FADS Polymorphisms Affect the Clinical and Biochemical Phenotypes of Metabolic Syndrome
Abstract
1. Introduction
2. Results
2.1. Clinical and Biochemical Parameters
2.2. Fatty Acid Profiles in Plasma Phospholipids
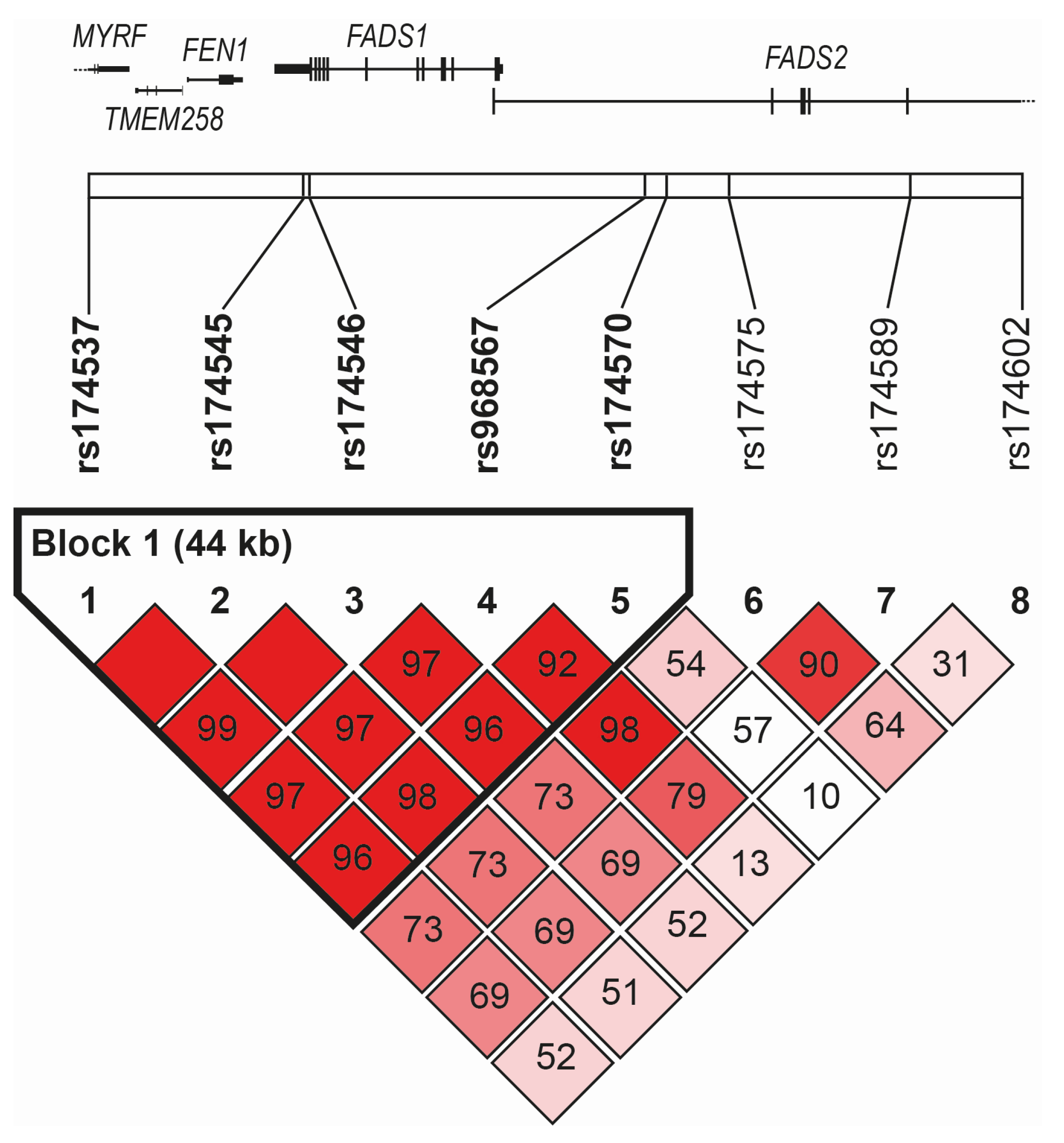
2.3. Genetic Analyses and Statistically Reconstructed Haplotypes
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Laboratory Measurements
4.3. Clustering
4.4. Genetic Analyses
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dizaji, B.F. The investigations of genetic determinants of the metabolic syndrome. Diabetes Metab. Syndr. 2018, 12, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Zafar, U.; Khaliq, S.; Ahmad, H.U.; Manzoor, S.; Lone, K.P. Metabolic syndrome: An update on diagnostic criteria, pathogenesis, and genetic links. Hormones 2018, 17, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Fahed, G.; Aoun, L.; Bou Zerdan, M.; Allam, S.; Bou Zerdan, M.; Bouferraa, Y.; Assi, H.I. Metabolic Syndrome: Updates on Pathophysiology and Management in 2021. Int. J. Mol. Sci. 2022, 23, 786. [Google Scholar] [CrossRef] [PubMed]
- Stančáková, A.; Laakso, M. Genetics of metabolic syndrome. Rev. Endocr. Metab. Disord. 2014, 15, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.E.; Walker, M. Genetics of Insulin Resistance and the Metabolic Syndrome. Curr. Cardiol. Rep. 2016, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- Panda, C.; Varadharaj, S.; Voruganti, V.S. PUFA, genotypes and risk for cardiovascular disease. Prostaglandins Leukot. Essent. Fat. Acids 2022, 176, 102377. [Google Scholar] [CrossRef]
- Das, U.N. Metabolic Syndrome Pathophysiology: The Role of Essential Fatty Acids, 1st ed.; Wiley-Blackwell: Ames, IA, USA, 2010; 268p. [Google Scholar]
- Vávrová, L.; Kodydková, J.; Zeman, M.; Dušejovská, M.; Macášek, J.; Staňková, B.; Tvrzická, E.; Žák, A. Altered Activities of Antioxidant Enzymes in Patients with Metabolic Syndrome. Obes. Facts 2013, 6, 39–47. [Google Scholar] [CrossRef]
- Žák, A.; Burda, M.; Vecka, M.; Zeman, M.; Tvrzická, E.; Staňková, B. Fatty Acid Composition Indicates Two Types of Metabolic Syndrome Independent of Clinical and Laboratory Parameters. Phys. Res. 2014, 63, S375–S385. [Google Scholar] [CrossRef]
- Mayneris-Perxachs, J.; Guerendiain, M.; Castellote, A.I.; Estruch, R.; Covas, M.I.; Fitó, M.; Salas-Salvadó, J.; Martínez-González, M.A.; Aros, F.; Lamuela-Raventós, R.M.; et al. Plasma fatty acid composition, estimated desaturase activities, and their relation with the metabolic syndrome in a population at high risk of cardiovascular disease. Clin. Nutr. 2014, 33, 90–97. [Google Scholar] [CrossRef]
- Zeman, M.; Vecka, M.; Burda, M.; Tvrzická, E.; Staňková, B.; Macášek, J.; Žák, A. Phosphatidylcholine Determines Body Fat Parameters in Subjects with Metabolic Syndrome-Related Traits. Metabol. Syndrome Rel. Disord. 2017, 15, 371–378. [Google Scholar] [CrossRef]
- Muzsik, A.; Jeleń, H.H.; Chmurzynska, A. Metabolic syndrome in postmenopausal women is associated with lower erythrocyte PUFA/MUFA and n-3/n-6 ratio: A case-control study. Prostaglandins Leukot. Essent. Fatty Acids 2020, 159, 102155. [Google Scholar] [CrossRef] [PubMed]
- Lattka, E.; Illig, T.; Koletzko, B.; Heinrich, J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr. Opin. Lipidol. 2010, 21, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.M.; Johnston, H.; Clarke, S.; Roke, K.; Nielsen, D.; Badawi, A.; El-Sohemy, A.; Ma, D.W.; Mutch, D.M. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol. Genet. Metab. 2011, 103, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Lattka, E.; Illig, T.; Heinrich, J.; Koletzko, B. FADS Gene Cluster Polymorphisms: Important Modulators of Fatty Acid Levels and Their Impact on Atopic Diseases. J. Nutrigenet. Nutr. 2009, 2, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Glaser, C.; Heinrich, J.; Koletzko, B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism 2010, 59, 993–999. [Google Scholar] [CrossRef]
- Reynolds, L.M.; Dutta, R.; Seeds, M.C.; Kirsten, N.; Lake, K.N.; Hallmark, B.; Mathias, R.A.; Timothy, D.; Howard, T.D.; Chilton, F.C. FADS genetic and metabolomic analyses identify the Δ5 desaturase (FADS1) step as a critical control point in the formation of biologically important lipids. Sci. Rep. 2020, 10, 15873. [Google Scholar] [CrossRef]
- Lankinen, M.; Uusitupa, M.; Schwab, U. Genes and Dietary Fatty Acids in Regulation of Fatty Acid Composition of Plasma and Erythrocyte Membranes. Nutrients 2018, 10, 1785. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Nara, T.Y. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 2004, 24, 345–376. [Google Scholar] [CrossRef]
