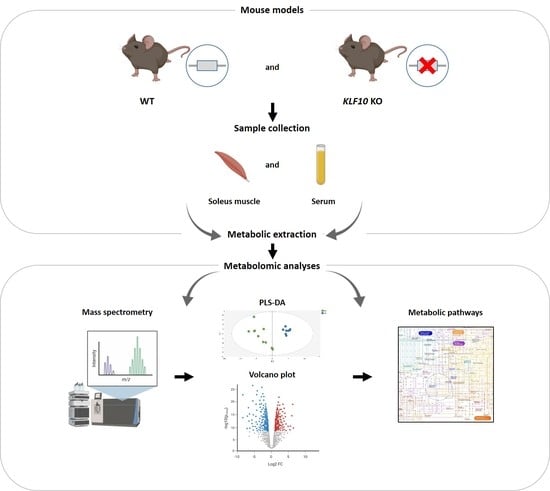
Serum and Soleus Metabolomics Signature of Klf10 Knockout Mice to Identify Potential Biomarkers
Abstract
:1. Introduction
2. Results
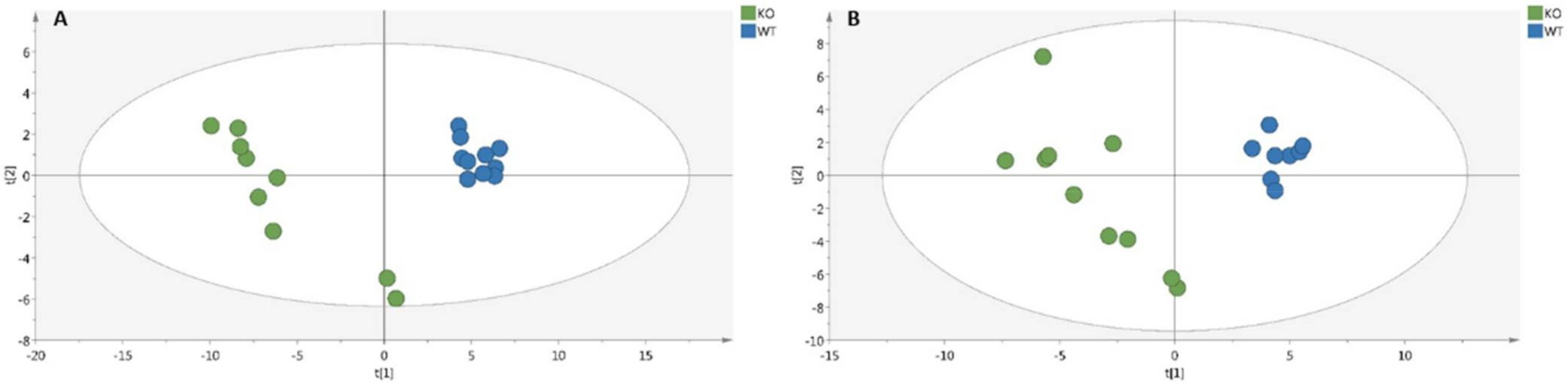
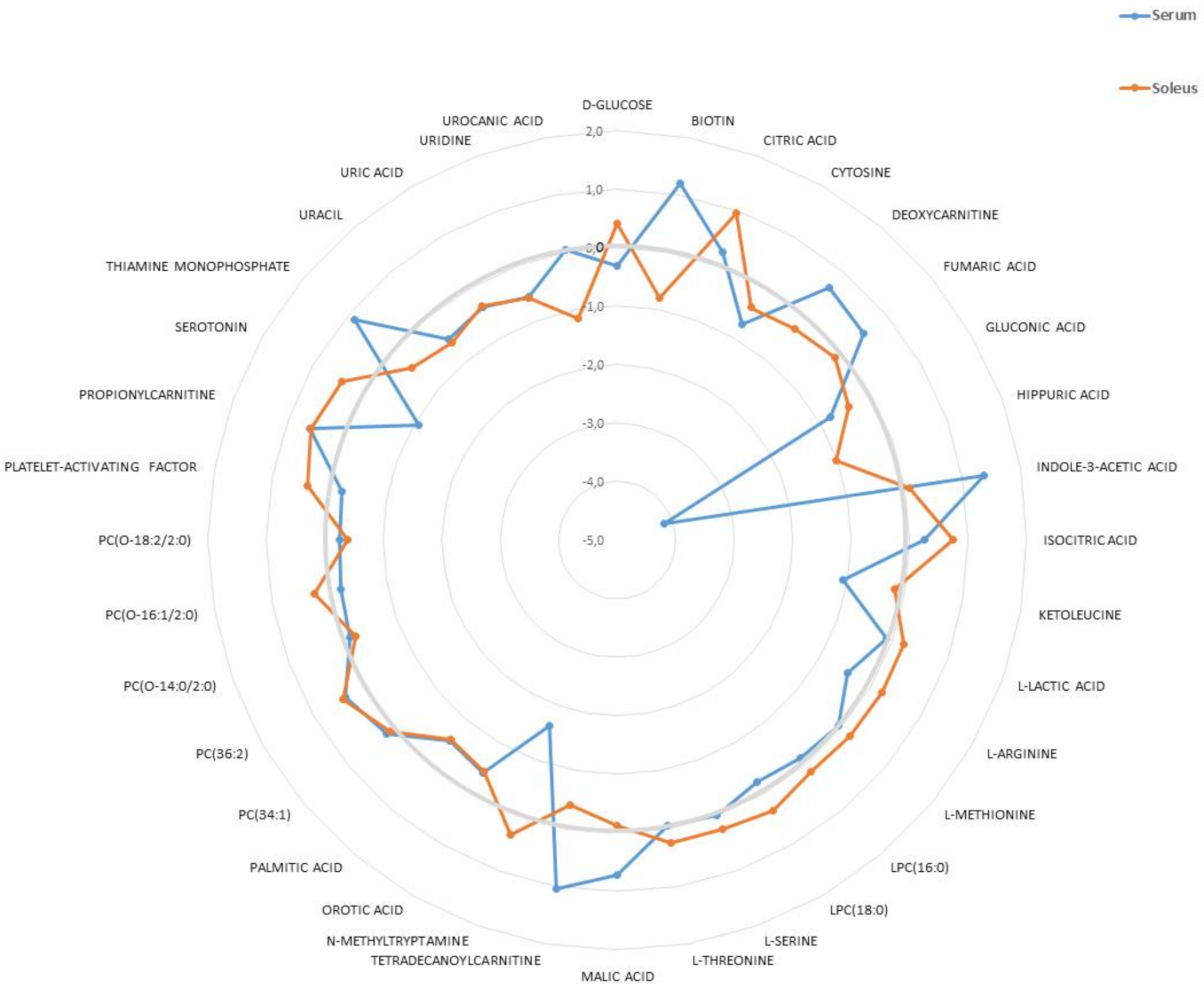
2.1. Metabolomic Analyses of Serum and Soleus
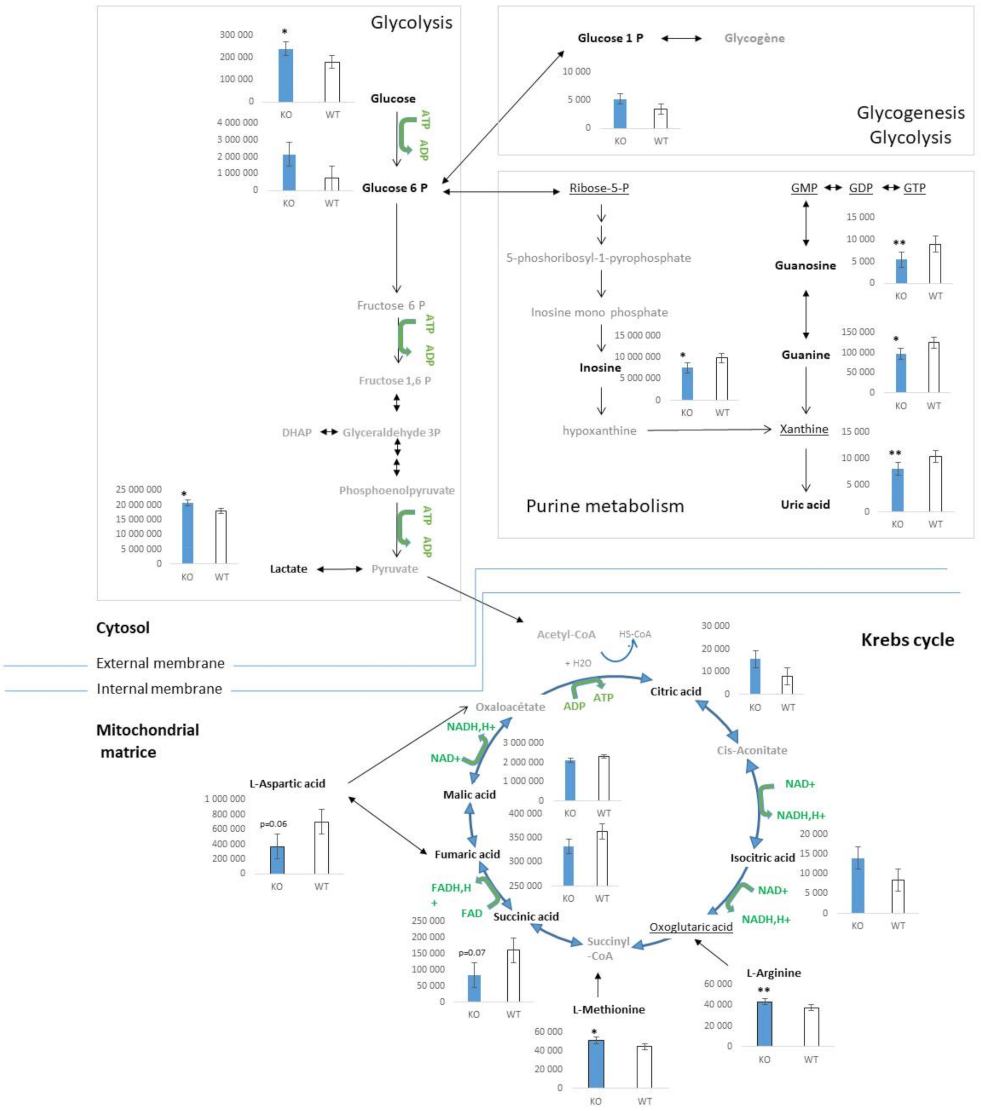
2.2. Characterization of Metabolic Changes in Klf10 KO Mice
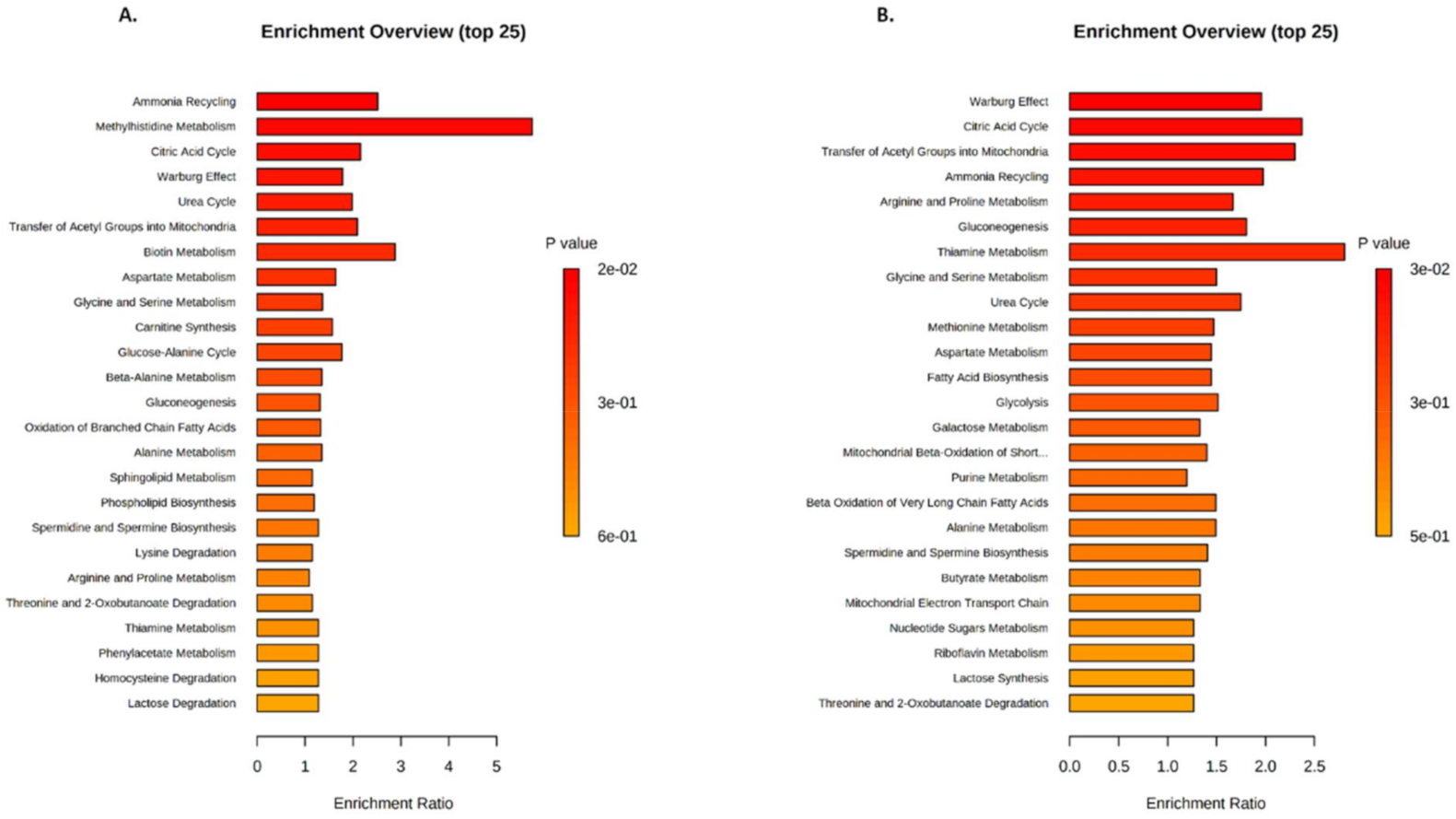
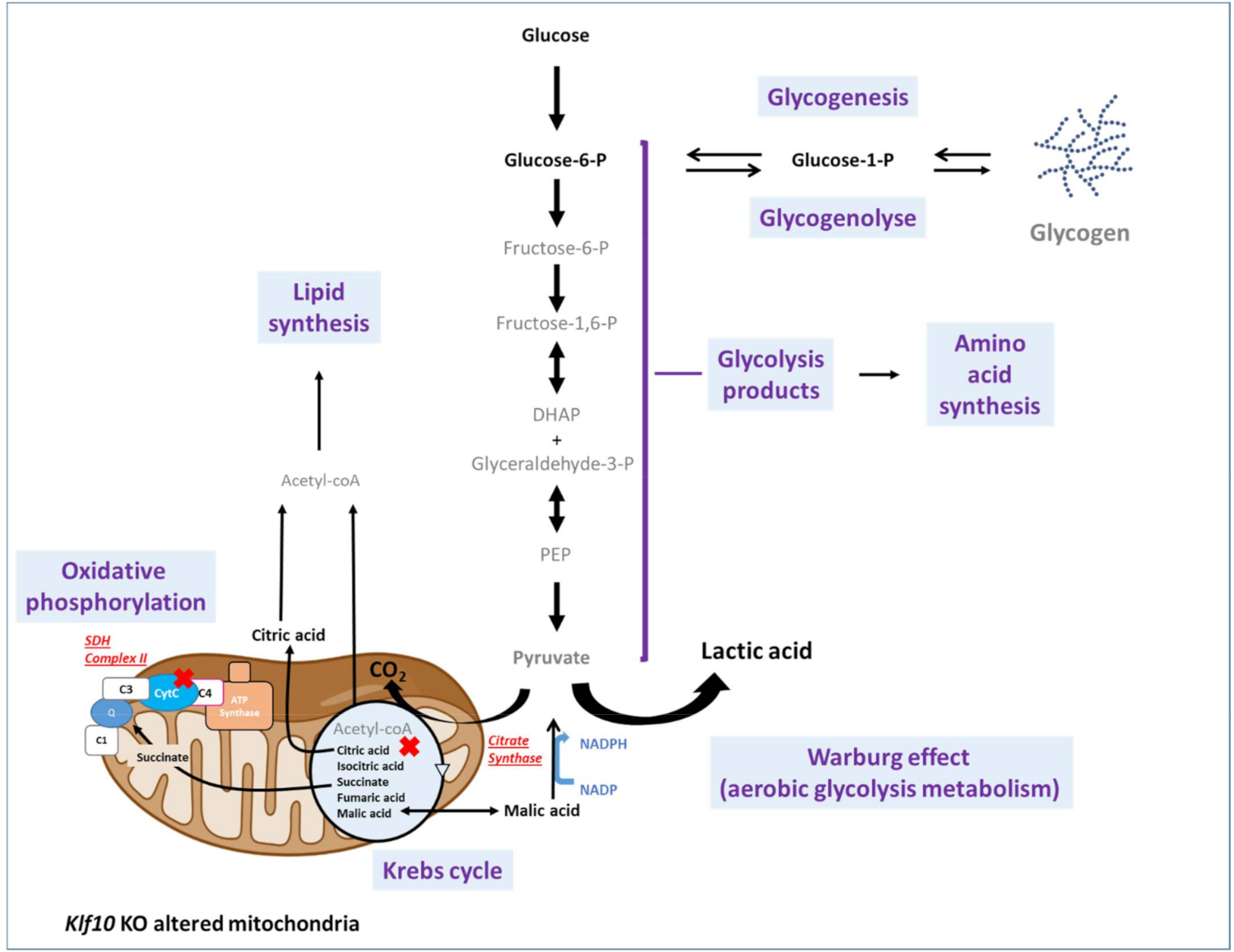
2.3. Identification of Metabolic Pathways Involved in Klf10 KO Mice
2.4. Identification of Metabolic Pathways Involved in Klf10 KO Mice
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Sample Collection and Preparation
4.3. Ultra-High-Performance Liquid Chromatography-Mass Spectroscopy (UHPLC-MS)
4.3.1. Data Acquisition
4.3.2. Data Processing
4.3.3. Data Analysis
4.4. Statistical Analysis
4.4.1. Multivariate Analysis
4.4.2. Univariate Analysis
4.4.3. Metabolites Set Enrichment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Subramaniam, M.; Hefferan, T.E.; Tau, K.; Peus, D.; Pittelkow, M.; Jalal, S.; Riggs, B.L.; Roche, P.; Spelsberg, T.C. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J. Cell. Biochem. 1998, 68, 226–236. [Google Scholar] [CrossRef]
- Rajamannan, N.M.; Subramaniam, M.; Abraham, T.P.; Vasile, V.C.; Ackerman, M.J.; Monroe, D.G.; Chew, T.L.; Spelsberg, T.C. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. J. Cell. Biochem. 2007, 100, 315–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConnell, B.B.; Yang, V.W. Mammalian Krüppel-like factors in health and diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef]
- Cao, Z.; Sun, X.; Icli, B.; Wara, A.K.; Feinberg, M.W. Role of Krüppel-like factors in leukocyte development, function, and disease. Blood 2010, 116, 4404–4414. [Google Scholar] [CrossRef] [Green Version]
