide | 282.28 | 41.20 | Quantifier | 9.84 | 6 | 104 | 69 | 7 | Positive |
| Oleic acid | 281.25 | 281.25 | Quantifier | 9.50 | 6 | 161 | 0 | 7 | Negative |
| Palmitic acid d31 | 286.23 | 286.23 | Quantifier | 10.13 | 6 | 156 | 0 | 7 | Negative |
| Palmitoleic acid | 253.23 | 253.23 | Quantifier | 9.66 | 6 | 109 | 0 | 7 | Negative |
| SPA-1P | 382.27 | 382.27 | Qualifier | 7.79 | 6 | 130 | 0 | 7 | Positive |
| SPA-1P | 382.27 | 284.10 | Quantifier | 7.79 | 6 | 130 | 12 | 7 | Positive |
| SPA-1P | 382.27 | 60.00 | Qualifier | 7.79 | 6 | 130 | 24 | 7 | Positive |
| Sphinganine-C17 | 288.29 | 288.29 | Quantifier | 6.53 | 6 | 100 | 0 | 7 | Positive |
| Sphinganine-C17 | 288.29 | 60.00 | Qualifier | 6.53 | 6 | 100 | 12 | 7 | Positive |
| Sphinganine-C17 | 288.29 | 30.10 | Qualifier | 6.53 | 6 | 100 | 80 | 7 | Positive |
| Sphingosine | 300.29 | 282.10 | Quantifier | 6.59 | 6 | 100 | 8 | 7 | Positive |
| Sphingosine | 300.29 | 55.40 | Qualifier | 6.59 | 6 | 100 | 40 | 7 | Positive |
| S1P | 380.26 | 264.00 | Quantifier | 7.62 | 6 | 100 | 16 | 7 | Positive |
| S1P | 380.26 | 81.90 | Qualifier | 7.62 | 6 | 100 | 36 | 7 | Positive |
| Sphingosine d7 | 307.34 | 289.10 | Quantifier | 6.72 | 6 | 100 | 12 | 7 | Positive |
| Sphingosine d7 | 307.34 | 30.10 | Qualifier | 6.72 | 6 | 100 | 84 | 7 | Positive |
| Metabolite | ISTD Used | Blank Signal (%) & | Standard Linearity (r) | Sample Linearity (r) | Matrix Effect (p-Value) | External Calibration Curve Range (µg/mL) |
|---|---|---|---|---|---|---|
| HILIC Method | ||||||
| Adenosine | Carnitine d3 | 3.01 | 0.992 | 0.987 | <0.05 | 0.000–0.001 |
| Arginine | Carnitine d3 | 4.51 | 0.993 | 0.995 | <0.05 | 0.325–3.273 |
| Betaine | Carnitine d3 | 37.95 | 0.990 | 0.996 | <0.05 | 0.282–1.028 |
| Carnitine | Carnitine d3 | 0.85 | 1.000 | 0.999 | <0.05 | 0.195–1.643 |
| Cortisol | Carnitine d3 | NA | 0.994 | 0.998 | <0.05 | 0.005–0.109 |
| Creatine | Carnitine d3 | 0.33 | 0.989 | 0.998 | <0.05 | 0.337–1.876 |
| Creatinine | Carnitine d3 | 0.02 | 0.995 | 0.998 | <0.05 | 0.400–1.256 |
| Hexanoylcarnitine | Carnitine d3 | 0.41 | 0.994 | 0.997 | <0.05 | 0.000–0.003 |
| Hippuric acid | Carnitine d3 | 0.02 | 0.992 | 0.998 | <0.05 | 0.010–0.099 |
| Hypoxanthine | Carnitine d3 | 1.00 | 0.997 | 0.999 | <0.05 | 0.027–0.501 |
| Leucine/isoleucine | Isoleucine d7 | NA | 0.999 | 0.999 | <0.05 | 0.570–3.599 |
| Phenylalanine | Isoleucine d7 | 0.04 | 0.998 | 0.999 | <0.05 | 0.163–2.289 |
| Proline | Isoleucine d7 | 0.53 | 0.999 | 0.994 | <0.05 | 1.003–3.422 |
| Propionylcarnitine | Carnitine d3 | 0.16 | 0.992 | 0.996 | <0.05 | 0.001–0.028 |
| Urea | Carnitine d3 | 0.01 | 0.998 | 0.996 | <0.05 | 11.40–36.41 |
