Ratios of Acetaminophen Metabolites Identify New Loci of Pharmacogenetic Relevance in a Genome-Wide Association Study
Abstract
:1. Introduction
2. Results
2.1. A GWAS with Ratios between Acetaminophen Metabolites
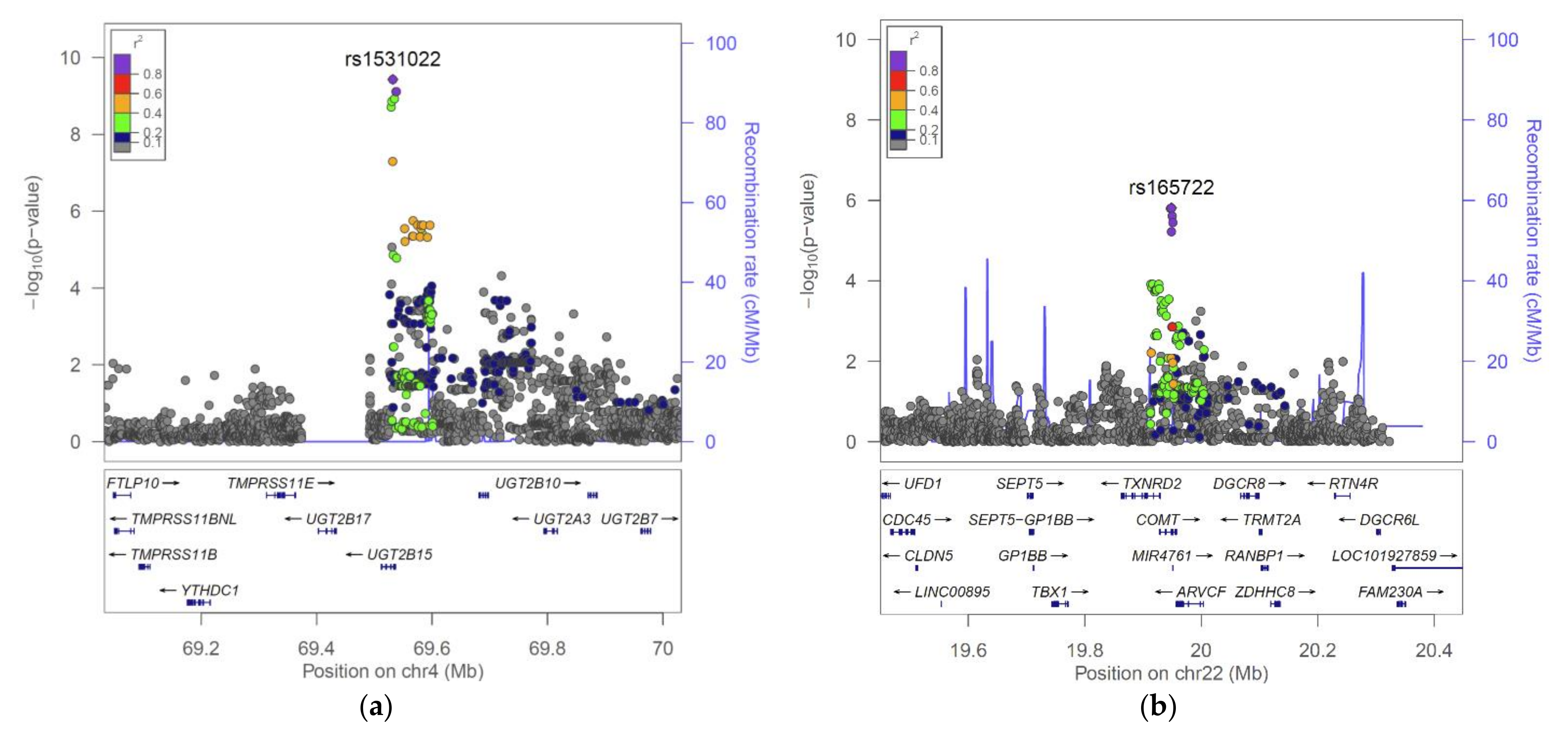
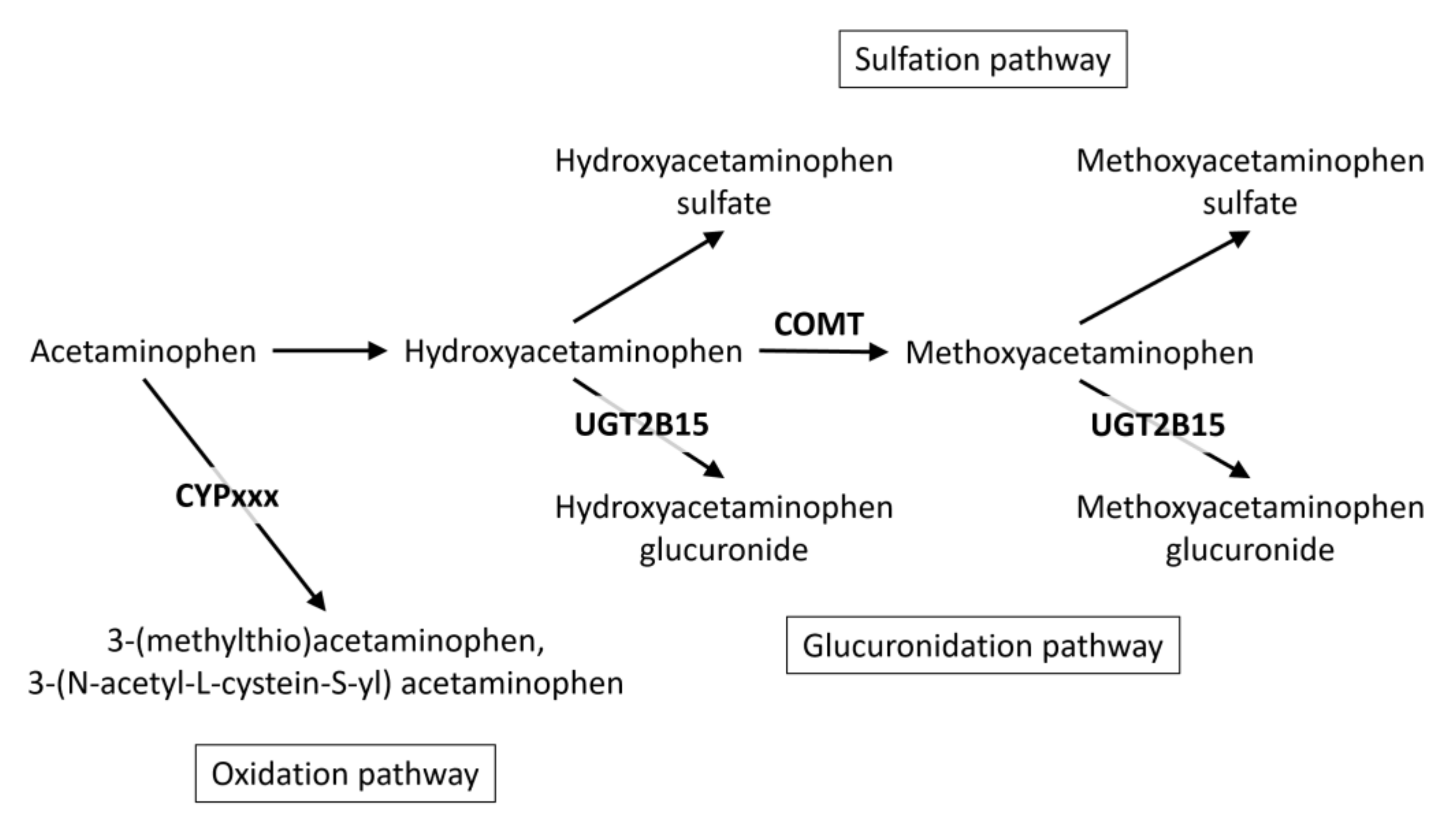
2.2. The UGT2B15 Locus
2.3. The COMT Locus
2.4. Other Loci
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Metabolomics Measurements
4.3. Synthesis of 3-Methoxyacetaminophen Sulfate and Glucuronide Conjugates
4.4. Genotyping
4.5. Locus Annotation
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graham, G.G.; Davies, M.J.; Day, R.O.; Mohamudally, A.; Scott, K.F. The modern pharmacology of paracetamol: Therapeutic actions, mechanism of action, metabolism, toxicity and recent pharmacological findings. Inflammopharmacology 2013, 21, 201–232. [Google Scholar] [CrossRef] [PubMed]
- Gemborys, M.W.; Mudge, G.H. Formation and disposition of the minor metabolites of acetaminophen in the hamster. Drug Metab. Dispos. 1981, 9, 340–351. [Google Scholar] [PubMed]
- Wilson, J.M.; Slattery, J.T.; Forte, A.J.; Nelson, S.D. Analysis of acetaminophen metabolites in urine by high-performance liquid chromatography with UV and amperometric detection. J. Chromatogr. 1982, 227, 453–462. [Google Scholar] [CrossRef]
- Rashed, M.S.; Myers, T.G.; Nelson, S.D. Hepatic protein arylation, glutathione depletion, and metabolite profiles of acetaminophen and a non-hepatotoxic regioisomer, 3’-hydroxyacetanilide, in the mouse. Drug Metab. Dispos. 1990, 18, 765–770. [Google Scholar]
- Spraul, M.; Freund, A.S.; Nast, R.E.; Withers, R.S.; Maas, W.E.; Corcoran, O. Advancing NMR sensitivity for LC-NMR-MS using a cryoflow probe: Application to the analysis of acetaminophen metabolites in urine. Anal. Chem. 2003, 75, 1536–1541. [Google Scholar] [CrossRef]
- Bessems, J.G.; Vermeulen, N.P. Paracetamol (acetaminophen)-induced toxicity: Molecular and biochemical mechanisms, analogues and protective approaches. Crit. Rev. Toxicol. 2001, 31, 55–138. [Google Scholar] [CrossRef]
- Kastenmüller, G.; Raffler, J.; Gieger, C.; Suhre, K. Genetics of human metabolism: An update. Hum. Mol. Genet. 2015, 24, R93–R101. [Google Scholar] [CrossRef] [Green Version]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; de Angelis, M.H.; Kronenberg, F.; Meitinger, T.; Mewes, H.-W.; Wichmann, H.E.; Weinberger, K.M.; Adamski, J.; et al. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef] [Green Version]