- Memon, A.; Lee, W.K. KLF10 as a Tumor Suppressor Gene and Its TGF-beta Signaling. Cancers 2018, 10, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsen, S.A.; Subramaniam, M.; Janknecht, R.; Spelsberg, T.C. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 2002, 21, 5783–5790. [Google Scholar] [CrossRef] [Green Version]
- Guillaumond, F.; Grechez-Cassiau, A.; Subramaniam, M.; Brangolo, S.; Peteri-Brunback, B.; Staels, B.; Fievet, C.; Spelsberg, T.C.; Delaunay, F.; Teboul, M. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell. Biol. 2010, 30, 3059–3070. [Google Scholar] [CrossRef] [Green Version]
- Ruberto, A.A.; Grechez-Cassiau, A.; Guerin, S.; Martin, L.; Revel, J.S.; Mehiri, M.; Subramaniam, M.; Delaunay, F.; Teboul, M. KLF10 integrates circadian timing and sugar signaling to coordinate hepatic metabolism. eLife 2021, 10, e65574. [Google Scholar] [CrossRef]
- Barbosa Vieira, T.K.; Jurema da Rocha Leao, M.; Pereira, L.X.; Alves da Silva, L.C.; Pereira da Paz, B.B.; Santos Ferreira, R.J.; Feitoza, C.C.; Fernandes Duarte, A.K.; Barros Ferreira Rodrigues, A.K.; Cavalcanti de Queiroz, A.; et al. Correlation between circadian rhythm related genes, type 2 diabetes, and cancer: Insights from metanalysis of transcriptomics data. Mol. Cell. Endocrinol. 2021, 526, 111214. [Google Scholar] [CrossRef]
- Reinholz, M.M.; An, M.W.; Johnsen, S.A.; Subramaniam, M.; Suman, V.J.; Ingle, J.N.; Roche, P.C.; Spelsberg, T.C. Differential gene expression of TGF beta inducible early gene (TIEG), Smad7, Smad2 and Bard1 in normal and malignant breast tissue. Breast Cancer Res. Treat. 2004, 86, 75–88. [Google Scholar] [CrossRef]
