Heritability of Urinary Amines, Organic Acids, and Steroid Hormones in Children
Abstract
:1. Introduction
2. Results
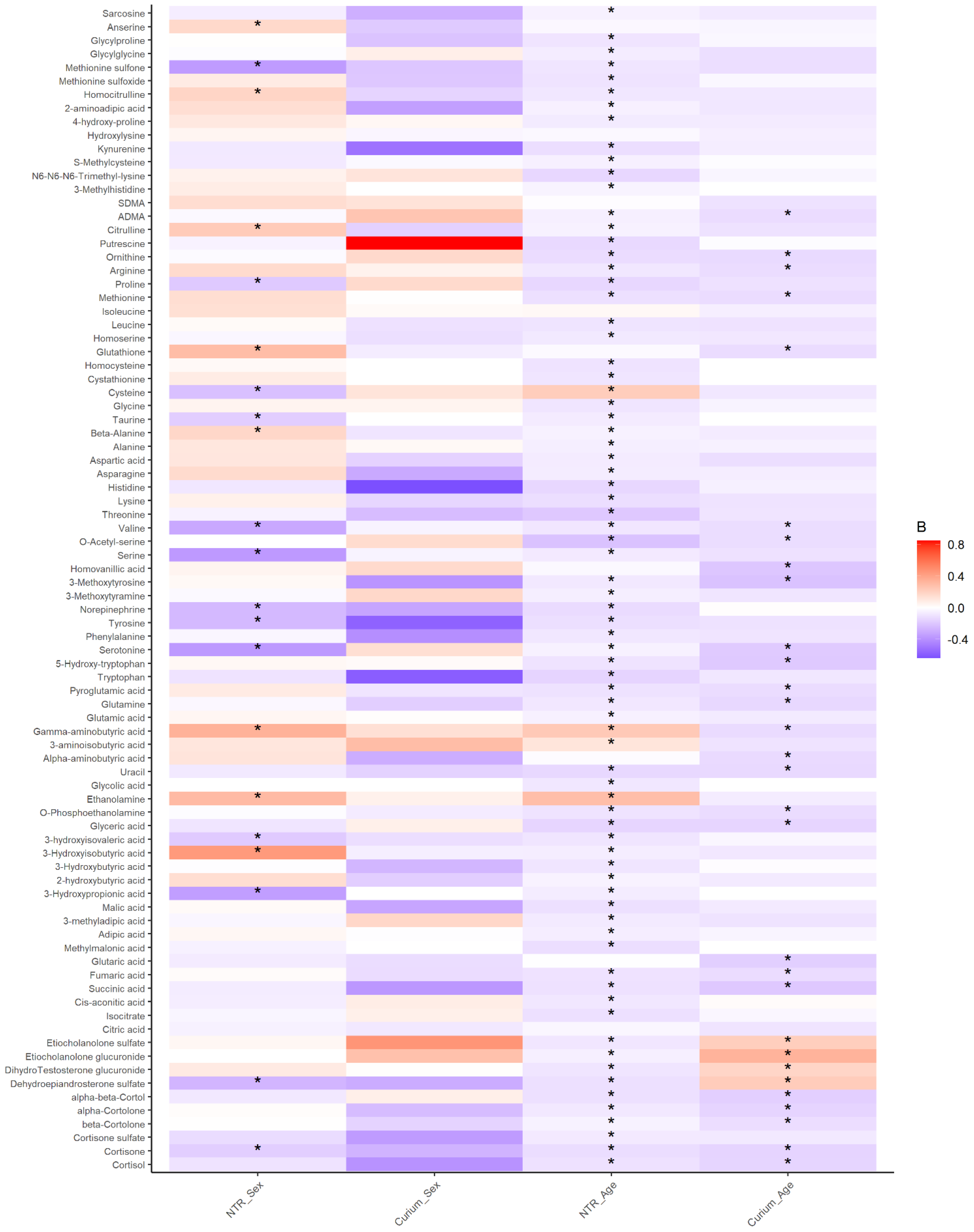
2.1. Urinary Metabolites in Children Have Robust Associations with Age Not with Sex
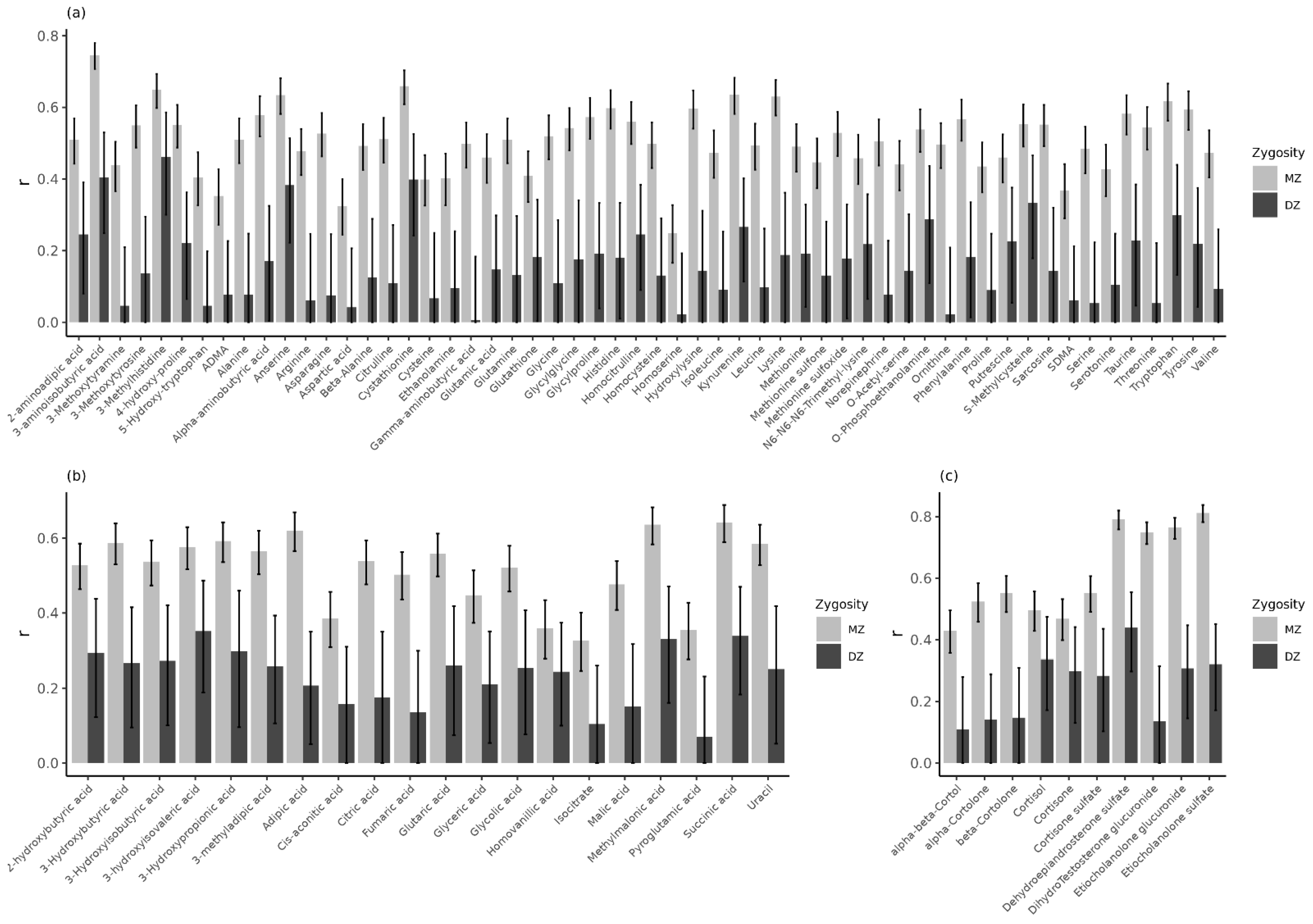
2.2. Ninety Percent of Urinary Metabolites in Children Have Moderate to High Reliability
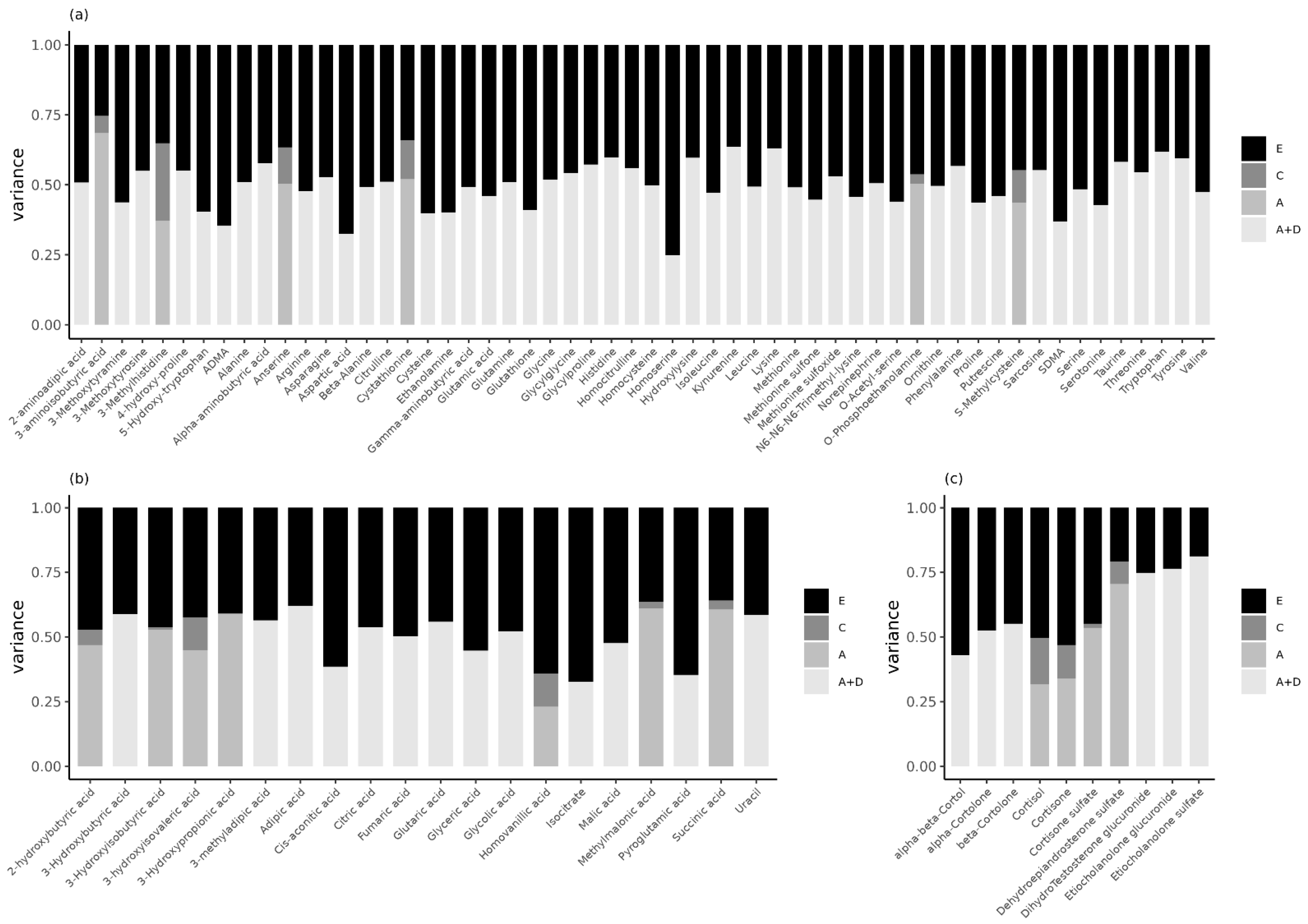
2.3. Genetic Inluences Explain Familial Resemblance in Urinary Metabolites in Children
3. Discussion
4. Materials and Methods
4.1. Study Population and Procedures
4.1.1. NTR-ACTION Cohort
4.1.2. LUMC-Curium Cohort
4.2. Creatinine Measurement
4.3. Metabolite Profiling
4.3.1. Measurement Protocol
4.3.2. LC-MS Amine Platform
4.3.3. GC-MS Organic Acid Platform
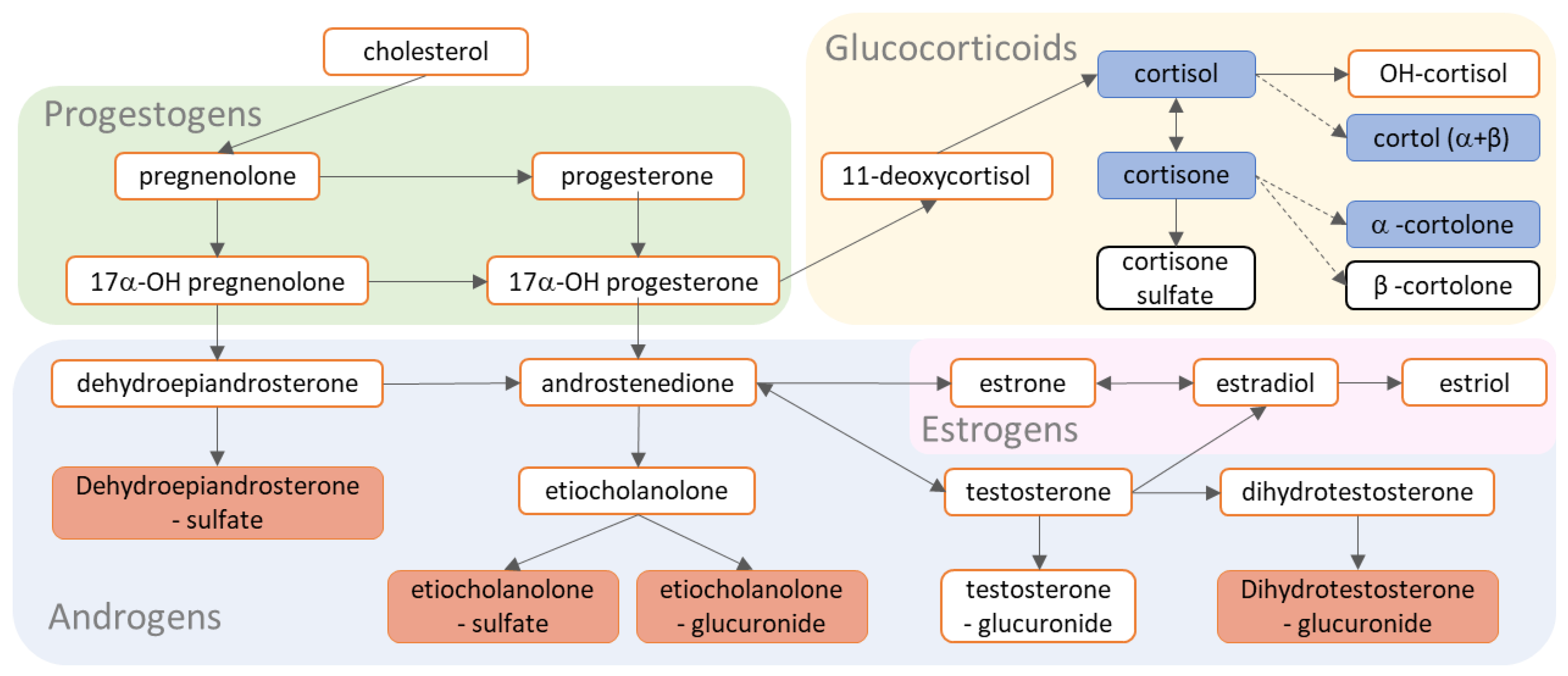
4.3.4. LC-MS Steroid Platform
4.3.5. Metabolomics Data Preprocessing
4.4. Statistical Analyses
4.4.1. Associations with Sex and Age
4.4.2. Genetic Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef] [PubMed]
- Chaleckis, R.; Murakami, I.; Takada, J.; Kondoh, H.; Yanagida, M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. USA 2016, 113, 4252–4259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, H.; Jia, W.; Hu, Z. Emerging Applications of Metabolomics in Clinical Pharmacology. Clin. Pharmacol. Ther. 2019, 106, 544–556. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics–the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Fauman, E.B.; Petersen, A.-K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.-P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Yet, I.; Menni, C.; Shin, S.-Y.; Mangino, M.; Soranzo, N.; Adamski, J.; Suhre, K.; Spector, T.D.; Kastenmüller, G.; Bell, J.T. Genetic Influences on Metabolite Levels: A Comparison across Metabolomic Platforms. PLoS ONE 2016, 11, e0153672. [Google Scholar] [CrossRef] [Green Version]