- Lee, J.M.; Lee, H.; Kang, S.; Park, W.J. Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances. Nutrients 2016, 8, 23. [Google Scholar] [CrossRef]
- Lankinen, M.A.; Fauland, A.; Shimizu, B.-I.; Agren, J.; Wheelock, C.E.; Laakso, M.; Schwab, U.; Pihlajamaki, J. Inflammatory response to dietary linoleic acid depends on FADS1 genotype. Am. J. Clin. Nutr. 2019, 109, 165–175. [Google Scholar] [CrossRef]
- Bokor, S.; Dumont, J.; Spinneker, A.; Gonzalez-Gross, M.; Nova, E.; Widhalm, W.; Moschonis, G.; Stehle, P.; Amouyel, P.; De Henauw, S.; et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J. Lipid Res. 2010, 51, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Ameur, A.; Enroth, S.; Johansson, A.; Zaboli, G.; Igl, W.; Johansson, A.C.; Rivas, M.A.; Daly, M.J.; Schmitz, G.; Hicks, A.A.; et al. Genetic adaptation of fatty-acid metabolism: A human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am. J. Hum. Genet. 2012, 90, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Malerba, G.; Schaeffer, L.; Xumerle, L.; Klopp, N.; Trabetti, E.; Biscuola, M.; Cavallari, U.; Galavotti, R.; Martinelli, N.; Guarini, P.; et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids 2008, 43, 289–299. [Google Scholar] [CrossRef]
- Schaeffer, L.; Gohlke, H.; Müller, M.; Heid, I.M.; Palmer, L.J.; Kompauer, I.; Demmelmair, H.; Illig, T.; Koletzko, B.; Heinrich, J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 2006, 15, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Czumaj, A.; Sledzinski, T. Biological Role of Unsaturated Fatty Acid Desaturases in Health and Disease. Nutrients 2020, 12, 356. [Google Scholar] [CrossRef]
- Brayner, B.; Kaur, G.; Keske, M.A.; Livingstone, K.M. FADS Polymorphism, Omega-3 Fatty Acids and Diabetes Risk: A Systematic Review. Nutrients 2018, 10, 758. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yin, R.X.; Cao, X.L.; Wu, D.F.; Chen, W.X.; Zhou, Y.J. Association of two polymorphisms in the FADS1/FADS2 gene cluster and the risk of coronary artery disease and ischemic stroke. Int. J. Clin. Exp. Pathol. 2015, 8, 7318–7331. [Google Scholar]
- Kathiresan, S.; Willer, C.J.; Peloso, G.M.; Demissie, S.; Musunuru, K.; Schadt, E.E.; Kaplan, L.; Bennett, D.; Li, Y.; Tanaka, T.; et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009, 41, 56–65. [Google Scholar] [CrossRef]
- Nakayama, K.; Bayasgalan, T.; Tazoe, F.; Yanagisawa, Y.; Gotoh, T.; Yamanaka, K.; Ogawa, A.; Munkhtulga, L.; Chimedregze, U.; Kagawa, Y.; et al. A single nucleotide polymorphism in the FADS1/FADS2 gene is associated with plasma lipid profiles in two genetically similar Asian ethnic groups with distinctive differences in lifestyle. Hum. Genet. 2010, 127, 685–690. [Google Scholar] [CrossRef]
- Standl, M.; Lattka, E.; Stach, B.; Koletzko, S.; Bauer, C.-P.; von Berg, A.; Berdel, D.; Krämer, U.; Schaaf, B.; Röder, S.; et al. FADS1 FADS2 Gene Cluster, PUFA Intake and Blood Lipids in Children: Results from the GINIplus and LISAplus Studies. PLoS ONE 2012, 7, e37780. [Google Scholar] [CrossRef]
- Park, S.; Kim, D.S.; Kang, S. Carrying minor allele of FADS1 and haplotype of FADS1 and FADS2 increased the risk of metabolic syndrome and moderate but not low fat diets lowered the risk in two Korean cohorts. Eur. J. Nutr. 2019, 58, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Marklund, M.; Morris, A.P.; Mahajan, A.; Ingelsson, E.; Lindgren, C.M.; Lind, L.; Risérus, U. Genome-Wide Association Studies of Estimated Fatty Acid Desaturase Activity in Serum and Adipose Tissue in Elderly Individuals: Associations with Insulin Sensitivity. Nutrients 2018, 10, 1791. [Google Scholar] [CrossRef] [PubMed]
- Ching, Y.K.; Chin, Y.S.; Appukutty, M.; Ramanchadran, V.; Yu, C.Y.; Ang, G.Y.; Gan, W.Y.; Chan, Y.M.; Teh, L.K.; Salleh, M.Z. Interaction of Dietary Linoleic Acid and α-Linolenic Acids with rs174547 in FADS1 Gene on Metabolic Syndrome Components among Vegetarians. Nutrients 2019, 11, 1686. [Google Scholar] [CrossRef]
- Shetty, S.S.; Kumari, N.S. Fatty acid desaturase 2 (FADS 2) rs174575 (C/G) polymorphism, circulating lipid levels and susceptibility to type-2 diabetes mellitus. Sci. Rep. 2021, 11, 13151. [Google Scholar] [CrossRef] [PubMed]