- Cook, T.; Urrutia, R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 278, G513–G521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, A.; Bronk, S.F.; Roberts, P.J.; Urrutia, R.; Gores, G.J. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology 1999, 30, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Spittau, G.; Happel, N.; Behrendt, M.; Chao, T.I.; Krieglstein, K.; Spittau, B. Tieg1/Klf10 is upregulated by NGF and attenuates cell cycle progression in the pheochromocytoma cell line PC12. J. Neurosci. Res. 2010, 88, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, I.; Imoto, M.; Adjei, P.N.; Gores, G.J.; Subramaniam, M.; Spelsberg, T.C.; Urrutia, R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Investig. 1997, 99, 2365–2374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.M.; Lee, Y.J.; Ko, P.Y.; Lin, Y.M.; Sung, W.W. High Expression of KLF10 Is Associated with Favorable Survival in Patients with Oral Squamous Cell Carcinoma. Medicina 2020, 57, 17. [Google Scholar] [CrossRef]
- Wara, A.K.; Wang, S.; Wu, C.; Fang, F.; Haemmig, S.; Weber, B.N.; Aydogan, C.O.; Tesmenitsky, Y.; Aliakbarian, H.; Hawse, J.R.; et al. KLF10 Deficiency in CD4(+) T Cells Triggers Obesity, Insulin Resistance, and Fatty Liver. Cell Rep. 2020, 33, 108550. [Google Scholar] [CrossRef]
- Hsieh, P.N.; Fan, L.; Sweet, D.R.; Jain, M.K. The Krüppel-Like Factors and Control of Energy Homeostasis. Endocr. Rev. 2019, 40, 137–152. [Google Scholar] [CrossRef] [Green Version]
- Oishi, Y.; Manabe, I. Kruppel-Like Factors in Metabolic Homeostasis and Cardiometabolic Disease. Front. Cardiovasc. Med. 2018, 5, 69. [Google Scholar] [CrossRef]
- Subramaniam, M.; Gorny, G.; Johnsen, S.A.; Monroe, D.G.; Evans, G.L.; Fraser, D.G.; Rickard, D.J.; Rasmussen, K.; van Deursen, J.M.; Turner, R.T.; et al. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol. Cell. Biol. 2005, 25, 1191–1199. [Google Scholar] [CrossRef] [Green Version]