- Kettunen, J.; Tukiainen, T.; Sarin, A.-P.; Ortega-Alonso, A.; Tikkanen, E.; Lyytikäinen, L.-P.; Kangas, A.J.; Soininen, P.; Würtz, P.; Silander, K.; et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012, 44, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Tsepilov, Y.; Shin, S.-Y.; Soranzo, N.; Spector, T.D.; Prehn, C.; Adamski, J.; Kastenmüller, G.; Wang-Sattler, R.; Strauch, K.; Gieger, C.; et al. Nonadditive Effects of Genes in Human Metabolomics. Genetics 2015, 200, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Pool, R.; Hagenbeek, F.A.; Hendriks, A.M.; van Dongen, J.; Willemsen, G.; de Geus, E.; Willems van Dijk, K.; Verhoeven, A.; Suchiman, H.E.; Beekman, M.; et al. Genetics and Not Shared Environment Explains Familial Resemblance in Adult Metabolomics Data. Twin Res. Hum. Genet. 2020, 23, 145–155. [Google Scholar] [CrossRef]
- Frahnow, T.; Osterhoff, M.A.; Hornemann, S.; Kruse, M.; Surma, M.A.; Klose, C.; Simons, K.; Pfeiffer, A.F.H. Heritability and responses to high fat diet of plasma lipidomics in a twin study. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.W.; Thalamuthu, A.; Braidy, N.; Mather, K.A.; Liu, Y.; Ciobanu, L.; Baune, B.T.; Armstrong, N.J.; Kwok, J.; Schofield, P.; et al. Genetic and environmental determinants of variation in the plasma lipidome of older Australian twins. eLife 2020, 9, e58954. [Google Scholar] [CrossRef]
- Bermingham, K.; Brennan, L.; Segurado, R.; Gray, I.; Barron, R.; Gibney, E.; Ryan, M.; Gibney, M.; Newman, J.; O’Sullivan, A. Genetic and environmental influences on serum oxylipins, endocannabinoids, bile acids and steroids. Prostaglandins Leukot. Essent. Fat. Acids 2021, 173, 102338. [Google Scholar] [CrossRef]
