Mendelian Randomization Analysis Identifies Blood Tyrosine Levels as a Biomarker of Non-Alcoholic Fatty Liver Disease
Abstract
:1. Introduction
2. Results
2.1. Mendelian Randomization Analysis on the Impact of Non-Alcoholic Fatty Liver Disease on the Blood Metabolome
2.2. Observational Analysis of the Impact of Non-Alcoholic Fatty Liver Disease on Blood Tyrosine Levels in the Estonian Biobank
2.3. Impact of Non-Alcoholic Steatohepatitis on Tyrosine Levels in Patients Undergoing Bariatric Surgery
3. Discussion
4. Materials and Methods
4.1. Mendelian Randomization Analyses on the Impact of NAFLD on the Blood Metabolome
4.2. Impact of NAFLD on Tyrosine Levels in the Estonian Biobank
4.3. Impact of NAFLD and NASH on Tyrosine Levels in Québec Bariatric Surgery Cohort
4.4. URLs
- GWAS summary statistics for NAFLD (study accession: GCST90091033) are available at: https://www.ebi.ac.uk/gwas/studies/GCST90091033 (accessed on 30 August 2021).
- GWAS summary statistics for the 123 metabolites are available on MR-Base GWAS Catalog (GWAS id: “met-c-838” to “met-c-960”): https://gwas.mrcieu.ac.uk/datasets/?gwas_id__icontains=met-c (accessed on 30 August 2021). Additional information about the metabolites can be accessed in the article from Kettunen et al., 2016 (Supplementary Table S4): https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4814583/ (accessed on 30 August 2021).
- The TwoSampleMR package is available at: https://github.com/MRCIEU/TwoSampleMR (accessed on 30 August 2021).
- The MendelianRandomization package is available at: https://github.com/cran/MendelianRandomization (accessed on 30 August 2021).
- The data.table package is available at https://github.com/Rdatatable/data.table (accessed on 30 August 2021).
- The tidyverse package collection is available at: https://github.com/tidyverse/tidyverse (accessed on 30 August 2021).
- The LDlinkR package is available at: https://github.com/CBIIT/LDlinkR (accessed on 30 August 2021).
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Targher, G.; Byrne, C.D.; Tilg, H. NAFLD and increased risk of cardiovascular disease: Clinical associations, pathophysiological mechanisms and pharmacological implications. Gut 2020, 69, 1691–1705. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62, S47–S64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, M.J.; Adams, L.A.; Canbay, A.; Syn, W.-K. Extrahepatic complications of nonalcoholic fatty liver disease. Hepatology 2014, 59, 1174–1197. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, M.; Gastaldelli, A.; Hyötyläinen, T.; Arretxe, E.; Alonso, C.; Gaggini, M.; Brosnan, J.; Anstee, Q.M.; Millet, O.; Ortiz, P.; et al. Metabolomics and lipidomics in NAFLD: Biomarkers and non-invasive diagnostic tests. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 835–856. [Google Scholar] [CrossRef]
- Estep, J.; Birerdinc, A.; Younossi, Z. Non-invasive diagnostic tests for non-alcoholic fatty liver disease. Curr. Mol. Med. 2010, 10, 166–172. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. (Clinical report). eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Mohammadi-Shemirani, P.; Sjaarda, J.; Gerstein, H.C.; Treleaven, D.J.; Walsh, M.; Mann, J.F.; McQueen, M.J.; Hess, S.; Paré, G. A Mendelian randomization-based approach to identify early and sensitive diagnostic biomarkers of disease. Clin. Chem. 2019, 65, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, S.C.; Liu, Y.; Lambert, S.A.; Teo, S.M.; Scepanovic, P.; Marten, J.; Zahid, S.; Chaffin, M.; Abraham, G.; Soranzo, N. Integrative analysis of the plasma proteome and polygenic risk of cardiometabolic diseases. Nat. Metab. 2021, 3, 1476–1483. [Google Scholar] [CrossRef]
- Ghodsian, N.; Abner, E.; Emdin, C.A.; Gobeil, É.; Taba, N.; Haas, M.E.; Perrot, N.; Manikpurage, H.D.; Gagnon, É.; Bourgault, J.; et al. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep. Med. 2021, 2, 100437. [Google Scholar] [CrossRef]
- Haycock, P.C.; Burgess, S.; Wade, K.H.; Bowden, J.; Relton, C.; Davey Smith, G. Best (but oft-forgotten) practices: The design, analysis, and interpretation of Mendelian randomization studies. Am. J. Clin. Nutr. 2016, 103, 965–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzych, G.; Vonghia, L.; Bout, M.-A.; Weyler, J.; Verrijken, A.; Dirinck, E.; Chevalier Curt, M.J.; Van Gaal, L.; Paumelle, R.; Francque, S. Plasma BCAA changes in Patients with NAFLD are Sex Dependent. J. Clin. Endocrinol. Metab. 2020, 105, dgaa175. [Google Scholar] [CrossRef]