- Bensamoun, S.F.; Tsubone, T.; Subramaniam, M.; Hawse, J.R.; Boumediene, E.; Spelsberg, T.C.; An, K.N.; Amadio, P.C. Age-dependent changes in the mechanical properties of tail tendons in TGF-beta inducible early gene-1 knockout mice. J. Appl. Physiol. 2006, 101, 1419–1424. [Google Scholar] [CrossRef] [Green Version]
- Gumez, L.; Bensamoun, S.F.; Doucet, J.; Haddad, O.; Hawse, J.R.; Subramaniam, M.; Spelsberg, T.C.; Pichon, C. Molecular structure of tail tendon fibers in TIEG1 knockout mice using synchrotron diffraction technology. J. Appl. Physiol. 2010, 108, 1706–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensamoun, S.F.; Hawse, J.R.; Subramaniam, M.; Ilharreborde, B.; Bassillais, A.; Benhamou, C.L.; Fraser, D.G.; Oursler, M.J.; Amadio, P.C.; An, K.N.; et al. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone 2006, 39, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Haddad, O.; Hawse, J.R.; Subramaniam, M.; Spelsberg, T.C.; Bensamoun, S.F. Tieg1-Null Osteocytes Display Defects in Their Morphology, Density and Surrounding Bone Matrix. J. Musculoskelet. Res. 2009, 12, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Hawse, J.R.; Iwaniec, U.T.; Bensamoun, S.F.; Monroe, D.G.; Peters, K.D.; Ilharreborde, B.; Rajamannan, N.M.; Oursler, M.J.; Turner, R.T.; Spelsberg, T.C.; et al. TIEG-null mice display an osteopenic gender-specific phenotype. Bone 2008, 42, 1025–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsubone, T.; Moran, S.L.; Subramaniam, M.; Amadio, P.C.; Spelsberg, T.C.; An, K.N. Effect of TGF-beta inducible early gene deficiency on flexor tendon healing. J. Orthop. Res. 2006, 24, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Kammoun, M.; Pouletaut, P.; Canon, F.; Subramaniam, M.; Hawse, J.R.; Vayssade, M.; Bensamoun, S.F. Impact of TIEG1 Deletion on the Passive Mechanical Properties of Fast and Slow Twitch Skeletal Muscles in Female Mice. PLoS ONE 2016, 11, e0164566. [Google Scholar] [CrossRef] [PubMed]