- Stančáková, A.; Paananen, J.; Soininen, P.; Kangas, A.J.; Bonnycastle, L.L.; Morken, M.A.; Collins, F.S.; Jackson, A.U.; Boehnke, M.L.; Kuusisto, J.; et al. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finish men. Diabetes 2011, 60, 1608–1616. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kwak, J.H.; Paik, J.K.; Kim, O.Y.; Jang, Y.; Lee, S.H.; Ordovas, J.M.; Lee, J.H. FADS gene polymorphisms in Koreans: Association with ω6 polyunsaturated fatty acids in serum phospholipids, lipid peroxides, and coronary artery disease. Atherosclerosis 2011, 214, 94–100. [Google Scholar] [CrossRef]
- Roke, K.; Ralston, J.C.; Abdelmagid, S.; Nielsen, D.E.; Badawi, A.; El-Sohemy, A.; Ma, D.W.; Mutch, D.M. Variation in the FADS1/2 gene cluster alters plasma n−6 PUFA and is weakly associated with hsCRP levels in healthy young adults. Prostaglandins Leukot. Essent. Fatty Acids 2013, 89, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Vaittinen, M.; Walle, P.; Kuosmanen, E.; Männistö, V.; Käkelä, P.; Ågren, J.; Schwab, U.; Pihlajamäki, J. FADS2 genotype regulates delta-6 desaturase aktivity and inflammation in human adipose tissue. J. Lipid Res. 2016, 57, 56–65. [Google Scholar] [CrossRef]
- Rifkin, S.B.; Shrubsole, M.J.; Cai, Q.; Smalley, W.E.; Ness, R.M.; Swift, L.L.; Milne, G.; Zheng, W.; Murff, H.J. Differences in erythrocyte phospholipid membrane long-chain polyunsaturated fatty acids and the prevalence of fatty acid desaturase genotype among African Americans and European Americans. Prostaglandins Leukot. Essent. Fatty Acids 2021, 164, 102216. [Google Scholar] [CrossRef]
- Meuronen, T.; Lankinen, M.A.; Kärkkäinen, O.; Laakso, M.; Pihlajamäki, J.; Hanhineva, K.; Schwab, U. FADS1 rs174550 genotype and high linoleic acid diet modify plasma PUFA phospholipids in a dietary intervention study. Eur. J. Nutr. 2022, 61, 1109–1120. [Google Scholar] [CrossRef]
- Hong, S.H.; Kwak, J.H.; Paik, J.K.; Chae, J.S.; Lee, J.H. Association of polymorphisms in FADS gene with age-related changes in serum phospholipid polyunsaturated fatty acids and oxidative stress markers in middle-aged nonobese men. Clin. Interv. Aging 2013, 8, 585–596. [Google Scholar] [CrossRef]
- Kröger, J.; Schulze, M.B. Recent insights into the relation of 5 desaturase and 6 desaturase activity to the development of type 2 diabetes. Curr. Opin. Lipidol. 2012, 23, 4–10. [Google Scholar] [CrossRef]
- Mansouri, V.; Javanmard, S.H.; Mahdavi, M.; Tajedini, M.H. Association of Polymorphism in Fatty Acid Desaturase Gene with the Risk of Type 2 Diabetes in Iranian Population. Adv. Biomed. Res. 2018, 7, 98. [Google Scholar] [CrossRef]
- Chen, Y.; Estampador, A.C.; Keller, M.; Poveda, A.; Dalla-Riva, J.; Johansson, I.; Renström, F.; Kurbasic, A.; Franks, P.W.; Varga, T.V. The combined effects of FADS gene variation and dietary fats in obesity-related traits in a population from the far north of Sweden: The GLACIER Study. Int. J. Obes. 2019, 43, 808–820. [Google Scholar] [CrossRef]
- Dumont, J.; Goumidi, L.; Grenier-Boley, B.; Cottel, D.; Marecaux, N.; Montaye, M.; Wagner, A.; Arveiler, D.; Simon, C.; Ferrieres, J.; et al. Dietary linoleic acid interacts with FADS1 genetic variability to modulate HDL-Cholesterol and obesity-related traits. Clin. Nutr. 2018, 37, 1683–1689. [Google Scholar] [CrossRef]
- Khamlaoui, W.; Mehri, S.; Hammami, S.; Hammouda, S.; Chraeif, I.; Elosua, R.; Hammami, M. Association Between Genetic Variants in FADS1-FADS2 and ELOVL2 and Obesity, Lipid Traits, and Fatty Acids in Tunisian Population. Clin. Appl. Thromb. Hemost. 2020, 26, 1–9. [Google Scholar] [CrossRef]
- Maguolo, A.; Zusi, C.; Giontella, A.; Miraglia Del Giudice, E.; Tagetti, A.; Fava, C.; Morandi, A.; Maffeis, C. Influence of genetic variants in FADS2 and ELOVL2 genes on BMI and PUFAs homeostasis in children and adolescents with obesity. Int. J. Obes. 2021, 45, 56–65. [Google Scholar] [CrossRef]
- Metelcová, T.; Vaňková, M.; Zamrazilová, H.; Hovhannisyan, M.; Staňková, B.; Tvrzická, E.; Hill, M.; Hainer, V.; Včelák, J.; Kunešová, M. FADS1 gene polymorphism(s) and fatty acid composition of serum lipids in adolescents. Lipids 2021, 56, 499–508. [Google Scholar] [CrossRef]
- de la Garza Puentes, A.; Montes Goyanes, R.; Chisaguano Tonato, A.M.; Torres-Espínola, F.J.; Arias García, M.; de Almeida, L.; Bonilla Aguirre, M.; Guerendiain, M.; Castellote Bargalló, A.I.; Segura Moreno, M.; et al. Association of maternal weight with FADS and ELOVL genetic variants and fatty acid levels- The PREOBE follow-up. PLoS ONE 2017, 12, e0179135. [Google Scholar] [CrossRef]