- Bellis, C.; Kulkarni, H.; Mamtani, M.; Kent, J.W.; Wong, G.; Weir, J.M.; Barlow, C.; Diego, V.; de Almeida, M.A.A.; Dyer, T.D.; et al. Human Plasma Lipidome Is Pleiotropically Associated with Cardiovascular Risk Factors and Death. Circ. Cardiovasc. Genet. 2014, 7, 854–863. [Google Scholar] [CrossRef]
- Darst, B.F.; Koscik, R.L.; Hogan, K.J.; Johnson, S.C.; Engelman, C.D. Longitudinal plasma metabolomics of aging and sex. Aging 2019, 11, 1262–1282. [Google Scholar] [CrossRef]
- Rhee, E.P.; Ho, J.; Chen, M.-H.; Shen, D.; Cheng, S.; Larson, M.; Ghorbani, A.; Shi, X.; Helenius, I.T.; O’Donnell, C.J.; et al. A Genome-wide Association Study of the Human Metabolome in a Community-Based Cohort. Cell Metab. 2013, 18, 130–143. [Google Scholar] [CrossRef] [Green Version]
- Cadby, G.; Melton, P.E.; McCarthy, N.S.; Giles, C.; Mellett, N.A.; Huynh, K.; Hung, J.; Beilby, J.; Dubé, M.-P.; Watts, G.F.; et al. Heritability of 596 lipid species and genetic correlation with cardiovascular traits in the Busselton Family Heart Study. J. Lipid Res. 2020, 61, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Hagenbeek, F.A.; Pool, R.; van Dongen, J.; Draisma, H.H.M.; Jan Hottenga, J.; Willemsen, G.; Abdellaoui, A.; Fedko, I.O.; den Braber, A.; Visser, P.J.; et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Feofanova, E.V.; Chen, H.; Dai, Y.; Jia, P.; Grove, M.L.; Morrison, A.C.; Qi, Q.; Daviglus, M.; Cai, J.; North, K.E.; et al. A Genome-wide Association Study Discovers 46 Loci of the Human Metabolome in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2020, 107, 849–863. [Google Scholar] [CrossRef]
- Rhee, E.P.; Yang, Q.; Yu, B.; Liu, X.; Cheng, S.; Deik, A.; Pierce, K.A.; Bullock, K.; Ho, J.; Levy, D.; et al. An exome array study of the plasma metabolome. Nat. Commun. 2016, 7, 12360. [Google Scholar] [CrossRef]