- Krumsiek, J.; Suhre, K.; Evans, A.M.; Mitchell, M.W.; Mohney, R.P.; Milburn, M.V.; Wägele, B.; Römisch-Margl, W.; Illig, T.; Adamski, J.; et al. Mining the unknown: A systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012, 8, e1003005. [Google Scholar] [CrossRef] [Green Version]
- Suhre, K.; Shin, S.-Y.; Petersen, A.-K.; Mohney, R.P.; Meredith, D.; Wägele, B.; Altmaier, E.; Deloukas, P.; Erdmann, J.; Grundberg, E.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Wulff, J.E.; Mitchell, M.W. A Comparison of Various Normalization Methods for LC/MS Metabolomics Data. Adv. Biosci. Biotechnol. 2018, 9, 339–351. [Google Scholar] [CrossRef] [Green Version]
- Do, K.T.; Wahl, S.; Raffler, J.; Molnos, S.; Laimighofer, M.; Adamski, J.; Suhre, K.; Strauch, K.; Peters, A.; Gieger, C.; et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics 2018, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhre, K.; Stephan, N.; Zaghlool, S.; Triggle, C.R.; Robinson, R.J.; Evans, A.M.; Halama, A. Matching Drug Metabolites from Non-Targeted Metabolomics to Self-Reported Medication in the Qatar Biobank Study. Metabolites 2022, 12, 249. [Google Scholar] [CrossRef] [PubMed]
- Illig, T.; Gieger, C.; Zhai, G.; Römisch-Margl, W.; Wang-Sattler, R.; Prehn, C.; Altmaier, E.; Kastenmüller, G.; Kato, B.S.; Mewes, H.-W.; et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010, 42, 137–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, A.K.; Krumsiek, J.; Wagele, B.; Theis, F.J.; Wichmann, H.E.; Gieger, C.; Suhre, K. On the hypothesis-free testing of metabolite ratios in genome-wide and metabolome-wide association studies. BMC Bioinform. 2012, 13, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Mrochek, J.E.; Katz, S.; Christie, W.H.; Dinsmore, S.R. Acetaminophen Metabolism in Man, as Determined by High-Resolution Liquid Chromatography. Clin. Chem. 1974, 20, 1086–1096. [Google Scholar] [CrossRef]
- Thareja, G.; Al-Sarraj, Y.; Belkadi, A.; Almotawa, M.; Suhre, K.; Albagha, O.M.E. Whole genome sequencing in the Middle Eastern Qatari population identifies genetic associations with 45 clinically relevant traits. Nat. Commun. 2021, 12, 1250. [Google Scholar] [CrossRef]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.; Raffler, J.; Pfeufer, A.; Suhre, K.; Kastenmuller, G. SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 2015, 31, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, E.; Beaulieu, M.; Green, M.D.; Tephly, T.R.; Bélanger, A.; Hum, D.W. Isolation and characterization of UGT2B15(Y85): A UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics 1997, 7, 317–325. [Google Scholar] [CrossRef]