| Reversed-phase method | ||||||
| Arachidonic acid | Palmitic acid d13 | NA | 0.995 | 0.996 | <0.05 | 0.034–0.245 |
| Bilirubin | Palmitic acid d13 | NA | 0.994 | 0.977 | <0.05 | 0.551–3.623 |
| Lactic acid | Palmitic acid d13 | 0.58 | 0.991 | 0.987 | <0.05 | 9.898–26.732 |
| Lauric acid | Palmitic acid d13 | 48.27 | 0.998 | 0.992 | <0.05 | 0.062–0.434 |
| LPC 14:0 | LPC 18:1 d7 | 0.65 | 0.999 | 0.999 | <0.05 | 0.0018–0.196 |
| LPC 16:0 | LPC 18:1 d7 | 0.69 | 0.999 | 1.000 | <0.05 | 1.436–7.038 |
| LPC 17:0 | LPC 18:1 d7 | NA | 0.999 | 0.999 | <0.05 | 0.020–0.193 |
| LPC 17:1 | LPC 18:1 d7 | 37.94 | 0.999 | 0.998 | <0.05 | 0.001–0.038 |
| LPC 18:0 | LPC 18:1 d7 | 1.29 | 0.999 | 0.999 | <0.05 | 0.407–4.013 |
| LPC 18:1 | LPC 18:1 d7 | NA | 0.999 | 1.000 | <0.05 | 0.301–3.378 |
| LPC 19:0 | LPC 18:1 d7 | 4.17 | 0.995 | 0.992 | <0.05 | 0.001–0.007 |
| LPE 18:0 | LPC 18:1 d7 | 1.78 | 0.993 | 0.998 | <0.05 | 0.032–0.218 |
| LPI 16:0 | LPC 18:1 d7 | NA | 0.997 | 0.995 | <0.05 | 0.000–1.304 |
| LPI 20:4 | LPC 18:1 d7 | 0.09 | 0.997 | 0.991 | <0.05 | 0.009–0.093 |
| Oleamide | LPC 18:1 d7 | 37.12 | 0.994 | 0.990 | <0.05 | 0.141–0.398 |
| Oleic acid | Palmitic acid d13 | 7.75 | 0.994 | 0.992 | <0.05 | 1.289–4.806 |
| Palmitoleic acid | Palmitic acid d13 | 17.03 | 0.997 | 0.997 | <0.05 | 0.067–0.426 |
| SPA-1P | Sphingosine d7 | 8.99 | 0.989 | 0.987 | <0.05 | 0.000–0.467 |
| Sphinganine-C17 | Sphingosine d7 | 1.35 | 0.999 | 0.999 | <0.05 | 0.015–1.337 |
| Sphingosine | Sphingosine d7 | 0.08 | 0.999 | 0.999 | <0.05 | 0.000–1.319 |
| S1P | Sphingosine d7 | 3.41 | 0.991 | 0.986 | <0.05 | 0.002–0.044 |
| Metabolite | Precision of the Method | Recovery of the Method | ||||
|---|---|---|---|---|---|---|
| Intra-Assay (n = 6) | Inter-Assay (n = 12) | % (n = 12) | ± | RSD (%) | ||
| RSD (%) Day1 | RSD (%) Day2 | RSD (%) | ||||
| HILIC Method | ||||||
| Adenosine | 2.66 | 0.93 | 3.62 | 88.33 | ± | 3.20 |
| Arginine | 4.52 | 6.99 | 17.17 | 81.42 | ± | 8.46 |
| Betaine | 1.14 | 1.54 | 6.89 | 118.02 | ± | 8.14 |
| Carnitine | 0.37 | 0.39 | 0.40 | 95.47 | ± | 0.38 |
| Cortisol | 1.88 | 1.92 | 1.95 | 105.11 | ± | 2.05 |
| Creatine | 4.03 | 1.16 | 8.94 | 99.37 | ± | 8.89 |
| Creatinine | 4.03 | 1.16 | 8.94 | 99.37 | ± | 8.89 |
| Hexanoylcarnitine | 3.33 | 1.34 | 3.08 | 93.80 | ± | 2.88 |
| Hippuric acid | 3.52 | 2.78 | 3.78 | 100.83 | ± | 3.81 |
| Hypoxanthine | 2.24 | 1.13 | 5.23 | 117.57 | ± | 6.15 |
| Leucine/isoleucine | 1.48 | 1.20 | 1.28 | 94.75 | ± | 1.22 |
| Phenylalanine Proline | 3.60 | 2.56 | 4.45 | 93.69 | ± | 4.17 |
| 4.06 | 2.73 | 4.87 | 99.17 | ± | 4.83 | |
| Propionylcarnitine | 0.87 | 0.72 | 1.15 | 96.86 | ± | 1.11 |