- Tabassum, R.; Project, F.; Rämö, J.T.; Ripatti, P.; Koskela, J.; Kurki, M.; Karjalainen, J.; Palta, P.; Hassan, S.; Nunez-Fontarnau, J.; et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat. Commun. 2019, 10, 4329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rietschel, L.; Streit, F.; Zhu, G.; McAloney, K.; Frank, J.; Couvy-Duchesne, B.; Witt, S.H.; Binz, T.M.; McGrath, J.; Hickie, I.B.; et al. Hair Cortisol in Twins: Heritability and Genetic Overlap with Psychological Variables and Stress-System Genes. Sci. Rep. 2017, 7, 15351. [Google Scholar] [CrossRef] [PubMed]
- Van Keulen, B.J.; Dolan, C.V.; Andrew, R.; Walker, B.R.; Pol, H.E.H.; Boomsma, D.I.; Rotteveel, J.; Finken, M.J.J. Heritability of Cortisol Production and Metabolism Throughout Adolescence. J. Clin. Endocrinol. Metab. 2019, 105, 443–452. [Google Scholar] [CrossRef]
- Coviello, A.D.; Zhuang, W.V.; Lunetta, K.L.; Bhasin, S.; Ulloor, J.; Zhang, A.; Karasik, D.; Kiel, D.P.; Vasan, R.S.; Murabito, J.M. Circulating Testosterone and SHBG Concentrations Are Heritable in Women: The Framingham Heart Study. J. Clin. Endocrinol. Metab. 2011, 96, E1491–E1495. [Google Scholar] [CrossRef] [PubMed]
- Travison, T.G.; Zhuang, W.V.; Lunetta, K.; Karasik, D.; Bhasin, S.; Kiel, D.; Coviello, A.D.; Murabito, J.M. The heritability of circulating testosterone, oestradiol, oestrone and sex hormone binding globulin concentrations in men: The Framingham Heart Study. Clin. Endocrinol. 