- Kammoun, M.; Pouletaut, P.; Morandat, S.; Subramaniam, M.; Hawse, J.R.; Bensamoun, S.F. Krüppel-like factor 10 regulates the contractile properties of skeletal muscle fibers in mice. Muscle Nerve 2021, 64, 765–769. [Google Scholar] [CrossRef]
- Kammoun, M.; Piquereau, J.; Nadal-Desbarats, L.; Meme, S.; Beuvin, M.; Bonne, G.; Veksler, V.; Le Fur, Y.; Pouletaut, P.; Meme, W.; et al. Novel role of Tieg1 in muscle metabolism and mitochondrial oxidative capacities. Acta Physiol. 2020, 228, e13394. [Google Scholar] [CrossRef]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Wishart, D.S.; Mandal, R.; Stanislaus, A.; Ramirez-Gaona, M. Cancer Metabolomics and the Human Metabolome Database. Metabolites 2016, 6, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Canto, C.; Jeninga, E.H.; Andreux, P.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The metabolic footprint of aging in mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Koves, T.R.; Ussher, J.R.; Noland, R.C.; Slentz, D.; Mosedale, M.; Ilkayeva, O.; Bain, J.; Stevens, R.; Dyck, J.R.; Newgard, C.B.; et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008, 7, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Mihalik, S.J.; Goodpaster, B.H.; Kelley, D.E.; Chace, D.H.; Vockley, J.; Toledo, F.G.; DeLany, J.P. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity 2010, 18, 1695–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, M.; Hawse, J.R.; Rajamannan, N.M.; Ingle, J.N.; Spelsberg, T.C. Functional role of KLF10 in multiple disease processes. Biofactors 2010, 36, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Garvey, S.M.; Dugle, J.E.; Kennedy, A.D.; McDunn, J.E.; Kline, W.; Guo, L.; Guttridge, D.C.; Pereira, S.L.; Edens, N.K. Metabolomic profiling reveals severe skeletal muscle group-specific perturbations of metabolism in aged FBN rats. Biogerontology 2014, 15, 217–232. [Google Scholar] [CrossRef] [Green Version]