| Urea | 1.94 | 1.05 | 3.83 | 101.77 | ± | 3.89 |
| Reversed-phase method | ||||||
| Arachidonic acid | 2.44 | 1.25 | 2.05 | 104.15 | ± | 2.13 |
| Bilirubin | 9.69 | 7.39 | 22.29 | 87.92 | ± | 21.60 |
| Lactic acid | 0.84 | 1.43 | 9.10 | 81.67 | ± | 7.43 |
| Lauric acid | 5.64 | 4.94 | 5.28 | 97.10 | ± | 5.12 |
| LPC 14:0 | 3.05 | 1.94 | 2.44 | 102.63 | ± | 2.50 |
| LPC 16:0 | 1.23 | 0.47 | 1.25 | 102.32 | ± | 1.28 |
| LPC 17:0 | 2.38 | 3.01 | 2.61 | 103.79 | ± | 2.71 |
| LPC 17:1 | 4.28 | 3.07 | 4.16 | 102.26 | ± | 4.25 |
| LPC 18:0 | 1.46 | 1.78 | 1.57 | 99.76 | ± | 1.56 |
| LPC 18:1 | 1.86 | 1.57 | 1.81 | 100.22 | ± | 1.81 |
| LPC 19:0 | 7.81 | 13.67 | 10.64 | 89.15 | ± | 9.48 |
| LPE 18:0 | 2.75 | 3.13 | 3.14 | 100.07 | ± | 3.14 |
| LPI 16:0 | 5.74 | 3.09 | 27.64 | 116.24 | ± | 9.75 |
| LPI 20:4 | 46.84 | 65.94 | 56.42 | 50.33 | ± | 28.19 |
| Oleamide | 4.32 | 2.18 | 6.83 | 33.98 | ± | 2.32 |
| Oleic acid | 2.25 | 1.94 | 2.02 | 101.38 | ± | 2.04 |
| Palmitoleic acid | 2.64 | 1.68 | 2.83 | 96.07 | ± | 2.72 |
| SPA-1P | 9.60 | 13.12 | 19.19 | 132.39 | ± | 4.81 |
| Sphinganine-C17 | 0.89 | 0.96 | 1.66 | 101.79 | ± | 1.69 |
| Sphingosine | 0.65 | 0.90 | 0.79 | 100.43 | ± | 0.79 |
| S1P | 7.08 | 8.33 | 14.31 | 79.31 | ± | 11.35 |
| Metabolite | HMDB Data Base (µM) | Quantified Concentrations (µM) | Included in the Range |
|---|---|---|---|
| HILIC Method | |||
| Adenosine | 0.01–1.71 | 0.04 | ✓ |
| Arginine | 10.00–140.00 | 8.30 | OR |
| Betaine | 20.00–144.00 | 83.07 | ✓ |
| Carnitine | 20.00–60.00 | 85.67 | OR |
| Cortisol | 0.01–0.70 | 204.02 | OR |
| Creatine | 32.22–80.00 | 73.42 | ✓ |
| Creatinine | 8.00–150.00 | 136.95 | ✓ |
| Hexanoylcarnitine | 0.06–0.13 | 0.07 | ✓ |
| Hippuric acid | 1.00–30.00 | 4.07 | ✓ |
| Hypoxanthine | 0.10–12.00 | 12.62 | OR |
| Leucine/isoleucine | 20.00–250.00 | 152.74 | ✓ |
| Phenylalanine | 16.00–166.00 | 79.25 | ✓ |
| Proline | 100.00–300.00 | 458.32 | OR |
| Propionylcarnitine | 0.10–0.50 | 0.75 | OR |
| Urea | 50.00–9000.00 | 113.32 | ✓ |
| Reversed-phase method | |||
| Arachidonic acid | 2.00–600.00 | 5.35 | ✓ |
| Bilirubin | 3.00–20.00 | 0.01 | OR |
| Lactic acid | 740.00–2400.00 | 146.91 | OR |
| Lauric acid | 1.00–12.00 | 3.02 | ✓ |
| LPC 14:0 | 2.00–5.00 | 2.59 | ✓ |
| LPC 16:0 | 40.00–140.00 | 173.24 | OR |
| LPC 17:0 | 0.70–3.00 | 2.65 | ✓ |
| LPC 17:1 | NA | 0.30 | NA |
| LPC 18:0 | NA | 51.10 | NA |
| LPC 18:1 | 10.00–40.00 | 34.84 | OR |
| LPC 19:0 | NA | 0.05 | NA |
| LPE 18:0 | NA | 3.14 | NA |
| LPI 16:0 | NA | 0.00 | NA |
| LPI 20:4 | NA | LA | NA |
| Oleamide | 1500.00–3000.00 | LA | NA |
| Oleic acid | 11.00–500.00 | 208.24 | ✓ |
| Palmitoleic acid | 11.00–300.00 | 12.88 | ✓ |