- Sun, C.; Southard, C.; Witonsky, D.B.; Olopade, O.I.; Di Rienzo, A. Allelic imbalance (AI) identifies novel tissue-specific cis-regulatory variation for human UGT2B15. Hum. Mutat. 2010, 31, 99–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegman, P.; Elingarami, S.; Carstensen, J.; Stål, O.; Nordenskjöld, B.; Wingren, S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007, 9, R7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowell, S.A.; Ahn, J.; Rae, J.M.; Scheys, J.O.; Trovato, A.; Sweeney, C.; MacLeod, S.L.; Kadlubar, F.F.; Ambrosone, C.B. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res. Treat. 2005, 91, 249–258. [Google Scholar] [CrossRef]
- Court, M.H.; Duan, S.X.; Guillemette, C.; Journault, K.; Krishnaswamy, S.; Von Moltke, L.L.; Greenblatt, D.J. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTS): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab. Dispos. 2002, 30, 1257–1265. [Google Scholar] [CrossRef] [Green Version]
- Divakaran, K.; Hines, R.N.; McCarver, D.G. Human Hepatic UGT2B15 Developmental Expression. Toxicol. Sci. 2014, 141, 292–299. [Google Scholar] [CrossRef] [Green Version]
- Männistö, P.T.; Kaakkola, S. Catechol-O-methyltransferase (COMT): Biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol. Rev. 1999, 51, 593–628. [Google Scholar]
- Chen, J.; Lipska, B.K.; Halim, N.; Ma, Q.D.; Matsumoto, M.; Melhem, S.; Kolachana, B.S.; Hyde, T.M.; Herman, M.M.; Apud, J.; et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): Effects on mRNA, protein, and enzyme activity in postmortem human brain. Am. J. Hum. Genet. 2004, 75, 807–821. [Google Scholar] [CrossRef] [Green Version]
- David, A.; Chaker, J.; Léger, T.; Al-Salhi, R.; Dalgaard, M.D.; Styrishave, B.; Bury, D.; Koch, H.M.; Jégou, B.; Kristensen, D.M. Acetaminophen metabolism revisited using non-targeted analyses: Implications for human biomonitoring. Environ. Int. 2021, 149, 106388. [Google Scholar] [CrossRef]
- Long, T.; Hicks, M.; Yu, H.C.; Biggs, W.H.; Kirkness, E.F.; Menni, C.; Zierer, J.; Small, K.S.; Mangino, M.; Messier, H.; et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017, 49, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Joehanes, R.; Zhang, X.; Huan, T.; Yao, C.; Ying, S.-X.; Nguyen, Q.T.; Demirkale, C.Y.; Feolo, M.L.; Sharopova, N.R.; Sturcke, A.; et al. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 2017, 18, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baer, B.R.; Rettie, A.E. CYP4B1: An Enigmatic P450 at the Interface between Xenobiotic and Endobiotic Metabolism. Drug Metab. Rev. 2006, 38, 451–476. [Google Scholar] [CrossRef] [PubMed]