2013, 80, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Grotzinger, A.D.; Briley, D.A.; Engelhardt, L.E.; Mann, F.D.; Patterson, M.W.; Tackett, J.L.; Tucker-Drob, E.M.; Harden, K.P. Genetic and environmental influences on pubertal hormones in human hair across development. Psychoneuroendocrinology 2018, 90, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Peters, S.R.; Overmyer, K.A.; Paulson, B.R.; Westphall, M.S.; Coon, J.J. Real-time health monitoring through urine metabolomics. NPJ Digit. Med. 2019, 2, 109. [Google Scholar] [CrossRef] [Green Version]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The Human Urine Metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef]
- Calvo-Serra, B.; Maitre, L.; Lau, C.-H.E.; Siskos, A.P.; Gützkow, K.B.; Andrušaitytė, S.; Casas, M.; Cadiou, S.; Chatzi, L.; González, J.R.; et al. Urinary metabolite quantitative trait loci in children and their interaction with dietary factors. Hum. Mol. Genet. 2020, 29, 3830–3844. [Google Scholar] [CrossRef]
- Bartels, M.; Hendriks, A.; Mauri, M.; Krapohl, E.; Whipp, A.; Bolhuis, K.; Conde, L.C.; Luningham, J.; Ip, H.F.; Hagenbeek, F.; et al. Childhood aggression and the co-occurrence of behavioural and emotional problems: Results across ages 3–16 years from multiple raters in six cohorts in the EU-ACTION project. Eur. Child Adolesc. Psychiatry 2018, 27, 1105–1121. [Google Scholar] [CrossRef] [Green Version]
- Boomsma, D.I. Aggression in children: Unravelling the interplay of genes and environment through (epi)genetics and metabolomics. J. Pediatr. Neonatal Individ. Med. 2015, 4, e040251. [Google Scholar] [CrossRef]
- Hagenbeek, F.A.; Roetman, P.J.; Pool, R.; Kluft, C.; Harms, A.C.; van Dongen, J.; Colins, O.F.; Talens, S.; van Beijsterveldt, T.; VandenBosch, M.M.L.J.Z.; et al. Urinary Amine and Organic Acid Metabolites Evaluated as Markers for Childhood Aggression: The ACTION Biomarker Study. Front. Psychiatry 2020, 11, 165. [Google Scholar] [CrossRef]