- Song, Z.; Cao, H.; Qin, L.; Jiang, Y. A Case-Control Study between Gene Polymorphisms of Polyunsaturated Fatty Acid Metabolic Rate-Limiting Enzymes and Acute Coronary Syndrome in Chinese Han Population. Biomed. Res. Int. 2013, 2013, 928178. [Google Scholar] [CrossRef]
- Li, S.-W.; Wang, J.; Yang, Y.; Liu, Z.-J.; Cheng, L.; Liu, H.-Y.; Ma, P.; Wan Luo, W.; Liu, S.-M. Polymorphisms in FADS1 and FADS2 alterplasma fatty acids and desaturase levels in type 2 diabetic patients with coronary artery disease. J. Transl. Med. 2016, 14, 79. [Google Scholar] [CrossRef]
- Yuan, S.; Bäck, M.; Bruzelius, M.; Mason, A.M.; Burgess, S.; Larsson, S. Plasma Phospholipid Fatty Acids, FADS1 and Risk of 15 Cardiovascular Diseases: A Mendelian Randomisation Study. Nutrients 2019, 11, 3001. [Google Scholar] [CrossRef]
- Sergeant, S.; Hugenschmidt, C.E.; Rudock, M.E.; Ziegler, J.T.; Ivester, P.; Ainsworth, H.C.; Vaidya, D.; Case, L.D.; Langefeld, C.D.; Freedman, B.I.; et al. Differences in Arachidonic Acid Levels and Fatty Acid Desaturase (FADS) Gene Variants in African Americans and European Americans with Diabetes/Metabolic Syndrome. Br. J. Nutr. 2012, 107, 547–555. [Google Scholar] [CrossRef]
- Truong, H.; DiBello, J.R.; Ruiz-Narvaez, E.; Kraft, P.; Campos, H.; Baylin, A. Does genetic variation in the D6-desaturase promoter modify the association between a-linolenic acid and the prevalence of metabolic syndrome? Am. J. Clin. Nutr. 2009, 89, 920–925. [Google Scholar] [CrossRef]
- Martinelli, N.; Girelli, D.; Malerba, G.; Guarini, P.; Illig, T.; Trabetti, E.; Sandri, M.; Friso, S.; Pizzolo, F.; Schaeffer, L.; et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am. J. Clin. Nutr. 2008, 88, 941–949. [Google Scholar] [CrossRef]
- Jump, D.B. Fatty acid regulation of hepatic lipid metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 115–120. [Google Scholar] [CrossRef]
- Tosi, F.; Sartori, F.; Guarini, P.; Olivieri, O.; Martinelli, N. Delta-5 and delta-6 desaturases: Crucial enzymes in polyunsaturated fatty acid-related pathways with pleiotropic influences in health and disease. Adv. Exp. Med. Biol. 2014, 824, 61–81. [Google Scholar] [CrossRef]
- Kremmyda, L.S.; Tvrzická, E.; Staňková, B.; Žák, A. Fatty acids as biocompounds: Their role in human metabolism, health and disease: A review. Part 2: Fatty acid physiological roles and applications in human health and disease. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2011, 155, 195–218. [Google Scholar] [CrossRef]
- Tintle, N.L.; Pottala, J.V.; Lacey, S.; Ramachandran, V.; Westra, J.; Rogers, A.; Clark, J.; Olthoff, B.; Larson, M.; Harris, W.; et al. A genome-wide association study of saturated, mono- and polyunsaturated red blood cell fatty acids in the Framingham heart offspring study. Prostaglandins Leukot. Essent. Fatty Acids 2015, 94, 65–72. [Google Scholar] [CrossRef]
- Tvrzická, E.; Kremmyda, L.S.; Staňková, B.; Žák, A. Fatty acids as biocompounds: Their role in human metabolism, health and disease--a review. Part 1: Classification, dietary sources and biological functions. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2011, 155, 117–130. [Google Scholar] [CrossRef]
- Solinas, G.; Borén, J.; Dulloo, A.G. De novo lipogenesis in metabolic homeostasis: More friend than foe? Mol. Metab. 2015, 4, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, C.; Gaggini, M.; Carli, F.; Gastaldelli, A. The Subtle Balance between Lipolysis and Lipogenesis: A Critical Point in Metabolic Homeostasis. Nutrients 2015, 7, 9453–9474. [Google Scholar] [CrossRef]
- AlJohani, A.M.; Syed, D.N.; Ntambi, J.M. Insights into Stearoyl-CoA Desaturase-1 Regulation of Systemic Metabolism. Trends Endocrinol. Metab. 2017, 28, 831–842. [Google Scholar] [CrossRef]
- Dron, J.S.; Hegele, R.A. Genetics of Hypertriglyceridemia. Front. Endocrinol. 2020, 11, 455. [Google Scholar] [CrossRef]
- Bauer, R.C.; Khetarpal, S.A.; Hand, N.J.; Rader, D.J. Therapeutic targets of triglyceride metabolism as informed by human genetics. Trends Mol. Med. 2016, 22, 328–340. [Google Scholar] [CrossRef]
- Žák, A.; Tvrzická, E.; Vecka, M.; Jáchymová, M.; Duffková, L.; Staňková, B.; Vávrová, L.; Kodydková, J.; Zeman, M. Severity of metabolic syndrome unfavorably influences oxidative stress and fatty acid metabolism in men. Tohoku J. Exp. Med. 2007, 212, 359–371. [Google Scholar] [CrossRef]
- Ahotupa, M.; Ruutu, M.; Mantyla, E. Simple methods of quantifying oxidation products and antioxidant potential of low density lipoproteins. Clin. Biochem. 1996, 29, 139–144. [Google Scholar] [CrossRef]