- Sun, N.; Youle, R.J.; Finkel, T. The Mitochondrial Basis of Aging. Mol. Cell 2016, 61, 654–666. [Google Scholar] [CrossRef] [Green Version]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [Green Version]
- Russell, S.J.; Kahn, C.R. Endocrine regulation of ageing. Nat. Rev. Mol. Cell. Biol. 2007, 8, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Tseng, Y.; White, M.F. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol. Metab. 2010, 21, 589–598. [Google Scholar] [CrossRef] [Green Version]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell. Biol. 2011, 12, 21–35. [Google Scholar] [CrossRef] [Green Version]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the adult human plasma metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef]
- Senoo, N.; Miyoshi, N.; Goto-Inoue, N.; Minami, K.; Yoshimura, R.; Morita, A.; Sawada, N.; Matsuda, J.; Ogawa, Y.; Setou, M.; et al. PGC-1alpha-mediated changes in phospholipid profiles of exercise-trained skeletal muscle. J. Lipid Res. 2015, 56, 2286–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchitomi, R.; Hatazawa, Y.; Senoo, N.; Yoshioka, K.; Fujita, M.; Shimizu, T.; Miura, S.; Ono, Y.; Kamei, Y. Metabolomic Analysis of Skeletal Muscle in Aged Mice. Sci. Rep. 2019, 9, 10425. [Google Scholar] [CrossRef] [Green Version]
- Suhre, K.; Meisinger, C.; Doring, A.; Altmaier, E.; Belcredi, P.; Gieger, C.; Chang, D.; Milburn, M.V.; Gall, W.E.; Weinberger, K.M.; et al. Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS ONE 2010, 5, e13953. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Zhang, R.; Jain, R.; Shi, H.; Zhang, L.; Zhou, G.; Sangwung, P.; Tugal, D.; Atkins, G.B.; Prosdocimo, D.A.; et al. Circadian control of bile acid synthesis by a KLF15-Fgf15 axis. Nat. Commun. 2015, 6, 7231. [Google Scholar] [CrossRef] [Green Version]