- Al Kuwari, H.; Al Thani, A.; Al Marri, A.; Al Kaabi, A.; Abderrahim, H.; Afifi, N.; Qafoud, F.; Chan, Q.; Tzoulaki, I.; Downey, P.; et al. The Qatar Biobank: Background and methods. BMC Public Health 2015, 15, 1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Thani, A.; Fthenou, E.; Paparrodopoulos, S.; Al Marri, A.; Shi, Z.; Qafoud, F.; Afifi, N. Qatar Biobank Cohort Study: Study Design and First Results. Am. J. Epidemiol. 2019, 188, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Mbarek, H.; Gandhi, G.D.; Selvaraj, S.; Al-Muftah, W.; Badji, R.; Al-Sarraj, Y.; Saad, C.; Darwish, D.; Alvi, M.; Fadl, T.; et al. Qatar genome: Insights on genomics from the Middle East. Hum. Mutat. 2022, 43, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Elfatih, A.; Da’as, S.I.; Abdelrahman, D.; Mbarek, H.; Mohammed, I.; Hasan, W.; Fakhro, K.A.; Estivill, X.; Mifsud, B. Analysis of incidental findings in Qatar genome participants reveals novel functional variants in LMNA and DSP. Hum. Mol. Genet. 2022, ddac073. [Google Scholar] [CrossRef]
- Elfatih, A.; Mifsud, B.; Syed, N.; Badii, R.; Mbarek, H.; Abbaszadeh, F.; Estivill, X. Actionable genomic variants in 6045 participants from the Qatar Genome Program. Hum. Mutat. 2021, 42, 1584–1601. [Google Scholar] [CrossRef]
- Edwards, D.R.; Lohman, D.C.; Wolfenden, R. Catalytic Proficiency: The Extreme Case of S–O Cleaving Sulfatases. J. Am. Chem. Soc. 2012, 134, 525–531. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zaitlen, N.A.; Goddard, M.E.; Visscher, P.M.; Price, A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014, 46, 100–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| BIOCHEMICAL | CAS | LC/MS Mode | Retention Index ** | Mass | N (Out of 520) |
|---|---|---|---|---|---|
| 4-acetamidophenol (paracetamol) | 103-90-2 | Neg | 2173.7 | 150.05605 | 497 |
| 4-acetamidophenyl glucuronide | 120595-80-4 | Neg | 1400 | 326.08814 | 510 |
| 2-methoxyacetaminophen glucuronide * | 53446-12-1 | Neg | 1633 | 356.0987 | 416 |
| 2-hydroxyacetaminophen sulfate * | 53446-14-3 | Neg | 1674 | 246.00778 | 497 |
| 2-methoxyacetaminophen sulfate * | 53446-13-2 | Neg | 1949 | 260.02343 | 405 |
| 4-acetaminophen sulfate | 10066-90-7 | Neg | 1792 | 230.01287 | 514 |