- Esterbauer, H.; Gebicki, J.; Puhl, H.; Jürgens, G. The role of lipid peroxidation and antioxidants in oxidative modification of LDL. Free Radic. Biol. Med. 1992, 13, 341–390. [Google Scholar] [CrossRef]
- Gromovsky, A.D.; Schugar, R.C.; Brown, A.L.; Helsley, R.N.; Burrows, A.C.; Ferguson, D.; Zhang, R.; Sansbury, B.E.; Lee, R.G.; Morton, R.E.; et al. The Δ-5 Fatty Acid Desaturase FADS1 Impacts Metabolic Disease by Balancing Pro-Inflammatory and Pro-Resolving Lipid Mediators. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 218–231. [Google Scholar] [CrossRef]
- Mazoochian, L.; Sadeghi, H.M.; Pourfarzam, M. The effect of FADS2 gene rs174583 polymorphism on desaturase activities, fatty acid profile, insulin resistance, biochemical indices, and incidence of type 2 diabetes. J. Res. Med. Sci. 2018, 23, 47. [Google Scholar] [CrossRef]
- Zec, M.M.; Krga, I.; Stojković, L.; Živković, M.; Pokimica, B.; Stanković, A.; Glibetic, M. Is There a FADS2-Modulated Link between Long-Chain Polyunsaturated Fatty Acids in Plasma Phospholipids and Polyphenol Intake in Adult Subjects Who Are Overweight? Nutrients 2021, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- Dumont, J.; Huybrechts, I.; Spinneker, A.; Gottrand, F.; Grammatikaki, E.; Bevilacqua, N.; Vyncke, K.; Widhalm, K.; Kafatos, A.; Molnar, D.; et al. FADS1 genetic variability interacts with dietary alpha-linolenic acid intake to affect serum Non-HDL-Cholesterol concentrations in European adolescents. J. Nutr. 2011, 141, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, A.; Sugawara, S.; Okita, M.; Akahane, T.; Fukui, K.; Hashiuchi, M.; Kataoka, C.; Tsukamoto, I. Plasma fatty acid composition, estimated desaturase activities, and intakes of energy and nutrient in Japanese Men with Abdominal Obesity or Metabolic syndrome. J. Nutr. Sci. Vitaminol. 2009, 55, 400–406. [Google Scholar] [CrossRef]
- GWAS Catalog. Available online: https://www.ebi.ac.uk/gwas/variants/rs174537 (accessed on 8 May 2022).
- He, Z.; Zhang, R.; Jiang, F.; Zhang, H.; Zhao, A.; Xu, B.; Jin, L.; Wang, T.; Jia, W.; Jia, W.; et al. FADS1-FADS2 genetic polymorphisms are associated with fatty acid metabolism through changes in DNA methylation and gene expression. Clin. Epigenetics 2018, 10, 113. [Google Scholar] [CrossRef]
- Lottenberg, A.M.; da Silva Alfonso, M.; Lavrador, M.S.; Machado, R.M.; Nakandakare, E.R. The role of dietary fatty acids in the pathology of metabolic syndrome. J. Nutr. Biochem. 2012, 23, 1027–1040. [Google Scholar] [CrossRef] [PubMed]
- Alberti, K.G.; Zimmet, P.; Shaw, J. Metabolic syndrome—A new world-wide definition. A consensus statement from the International Diabetes Federation. Diabet. Med. 2006, 23, 469–480. [Google Scholar] [CrossRef]
- Vecka, M.; Dušejovská, M.; Staňková, B.; Rychlík, I.; Žák, A. A Matched Case-Control Study of Noncholesterol Sterols and Fatty Acids in Chronic Hemodialysis Patients. Metabolites 2021, 11, 774. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Amer. Statist. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Phan, L.; Jin, Y.; Zhang, H.; Qiang, W.; Shekhtman, E.; Shao, D.; Revoe, D.; Villamarin, R.; Ivanchenko, E.; Kimura, M.; et al. ALFA: Allele Frequency Aggregator. National Center for Biotechnology Information. U.S. National Library of Medicine. 2020. Available online: www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa (accessed on 8 May 2022).
- Mathias, R.A.; Pani, V.; Chilton, F.H. Genetic Variants in the FADS Gene: Implications for Dietary Recommendations for Fatty Acid Intake. Curr. Nutr. Rep. 2014, 3, 139–148. [Google Scholar] [CrossRef]
- Cormier, H.; Rudkowska, I.; Paradis, A.M.; Thifault, E.; Garneau, V.; Lemieux, S.; Couture, P.; Vohl, M.C. Association between polymorphisms in the fatty acid desaturase gene cluster and the plasma triacylglycerol response to an n-3 PUFA supplementation. Nutrients 2012, 4, 1026–1041. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, G.; Shamir, R. Maximum Likelihood Resolution of Multi-block Genotypes. In Proceedings of the Eighth Annual International Conference on Research in Computational Molecular Biology—RECOMB 04: San Diego, CA USA, New York, NY, USA, 27–31 March 2004; pp. 2–9. [Google Scholar] [CrossRef]
- Kimmel, G.; Shamir, R. GERBIL: Genotype resolution and block identification using likelihood. Proc. Natl. Acad. Sci. USA 2005, 102, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- The R Core Team: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. 2022. Available online: https://www.R-project.org/ (accessed on 16 April 2022).