- Leclere, P.S.; Rousseau, D.; Patouraux, S.; Guerin, S.; Bonnafous, S.; Grechez-Cassiau, A.; Ruberto, A.A.; Luci, C.; Subramaniam, M.; Tran, A.; et al. MCD diet-induced steatohepatitis generates a diurnal rhythm of associated biomarkers and worsens liver injury in Klf10 deficient mice. Sci. Rep. 2020, 10, 12139. [Google Scholar] [CrossRef]
- Lee, J.; Oh, A.R.; Lee, H.Y.; Moon, Y.A.; Lee, H.J.; Cha, J.Y. Deletion of KLF10 Leads to Stress-Induced Liver Fibrosis upon High Sucrose Feeding. Int. J. Mol. Sci. 2020, 22, 331. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, H.; Wang, X.; Guo, J.; Wei, L.; Song, Y.; Luo, Y.; Zhao, Y.; Subramaniam, M.; Spelsberg, T.C.; et al. Kruppel-like factor 10 protects against acute viral myocarditis by negatively regulating cardiac MCP-1 expression. Cell. Mol. Immunol. 2021, 18, 2236–2248. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, A.; Mavel, S.; Nadal-Desbarats, L.; Galineau, L.; Attucci, S.; Dufour, D.; Sokol, H.; Emond, P. Validation of a global quantitative analysis methodology of tryptophan metabolites in mice using LC-MS. Talanta 2019, 195, 593–598. [Google Scholar] [CrossRef]
- Martias, C.; Baroukh, N.; Mavel, S.; Blasco, H.; Lefevre, A.; Roch, L.; Montigny, F.; Gatien, J.; Schibler, L.; Dufour-Rainfray, D.; et al. Optimization of Sample Preparation for Metabolomics Exploration of Urine, Feces, Blood and Saliva in Humans Using Combined NMR and UHPLC-HRMS Platforms. Molecules 2021, 26, 4111. [Google Scholar] [CrossRef]
- Rathahao-Paris, E.; Alves, S.; Junot, C.; Tabet, J.-C. High resolution mass spectrometry for structural identification of metabolites in metabolomics. Metabolomics 2015, 12, 1–15. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [Green Version]