| 3-(N-acetyl-L-cystein-S-yl) acetaminophen | 52372-86-8 | Neg | 2094 | 311.07072 | 363 |
| 3-(methylthio) acetaminophen sulfate * | 78194-51-1 | Neg | 2265 | 276.00059 | 475 |
| 3-(cystein-S-yl) acetaminophen * | 53446-10-9 | Pos Early | 2420 | 271.07471 | 363 |
| Locus | Trait | SNP | Chr | Pos | MAF | Beta | p-Value | P-Gain |
|---|---|---|---|---|---|---|---|---|
| UGT2B15 | 2-methoxyacetaminophen glucuronide*/ 2-methoxyacetaminophen sulfate* | rs1531022 | 4 | 69,532,128 | 48.4% | 0.150 | 3.7 × 10−10 | 4.6 × 107 |
| rs72607822 | 4 | 70,331,309 | 5.8% | −0.253 | 1.2 × 10−6 | 1.7 × 104 | ||
| rs7688257 | 4 | 68,126,425 | 8.5% | 0.199 | 3.0 × 10−6 | 6.9 × 103 | ||
| rs1968718 | 4 | 69,530,496 | 37.3% | −0.107 | 8.5 × 10−6 | 3.7 × 104 | ||
| COMT | 2-methoxyacetaminophen sulfate*/ 2-hydroxyacetaminophen sulfate* | rs165722 | 22 | 19,949,013 | 45.0% | 0.112 | 1.5 × 10−6 | 4.4 × 104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thareja, G.; Evans, A.M.; Wood, S.D.; Stephan, N.; Zaghlool, S.; Halama, A.; Kastenmüller, G.; Belkadi, A.; Albagha, O.M.E.; The Qatar Genome Program Research Consortium; et al. Ratios of Acetaminophen Metabolites Identify New Loci of Pharmacogenetic Relevance in a Genome-Wide Association Study. Metabolites 2022, 12, 496. https://doi.org/10.3390/metabo12060496
Thareja G, Evans AM, Wood SD, Stephan N, Zaghlool S, Halama A, Kastenmüller G, Belkadi A, Albagha OME, The Qatar Genome Program Research Consortium, et al. Ratios of Acetaminophen Metabolites Identify New Loci of Pharmacogenetic Relevance in a Genome-Wide Association Study. Metabolites. 2022; 12(6):496. https://doi.org/10.3390/metabo12060496
Chicago/Turabian StyleThareja, Gaurav, Anne M. Evans, Spencer D. Wood, Nisha Stephan, Shaza Zaghlool, Anna Halama, Gabi Kastenmüller, Aziz Belkadi, Omar M. E. Albagha, The Qatar Genome Program Research Consortium, and et al. 2022. "Ratios of Acetaminophen Metabolites Identify New Loci of Pharmacogenetic Relevance in a Genome-Wide Association Study" Metabolites 12, no. 6: 496. https://doi.org/10.3390/metabo12060496
APA StyleThareja, G., Evans, A. M., Wood, S. D., Stephan, N., Zaghlool, S., Halama, A., Kastenmüller, G., Belkadi, A., Albagha, O. M. E., The Qatar Genome Program Research Consortium, & Suhre, K. (2022). Ratios of Acetaminophen Metabolites Identify New Loci of Pharmacogenetic Relevance in a Genome-Wide Association Study. Metabolites, 12(6), 496. https://doi.org/10.3390/metabo12060496