| Parameter | MetS | CON |
|---|---|---|
| Number of persons | 166 | 188 |
| Gender (M/F) | 98/68 a | 101/87NS b |
| Age (years) | 55.2 ± 10.6 | 54.5 ± 11.9 |
| Body weight (kg) | 88.0/19.9 *** | 75.7/17.2 |
| BMI (kg·m−2) | 29.3/4.3 *** | 26.0/4.9 |
| Waist circumference (cm) | 103 ± 10 *** | 91 ± 12 |
| Systolic BP (mm Hg) | 140/20 *** | 130/20 |
| Diastolic BP (mm Hg) | 90/14 *** | 80/10 |
| Relative fat mass (%) | 35.4/11.1 *** | 30.0/11.2 |
| Fat mass (kg) | 28.5/9.6 *** | 21.9/10.3 |
| Glucose (mmol/L) | 5.60/1.5 *** | 4.90/0.7 |
| Insulin (mU/L) | 10.70/7.24 *** | 7.70/5.56 |
| HOMA-IR (ratio) | 2.593/2.142 *** | 1.622/1.203 |
| TC (mmol/L) | 6.29/1.68 * | 5.88/1.98 |
| TAG (mmol/L) | 2.69/2.09 *** | 1.40/0.83 |
| HDL-C (mmol/L) | 1.23/050 *** | 1.50/0.54 |
| NEFA (mmol/L) | 0.600/0.530 ** | 0.530/0.360 |
| Apo B (g/L) | 1.33/041 *** | 1.17/0.52 |
| CD-LDL (μmol/L) | 66.7/23.7 *** | 56.4/22.9 |
| Parameter | MetS—Cluster 1 | MetS—Cluster 2 |
|---|---|---|
| Number of persons | 109 | 57 |
| Gender (M/F) | 67/42 a | 31/26 |
| Age (years) | 54.6 ± 11.1 | 56.3 ± 9.5 |
| Body weight (kg) | 90.0/19.0 | 85.8/20.3 |
| BMI (kg·m−2) | 29.7/4.3 | 28.4/4.4 |
| Waist circumference (cm) | 105 ± 11 * | 101 ± 9 |
| Systolic BP (mm Hg) | 140/20 | 140/20 |
| Diastolic BP (mm Hg) | 90/10 | 89/10 |
| Relative fat mass (%) | 33.7/10.3 | 37.5/11.6 |
| Fat mass (kg) | 28.5/8.9 | 28.6/11.2 |
| Glucose (mmol/L) | 5.7/1.8 | 5.3/1.1 |
| Insulin (mU/L) | 11.75/7.17 | 9.40/5.83 |
| HOMA-IR (ratio) | 3.03/2.30 * | 2.07/1.94 |
| TC (mmol/L) | 6.40/1.89 | 6.10/1.46 |
| TAG (mmol/L) | 2.86/3.09 | 2.43/1.60 |
| HDL-C (mmol/L) | 1.21/0.48 | 1.24/0.48 |
| NEFA (mmol/L) | 0.690/0.730 *** | 0.440/0.398 |
| Apo B (g/L) | 1.32/0.44 | 1.36/0.38 |
| CD-LDL (μmol/L) | 70.9/34.9 * | 61.0/19.7 |
| Parameter | CON—Cluster 1 | CON—Cluster 2 |
|---|---|---|
| Number of persons | 71 | 117 |
| Gender (M/F) | 43/28 | 58/59 |
| Age (years) | 53.8 ± 10.7 | 54.9 ± 12.6 |
| Body weight (kg) | 80.6/23.8 ** | 73.7/15.7 |
| BMI (kg·m−2) | 26.7/5.1 * | 25.3/4.5 |
| Waist circumference (cm) | 95.5 ± 12.3 *** | 88.9 ± 10.6 |
| Systolic BP (mm Hg) | 130/20 | 130/20 |
| Diastolic BP (mm Hg) | 80/10 | 80/5 |
| Relative fat mass (%) | 31.2/11.3 | 29.5/11.4 |
| Fat mass (kg) | 24.0/10.9 | 20.8/9.7 |
| Glucose (mmol/L) | 5.00/0.60 | 4.90/0.80 |
| Insulin (mU/L) | 8.59/6.00 | 7.43/5.3.0 |
| HOMA-IR (ratio) | 1.820/1.412 * | 1.568/1.226 |
| TC (mmol/L) | 6.09/2.21 | 5.73/1.94 |
| TAG (mmol/L) | 1.57/1.10 **++ | 1.27/0.7 |
| HDL-C (mmol/L) | 1.49/0.41 | 1.52/0.58 |
| NEFA (mmol/L) | 0.535/0.403 | 0.520/0.300 |
| Apo B (g/L) | 1.22/0.55 | 1.13/0.46 |
| CD-LDL (μmol/L) | 62.3/23.9 | 55.5/24.8 |
| Fatty Acid | MetS (n = 166) | CON (n = 188) |
|---|---|---|
| 14:0 a | 0.266/0.104 | 0.276/0.105 |
| 16:0 | 29.683/1.974 | 29.364/1.755 |
| 16:1n-9 | 0.102/0.038 | 0.111/0.043 |
| 16:1n-7 | 0.593/0.243 ** | 0.522/0.197 |
| 18:0 | 14.43 ± 1.28 *** | 13.84 ± 1.14 |
| 18:1n-9 | 9.850/1.971 | 9.795/2.050 |
| 18:1n-7 | 1.490/0.422 | 1.549/0.372 |
| 18:2n-6 | 21.94 ± 0.16 *** | 23.54 ± 3.00 |
| 18:3n-6 | 0.084/0.052 | 0.076/0.046 |
| 18:3n-3 | 0.191/0.081 | 0.209/0.096 |
| 20:2n-6 | 0.401/0.138 | 0.398/0.141 |
| 20:3n-6 | 3.351/0.803 *** | 3.011/0.764 |
| 20:4n-6 | 10.99 ± 2.05 | 10.91 ± 1.83 |
| 20:5n-3 | 0.943/0.497 | 0.924/0.483 |
| 22:4n-6 | 0.310/0.092 | 0.312/0.078 |
| 22:5n-6 | 0.193/0.076 | 0.194/0.063 |
| 22:5n-3 | 0.892/0.200 | 0.891/0.207 |
| 22:6n-3 | 3.441/1.250 | 3.243/1.157 |
| ∑satur | 44.368/1.640 *** | 43.552/1.955 |
| ∑MFA | 12.197/2.407 | 12.039/2.614 |
| ∑n-6 | 37.251/3.772 *** | 38.641/3.285 |
| ∑n-3 | 5.524/1.885 | 5.308/1.623 |