- Beauclercq, S.; Nadal-Desbarats, L.; Hennequet-Antier, C.; Collin, A.; Tesseraud, S.; Bourin, M.; Le Bihan-Duval, E.; Berri, C. Serum and Muscle Metabolomics for the Prediction of Ultimate pH, a Key Factor for Chicken-Meat Quality. J. Proteome Res. 2016, 15, 1168–1178. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell. Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baroukh, N.; Canteleux, N.; Lefèvre, A.; Dupuy, C.; Martias, C.; Presset, A.; Subramaniam, M.; Hawse, J.R.; Emond, P.; Pouletaut, P.; et al. Serum and Soleus Metabolomics Signature of Klf10 Knockout Mice to Identify Potential Biomarkers. Metabolites 2022, 12, 556. https://doi.org/10.3390/metabo12060556
Baroukh N, Canteleux N, Lefèvre A, Dupuy C, Martias C, Presset A, Subramaniam M, Hawse JR, Emond P, Pouletaut P, et al. Serum and Soleus Metabolomics Signature of Klf10 Knockout Mice to Identify Potential Biomarkers. Metabolites. 2022; 12(6):556. https://doi.org/10.3390/metabo12060556
Chicago/Turabian StyleBaroukh, Nadine, Nathan Canteleux, Antoine Lefèvre, Camille Dupuy, Cécile Martias, Antoine Presset, Malayannan Subramaniam, John R. Hawse, Patrick Emond, Philippe Pouletaut, and et al. 2022. "Serum and Soleus Metabolomics Signature of Klf10 Knockout Mice to Identify Potential Biomarkers" Metabolites 12, no. 6: 556. https://doi.org/10.3390/metabo12060556
APA StyleBaroukh, N., Canteleux, N., Lefèvre, A., Dupuy, C., Martias, C., Presset, A., Subramaniam, M., Hawse, J. R., Emond, P., Pouletaut, P., Morandat, S., Bensamoun, S. F., & Nadal-Desbarats, L. (2022). Serum and Soleus Metabolomics Signature of Klf10 Knockout Mice to Identify Potential Biomarkers. Metabolites, 12(6), 556. https://doi.org/10.3390/metabo12060556