- Hagenbeek, F.A.; Kluft, C.; Hankemeier, T.; Bartels, M.; Draisma, H.; Middeldorp, C.; Berger, R.; Noto, A.; Lussu, M.; Pool, R.; et al. Discovery of biochemical biomarkers for aggression: A role for metabolomics in psychiatry. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2016, 171, 719–732. [Google Scholar] [CrossRef] [Green Version]
- Ligthart, L.; van Beijsterveldt, C.E.; Kevenaar, S.T.; de Zeeuw, E.; van Bergen, E.; Bruins, S.; Pool, R.; Helmer, Q.; van Dongen, J.; Hottenga, J.-J.; et al. The Netherlands Twin Register: Longitudinal Research Based on Twin and Twin-Family Designs. Twin Res. Hum. Genet. 2019, 22, 623–636. [Google Scholar] [CrossRef] [Green Version]
- Lykken, D.T. Research with Twins: The Concept of Emergenesis. Psychophysiology 1982, 19, 361–372. [Google Scholar] [CrossRef] [Green Version]
- Cicchetti, D.V. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol. Assess. 1994, 6, 284–290. [Google Scholar] [CrossRef]
- Posthuma, D.; Beem, A.L.; de Geus, E.; Van Baal, G.C.M.; Hjelmborg, J.; Iachine, I.; Boomsma, D.I. Theory and Practice in Quantitative Genetics. Twin Res. Hum. Genet. 2003, 6, 361–376. [Google Scholar] [CrossRef]
- Visscher, P.M. Power of the Classical Twin Design Revisited. Twin Res. Hum. Genet. 2004, 7, 505–512. [Google Scholar] [CrossRef]
- Visscher, P.M.; Gordon, S.; Neale, M.C. Power of the Classical Twin Design Revisited: II Detection of Common Environmental Variance. Twin Res. Hum. Genet. 2008, 11, 48–54. [Google Scholar] [CrossRef] [Green Version]
- Koenis, M.M.G.; Brouwer, R.M.; Van Baal, G.C.M.; Van Soelen, I.L.C.; Peper, J.S.; van Leeuwen, M.; De Waal, H.A.D.-V.; Boomsma, D.I.; Pol, H.H. Longitudinal Study of Hormonal and Physical Development in Young Twins. J. Clin. Endocrinol. Metab. 2013, 98, E518–E527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettling, A.C.; Gunnar, M.R.; Donzella, B. Cortisol levels of young children in full-day childcare centers: Relations with age and temperament. Psychoneuroendocrinology 1999, 24, 519–536. [Google Scholar] [CrossRef]