| D9D 16 (16:1n-7/16:0) | 0.020/0.008 ** | 0.018/0.007 |
| D9D 18 (18:1n-9/18:0) | 0.678/0.171 | 0.709/0.154 |
| D6D n-6 (18:3n-6/18:2n-6) | 0.004/0.003 * | 0.003/0.002 |
| D5D n-6 (20:4n-6/20:3n-6) | 3.117/1.251 ** | 3.605/1.318 |
| Fatty Acid | MetS—Cluster 1 (n = 109) | MetS—Cluster 2 (n = 57) |
|---|---|---|
| 14:0 a | 0.268/0.110 | 0.264/0.105 |
| 16:0 | 29.752/1.903 | 29.091/2.072 |
| 16:1n-9 | 0.105/0.041 | 0.098/0.037 |
| 16:1n-7 | 0.634/0.284 *** | 0.484/0.183 |
| 18:0 | 14.59 ± 1.34 | 14.16 ± 1.12 |
| 18:1n-9 | 10.154/1.945 *** | 8.930/1.453 |
| 18:1n-7 | 1.556/0.467 ** | 1.382/0.291 |
| 18:2n-6 | 20.17 ± 2.07 *** | 25.31 ± 1.88 |
| 18:3n-6 | 0.089/0.053 * | 0.074/0.036 |
| 18:3n-3 | 0.198/0.081 | 0.186/0.078 |
| 20:2n-6 | 0.408/0.149 | 0.381/0.121 |
| 20:3n-6 | 3.363/0.708 * | 3.036/0.893 |
| 20:4n-6 | 11.34 ± 2.03 ** | 10.34 ± 1.95 |
| 20:5n-3 | 1.091/0.492 *** | 0.801/0.284 |
| 22:4n-6 | 0.312/0.103 * | 0.284/0.098 |
| 22:5n-6 | 0.199/0.075 | 0.181/0.072 |
| 22:5n-3 | 0.909/0.192 *** | 0.818/0.164 |
| 22:6n-3 | 3.574/1.251 ** | 3.018/0.973 |
| ∑satur | 44.722/1.698 *** | 43.665/.1,225 |
| ∑mono | 12.812/2.555 *** | 11.034/1.957 |
| ∑n-6 | 36.428/3.163 *** | 39.821/3.098 |
| ∑n-3 | 5.817/1.497 *** | 4.931/1.101 |
| D9D 16 (16:1n-7/16:0) | 0.021/0.010 *** | 0.016/0.005 |
| D9D 18 (18:1n-9/18:0) | 0.736/0.175 ** | 0.646/0.150 |
| D6D n-6 (18:3n-6/18:2n-6) | 0.005/0.003 *** | 0.003/0.002 |
| D5D n-6 (20:4n-6/20:3n-6) | 3.383/1.182 | 3.214/1.595 |
| Polymorphism | Group (Size) | A | a | AA | Aa | aa |
|---|---|---|---|---|---|---|
| FADS1 rs174537a | MetS (150) | G 204 (68.0) | T 96 (32.0) | GG 70 (46.7) | GT 64 (42.7) | TT 16 (10.6) |
| CON (180) | G 239 (66.4) | T 121 (33.6) | GG 74 (41.1) | GT 91 (50.6) | TT 15 (8.3) | |
| CON1 (68) | G 99 (72.8) | T 37 (27.2) | GG 34 (50.0) | GT 31 (45.6) | TT 3 (4.4) | |
| CON2 (112) | G 140 (62.5) | T 84 (37.5) | GG 40 (35.7) | GT 60 (53.6) | TT 12 (10.7) | |
| FADS2 rs174570 | MetS (150) | C 257 (85.7) | T 43 (14.3) | CC 110 (73.3) | CT 37 (24.7) | TT 3 (2.0) |
| CON (180) | C 314 (87.2) | T 46 (12.8) | CC 135 (75.0) | CT 44 (24.4) | TT 1 (0.6) | |
| CON1 (68) | C 124 (91.2) | T 12 (8.8) | CC 56 (82.4) | CT 12 (17.6) | TT 0 (0) | |
| CON2 (112) | C 190 (84.8) | T 34 (15.2) | CC 79 (70.5) | CT 32 (28.6) | TT 1 0.9) | |
| FADS2 rs174575 | MetS (150) | C 234 (78.0) | G 66 (22) | CC 90 (60.0) | CG 54 (36.0) | GG 6 (4.0) |
| CON (180) | C 264 (73.3) | G 96 (26.7) | CC 95 (52.8) | CG 74 (41.1) | GG 11 (6.1) | |
| CON1 (68) | C 105 (77.2) | G 31 (22.8) | CC 41 (60.3) | CG 23 (33.8) | GG 4 (5.9) | |
| CON2 (112) | C 159 (71.0) | G 65 (29.0) | CC 54 (48.2) | CG 51 (45.5) | GG 7 (6.3) | |
| FADS2 rs174602 | MetS (150) | T 247 (82.3) | C 53 (17.7) | TT 102 (68.0) | TC 43 (28.7) | CC 5 (3.3) |
| CON (180) | T 298 (82.8) | C 62 (17.2) | TT 122 (67.8) | TC 54 (30.0) | CC 4 2.2) | |
| CON1 (68) | T 118 (86.8) | C18 (13.2) | TT 51 (75.0) | TC 16 (23.5) | CC 1 (1.5) | |
| CON2 (112) | T 180 (80.4) | C 44 (19.6) | TT 71 (63.4) | TC 38 33.9) | CC 3 (2.7) | |
| FADS2 rs174589 | MetS (150) | C 244 (81.3) | G 56 (18.7) | CC 99 (66.0) | CG 46 (30.7) | GG 5 (3.3) |
| CON (180) | C 295 (81.9) | G 65 (18.1) | CC 117 (65.0) | CG 61 (33.9) | GG 2 (1.1) | |
| CON1 (68) | C 118 (86.8) | G 18 (13.2) | CC 50 (73.5) | CG 18 (26.5) | GG 0 (0) | |
| CON2 (112) | C 177 (79.0) | G 47 (21.0) | CC 67 (59.8) | CG 43 (38.4) | GG 2 (1.8) | |
| FADS2 rs968567 | MetS (150) | C 259 (86.3) | T 41 (13.7) | CC 111 (74.0) | CT 37 (24.7) | TT 2 (1.3) |
| CON (180) | C 300 (83.3) | T 60 (16.7) | CC 123 (68.3) | CT 54 (30.0) | TT 3 (1.7) | |
| CON1 (68) | C 117 (86.0) | T 19 (14.0) | CC 50 (73.5) | CT 17 (25.0) | TT 1 (1.5) | |