| SPA-1P | 0.01–0.10 | LA | NA |
| Sphinganine-C17 | NA | LA | NA |
| Sphingosine | 0.05 | 0.01 | OR |
| S1P | 0.04–0.40 | 0.92 | OR |
| Model 1 | Model 2 | |||||
|---|---|---|---|---|---|---|
| p-Value | ICS | UC | p-Value | IT | BIO | |
| N | NA | 6 | 4 | NA | 23 | 12 |
| Gender (%F/%M) | NA | 100.00/0.00 | 100.00/0.00 | 0.236 | 65.22/34.78 | 83.33/16.67 |
| Age (years) | 0.073 | 31.17 ± 12.09 | 52.25 ± 9.95 | <0.001 | 36.57 ± 9.82 | 46.58 ± 7.87 |
| BMI | 0.944 | 28.18 ± 5.86 | 27.87 ± 7.59 | 0.464 | 26.28 ± 4.83 | 27.58 ± 3.56 |
| Smoker (%) | 0.335 | 16.67 | 0.00 | 0.131 | 0.00 | 8.33 |
| Non-smoker (%) | NA | 83.33 | 75.00 | NA | 100.00 | 83.33 |
| Ex-smoker (%) | NA | 0.00 | 25.00 | NA | 0.00 | 8.33 |
| Onset age (years) | 0.790 | 17.00 ± 13.36 | 14.77 ± 11.00 | 0.419 | 15.39 ± 11.32 | 12.27 ± 3.56 |
| Total IgE (U) | 0.800 | 601.50 ± 945.92 | 226.25 ± 176.13 | 0.689 | 464.83 ± 500.87 | 565.73 ± 728.78 |
| AC (%) | 0.400 | 0.00 | 25.00 | 0.002 | 0.00 | 41.67 |
| AH (%) | 0.133 | 100.00 | 50.00 | 0.003 | 91.30 | 41.67 |
| BD (%) | NA | 0.00 | 0.00 | NA | 0.00 | 0.00 |
| Inhaled CS/LABA (%) | NA | 100.00 | 100.00 | <0.001 | 26.09 | 100.00 |
| Inhaled CS (%) | 0.133 | 0.00 | 0.00 | 0.009 | 4.35 | 0.00 |
| Topic CS (%) | 0.119 | 100.00 | 50.00 | <0.001 | 100.00 | 66.67 |
| S (%) | 0.024 | 16.67 | 75.00 | 0.104 | 13.04 | 75.00 |
| SABA (%) | 0.400 | 16.67 | 100.00 | NA | 78.26 | 100.00 |
| T (%) | NA | 0.00 | 25.00 | 0.657 | 0.00 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obeso, D.; Contreras, N.; Dolores-Hernández, M.; Carrillo, T.; Barbas, C.; Escribese, M.M.; Villaseñor, A.; Barber, D. Development of a Novel Targeted Metabolomic LC-QqQ-MS Method in Allergic Inflammation. Metabolites 2022, 12, 592. https://doi.org/10.3390/metabo12070592
Obeso D, Contreras N, Dolores-Hernández M, Carrillo T, Barbas C, Escribese MM, Villaseñor A, Barber D. Development of a Novel Targeted Metabolomic LC-QqQ-MS Method in Allergic Inflammation. Metabolites. 2022; 12(7):592. https://doi.org/10.3390/metabo12070592
Chicago/Turabian StyleObeso, David, Nuria Contreras, Mariana Dolores-Hernández, Teresa Carrillo, Coral Barbas, María M. Escribese, Alma Villaseñor, and Domingo Barber. 2022. "Development of a Novel Targeted Metabolomic LC-QqQ-MS Method in Allergic Inflammation" Metabolites 12, no. 7: 592. https://doi.org/10.3390/metabo12070592
APA StyleObeso, D., Contreras, N., Dolores-Hernández, M., Carrillo, T., Barbas, C., Escribese, M. M., Villaseñor, A., & Barber, D. (2022). Development of a Novel Targeted Metabolomic LC-QqQ-MS Method in Allergic Inflammation. Metabolites, 12(7), 592. https://doi.org/10.3390/metabo12070592