- Shi, L.; Wudy, S.A.; Buyken, A.E.; Maser-Gluth, C.; Hartmann, M.F.; Remer, T. Prepubertal Glucocorticoid Status and Pubertal Timing. J. Clin. Endocrinol. Metab. 2011, 96, E891–E898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Keulen, B.J.; Dolan, C.V.; van der Voorn, B.; Andrew, R.; Walker, B.R.; Pol, H.H.; Boomsma, D.I.; Rotteveel, J.; Finken, M.J.J. Sexual dimorphism in cortisol metabolism throughout pubertal development: A longitudinal study. Endocr. Connect. 2020, 9, 542–551. [Google Scholar] [CrossRef]
- Akalestou, E.; Genser, L.; Rutter, G.A. Glucocorticoid Metabolism in Obesity and Following Weight Loss. Front. Endocrinol. 2020, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Guneral, F.; Bachmann, C. Age-related reference values for urinary organic acids in a healthy Turkish pediatric population. Clin. Chem. 1994, 40, 862–868. [Google Scholar] [CrossRef]
- Chugani, D.C.; Muzik, O.; Behen, M.; Rothermel, R.; Ma, J.J.J.; Lee, J.; Chugani, H.T. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Ann. Neurol. 1999, 45, 287–295. [Google Scholar] [CrossRef]
- Gu, H.; Pan, Z.; Xi, B.; Hainline, B.E.; Shanaiah, N.; Asiago, V.; Gowda, G.A.N.; Raftery, D. 1H NMR metabolomics study of age profiling in children. NMR Biomed. 2009, 22, 826–833. [Google Scholar] [CrossRef] [Green Version]
- Mayo Clinic Laboratories Amino Acids, Quantitative, Random. Available online: https://www.mayocliniclabs.com/test-catalog/overview/60475#Clinical-and-Interpretive (accessed on 25 April 2022).