| CON2 (112) | C 183 (81.7) | T 41 (18.3) | CC 73 (65.2) | CT 37 (33.0) | TT 2 (1.8) |
| Polymorphism | MetS—Cluster 1 (n= 94) | MetS—Cluster 2 (n = 56) | χ2 Test a | ||
|---|---|---|---|---|---|
| Number | % | Number | % | ||
| FADS1 (rs174537 G/T) e | |||||
| GG | 52 | 55.3 | 18 | 32.1 | χ2 = 14.039 b p = 0.0024 d |
| GT | 38 | 40.4 | 26 | 46.5 | |
| TT | 4 | 4.3 | 12 | 21.4 | |
| G | 142 | 75.5 | 62 | 55.4 | χ2 = 12.218 c p = 0.0024 |
| T | 46 | 24.5 | 50 | 44.6 | |
| FADS2 (rs 174570 C/T) | |||||
| CC | 76 | 80.9 | 34 | 60.7 | χ2 = 10.084 p = 0.014 |
| CT | 18 | 19.1 | 19 | 33.9 | |
| TT | 0 | 0 | 3 | 5.4 | |
| C | 170 | 90.4 | 87 | 77.7 | χ2 = 8.279 p = 0.009 |
| T | 18 | 9.6 | 25 | 22.3 | |
| FADS2 (rs174575 C/G) | |||||
| CC | 62 | 66.0 | 28 | 50.0 | χ2 = 4.863 p = 0.105 |
| CG | 30 | 31.9 | 24 | 42.9 | |
| GG | 2 | 2.1 | 4 | 7.1 | |
| C | 154 | 81.9 | 80 | 71.4 | χ2 = 3.907 p = 0.064 |
| G | 34 | 18.1 | 32 | 28.6 | |
| FADS2 (rs174602 T/C) | |||||
| TT | 70 | 74.5 | 32 | 57.2 | χ2 = 6.988 p = 0.048 |
| TC | 23 | 24.5 | 20 | 35.7 | |
| CC | 1 | 1.0 | 4 | 7.1 | |
| T | 163 | 86.7 | 84 | 75.0 | χ2 = 5.828 p = 0.0284 |
| C | 25 | 13.3 | 28 | 25.0 | |
| FADS2 (rs174589 C/G) | |||||
| CC | 67 | 71.3 | 32 | 57.2 | χ2 = 5.695 p = 0.0713 |
| CG | 26 | 27.7 | 20 | 35.7 | |
| GG | 1 | 1.0 | 4 | 7.1 | |
| C | 160 | 85.1 | 84 | 75.0 | χ2 = 4.080 p = 0.0625 |
| G | 28 | 14.9 | 28 | 25.0 | |
| FADS2 (rs968567 C/T) | |||||
| CC | 73 | 77.7 | 38 | 67.9 | χ2 = 4.365 p = 0.1205 |
| CT | 21 | 22.3 | 16 | 28.6 | |
| TT | 0 | 0 | 2 | 3.5 | |
| C | 167 | 88.8 | 92 | 82.1 | χ2 = 2.123 p = 0.145 |
| T | 21 | 11.2 | 20 | 17.9 | |
| Genes | Polymorphisms 1 | Forward Primers 5′→ 3′ Reverse Primers 5′→ 3′ | Annealing (°C) | Methods RFLP, Direct Sequencing | |
|---|---|---|---|---|---|
| Restrictase | Sequencing | ||||
| FADS1 | rs174537 G > T | caggggagagaggtggagta aggtctgtctggctgtctcc | 59.3 | AvaII | |
| rs174545 G > C | ccatcctcatttgcaaacct cagcagcctaaggcagacat | 60.2 | CviKI-1 | ||
| rs174546 G > A | gccttaacctcactgctcca aggctttatgtccccaaacc | 60.3 | BsaJI | ||
| FADS2 | rs174570 C > T | agaggcaaggagggaagaaa cgggcctacacagcttagag | 60.2 | BsaBI | |
| rs174575 C > G | ctcagaagttggggcttgag actccaagggagcagacaga | 60.0 | BlpI | Direct sequencing | |
| rs174602 T > C | aggaaagggacagtggtgtg ctggtgattgtagggcaggt | 60.0 | BtsCI | ||
| rs174589 C > G | gccaagcctaacatcttcca ctaggcttccttccctgctc | 60.3 | - | Direct sequencing | |
| rs968567 C > T, A, G | aagatcctcctgggccaat gctatggacttttgcctcca | 60.5 | SacI | Direct sequencing | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Žák, A.; Jáchymová, M.; Burda, M.; Staňková, B.; Zeman, M.; Slabý, A.; Vecka, M.; Šeda, O. FADS Polymorphisms Affect the Clinical and Biochemical Phenotypes of Metabolic Syndrome. Metabolites 2022, 12, 568. https://doi.org/10.3390/metabo12060568
Žák A, Jáchymová M, Burda M, Staňková B, Zeman M, Slabý A, Vecka M, Šeda O. FADS Polymorphisms Affect the Clinical and Biochemical Phenotypes of Metabolic Syndrome. Metabolites. 2022; 12(6):568. https://doi.org/10.3390/metabo12060568
Chicago/Turabian StyleŽák, Aleš, Marie Jáchymová, Michal Burda, Barbora Staňková, Miroslav Zeman, Adolf Slabý, Marek Vecka, and Ondřej Šeda. 2022. "FADS Polymorphisms Affect the Clinical and Biochemical Phenotypes of Metabolic Syndrome" Metabolites 12, no. 6: 568. https://doi.org/10.3390/metabo12060568
APA StyleŽák, A., Jáchymová, M., Burda, M., Staňková, B., Zeman, M., Slabý, A., Vecka, M., & Šeda, O. (2022). FADS Polymorphisms Affect the Clinical and Biochemical Phenotypes of Metabolic Syndrome. Metabolites, 12(6), 568. https://doi.org/10.3390/metabo12060568