- Lau, C.-H.E.; Siskos, A.; Maitre, L.; Robinson, O.; Athersuch, T.J.; Want, E.J.; Urquiza, J.; Casas, M.; Vafeiadi, M.; Roumeliotaki, T.; et al. Determinants of the urinary and serum metabolome in children from six European populations. BMC Med. 2018, 16, 202. [Google Scholar] [CrossRef] [Green Version]
- Bermingham, K.M.; Brennan, L.; Segurado, R.; Barron, R.E.; Gibney, E.R.; Ryan, M.F.; Gibney, M.J.; O’Sullivan, A.M. Genetic and environmental influences on covariation in reproducible diet–metabolite associations. Am. J. Clin. Nutr. 2021, 113, 1232–1240. [Google Scholar] [CrossRef]
- Bermingham, K.M.; Brennan, L.; Segurado, R.; Barron, R.E.; Gibney, E.R.; Ryan, M.F.; Gibney, M.J.; O’Sullivan, A.M. Genetic and Environmental Contributions to Variation in the Stable Urinary NMR Metabolome over Time: A Classic Twin Study. J. Proteome Res. 2021, 20, 3992–4000. [Google Scholar] [CrossRef]
- Verhulst, B.; Prom-Wormley, E.; Keller, M.; Medland, S.; Neale, M.C. Type I Error Rates and Parameter Bias in Multivariate Behavioral Genetic Models. Behav. Genet. 2018, 49, 99–111. [Google Scholar] [CrossRef]
- Lykken, D.T. The mechanism of emergenesis. Genes Brain Behav. 2006, 5, 306–310. [Google Scholar] [CrossRef]
- Eaves, L.J. Dominance alone is not enough. Behav. Genet. 1988, 18, 27–33. [Google Scholar] [CrossRef]
- Odintsova, V.; Willemsen, G.; Dolan, C.V.; Hottenga, J.-J.; Martin, N.G.; Slagboom, P.E.; Ordoñana, J.R.; Boomsma, D.I. Establishing a Twin Register: An Invaluable Resource for (Behavior) Genetic, Epidemiological, Biomarker, and ‘Omics’ Studies. Twin Res. Hum. Genet. 2018, 21, 239–252. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/ (accessed on 28 April 2022).
- Lindstrom, M.J.; Bates, D. Nonlinear Mixed Effects Models for Repeated Measures Data. Biometrics 1990, 46, 673. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Boomsma, D.I.; Busjahn, A.; Peltonen, L. Classical twin studies and beyond. Nat. Rev. Genet. 2002, 3, 872–882. [Google Scholar] [CrossRef]
- Rijsdijk, F.V.; Sham, P.C. Analytic approaches to twin data using structural equation models. Brief. Bioinform. 2002, 3, 119–133. [Google Scholar] [CrossRef]
- Boker, S.; Neale, M.; Maes, H.; Wilde, M.; Spiegel, M.; Brick, T.; Spies, J.; Estabrook, R.; Kenny, S.; Bates, T.; et al. OpenMx: An Open Source Extended Structural Equation Modeling Framework. Psychometrika 2011, 76, 306–317. [Google Scholar] [CrossRef] [Green Version]
| Cohort | N (N Pairs) | Mean (SD) [Range] Age | N (%) Females | |
|---|---|---|---|---|
| NTR | Total | 1300 (645) | 9.6 (1.8) [5.7–12.9] | 626 (48.2%) |
| MZ | 1068 (531) | 9.6 (1.9) [6.0–12.9] | 506 (47.4%) | |
| DZ | 232 (114) | 9.9 (1.6) [5.7–12.0] | 120 (51.7%) | |
| LUMC-Curium | Total | 179 | 10.2 (1.8) [6.3–13.4] | 45 (25.1%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hagenbeek, F.A.; van Dongen, J.; Pool, R.; Harms, A.C.; Roetman, P.J.; Fanos, V.; van Keulen, B.J.; Walker, B.R.; Karu, N.; Hulshoff Pol, H.E.; et al. Heritability of Urinary Amines, Organic Acids, and Steroid Hormones in Children. Metabolites 2022, 12, 474. https://doi.org/10.3390/metabo12060474
Hagenbeek FA, van Dongen J, Pool R, Harms AC, Roetman PJ, Fanos V, van Keulen BJ, Walker BR, Karu N, Hulshoff Pol HE, et al. Heritability of Urinary Amines, Organic Acids, and Steroid Hormones in Children. Metabolites. 2022; 12(6):474. https://doi.org/10.3390/metabo12060474
Chicago/Turabian StyleHagenbeek, Fiona A., Jenny van Dongen, René Pool, Amy C. Harms, Peter J. Roetman, Vassilios Fanos, Britt J. van Keulen, Brian R. Walker, Naama Karu, Hilleke E. Hulshoff Pol, and et al. 2022. "Heritability of Urinary Amines, Organic Acids, and Steroid Hormones in Children" Metabolites 12, no. 6: 474. https://doi.org/10.3390/metabo12060474
APA StyleHagenbeek, F. A., van Dongen, J., Pool, R., Harms, A. C., Roetman, P. J., Fanos, V., van Keulen, B. J., Walker, B. R., Karu, N., Hulshoff Pol, H. E., Rotteveel, J., Finken, M. J. J., Vermeiren, R. R. J. M., Kluft, C., Bartels, M., Hankemeier, T., & Boomsma, D. I. (2022). Heritability of Urinary Amines, Organic Acids, and Steroid Hormones in Children. Metabolites, 12(6), 474. https://doi.org/10.3390/metabo12060474