- Lovric, A.; Granér, M.; Bjornson, E.; Arif, M.; Benfeitas, R.; Nyman, K.; Ståhlman, M.; Pentikäinen, M.O.; Lundbom, J.; Hakkarainen, A. Characterization of different fat depots in NAFLD using inflammation-associated proteome, lipidome and metabolome. Sci. Rep. 2018, 8, 14200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Taskinen, M.R.; Borén, J. Crosstalk between nonalcoholic fatty liver disease and cardiometabolic syndrome. Obes. Rev. 2019, 20, 599–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, R.; Banton, S.; Tran, V.T.; Konomi, J.V.; Li, S.; Jones, D.P.; Vos, M.B. Amino acid metabolism is altered in adolescents with nonalcoholic fatty liver disease—An untargeted, high resolution metabolomics study. J. Pediatrics 2016, 172, 14–19.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lake, A.D.; Novak, P.; Shipkova, P.; Aranibar, N.; Robertson, D.G.; Reily, M.D.; Lehman-McKeeman, L.D.; Vaillancourt, R.R.; Cherrington, N.J. Branched chain amino acid metabolism profiles in progressive human nonalcoholic fatty liver disease. Amino Acids 2015, 47, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Andersson, S.M.; Salaspuro, M.; Ohisalo, J.J. Metabolic basis of hypertyrosinemia in liver disease. Gastroenterology 1982, 82, 554–557. [Google Scholar] [CrossRef]
- Kimberly, W.T.; O’Sullivan, J.F.; Nath, A.K.; Keyes, M.; Shi, X.; Larson, M.G.; Yang, Q.; Long, M.T.; Vasan, R.; Peterson, R.T.; et al. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight 2017, 2, e92989. [Google Scholar] [CrossRef]
- Wewer Albrechtsen, N.J.; Junker, A.E.; Christensen, M.; Hædersdal, S.; Wibrand, F.; Lund, A.M.; Galsgaard, K.D.; Holst, J.J.; Knop, F.K.; Vilsbøll, T. Hyperglucagonemia correlates with plasma levels of non-branched-chain amino acids in patients with liver disease independent of type 2 diabetes. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G91–G96. [Google Scholar] [CrossRef]
- Boulet, M.M.; Chevrier, G.; Grenier-Larouche, T.; Pelletier, M.; Nadeau, M.; Scarpa, J.; Prehn, C.; Marette, A.; Adamski, J.; Tchernof, A. Alterations of plasma metabolite profiles related to adipose tissue distribution and cardiometabolic risk. Am. J. Physiol.-Endocrinol. Metab. 2015, 309, E736–E746. [Google Scholar] [CrossRef] [Green Version]
- Brennan, A.M.; Tchernof, A.; Gerszten, R.E.; Cowan, T.E.; Ross, R. Depot-Specific Adipose Tissue Metabolite Profiles and Corresponding Changes Following Aerobic Exercise. Front. Endocrinol. 2018, 9, 759. [Google Scholar] [CrossRef] [PubMed]
- Winther-Sørensen, M.; Galsgaard, K.D.; Santos, A.; Trammell, S.A.J.; Sulek, K.; Kuhre, R.E.; Pedersen, J.; Andersen, D.B.; Hassing, A.S.; Dall, M.; et al. Glucagon acutely regulates hepatic amino acid catabolism and the effect may be disturbed by steatosis. Mol. Metab. 2020, 42, 101080. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Castaño, G.O.; Scian, R.; Fernández Gianotti, T.; Dopazo, H.; Rohr, C.; Gaj, G.; San Martino, J.; Sevic, I.; Flichman, D.; et al. Serum aminotransferases in nonalcoholic fatty liver disease are a signature of liver metabolic perturbations at the amino acid and Krebs cycle level1,2. Am. J. Clin. Nutr. 2016, 103, 422–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namjou, B.; Lingren, T.; Huang, Y.; Parameswaran, S.; Cobb, B.L.; Stanaway, I.B.; Connolly, J.J.; Mentch, F.D.; Benoit, B.; Niu, X.; et al. GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network. BMC Med. 2019, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G.; Collaboration, C.C.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, T.W.; Day, F.R.; Croteau-Chonka, D.C.; Wood, A.R.; Locke, A.E.; Mägi, R.; Ferreira, T.; Fall, T.; Graff, M.; Justice, A.E.; et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014, 9, 1192–1212. [Google Scholar] [CrossRef] [Green Version]
- Kettunen, J.; Demirkan, A.; Würtz, P.; Draisma, H.H.; Haller, T.; Rawal, R.; Vaarhorst, A.; Kangas, A.J.; Lyytikäinen, L.-P.; Pirinen, M. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 2016, 7, 11122. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [Green Version]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Brunt, E.M.; Janney, C.G.; Di Bisceglie, A.M.; Neuschwander-Tetri, B.A.; Bacon, B.R. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am. J. Gastroenterol. 1999, 94, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
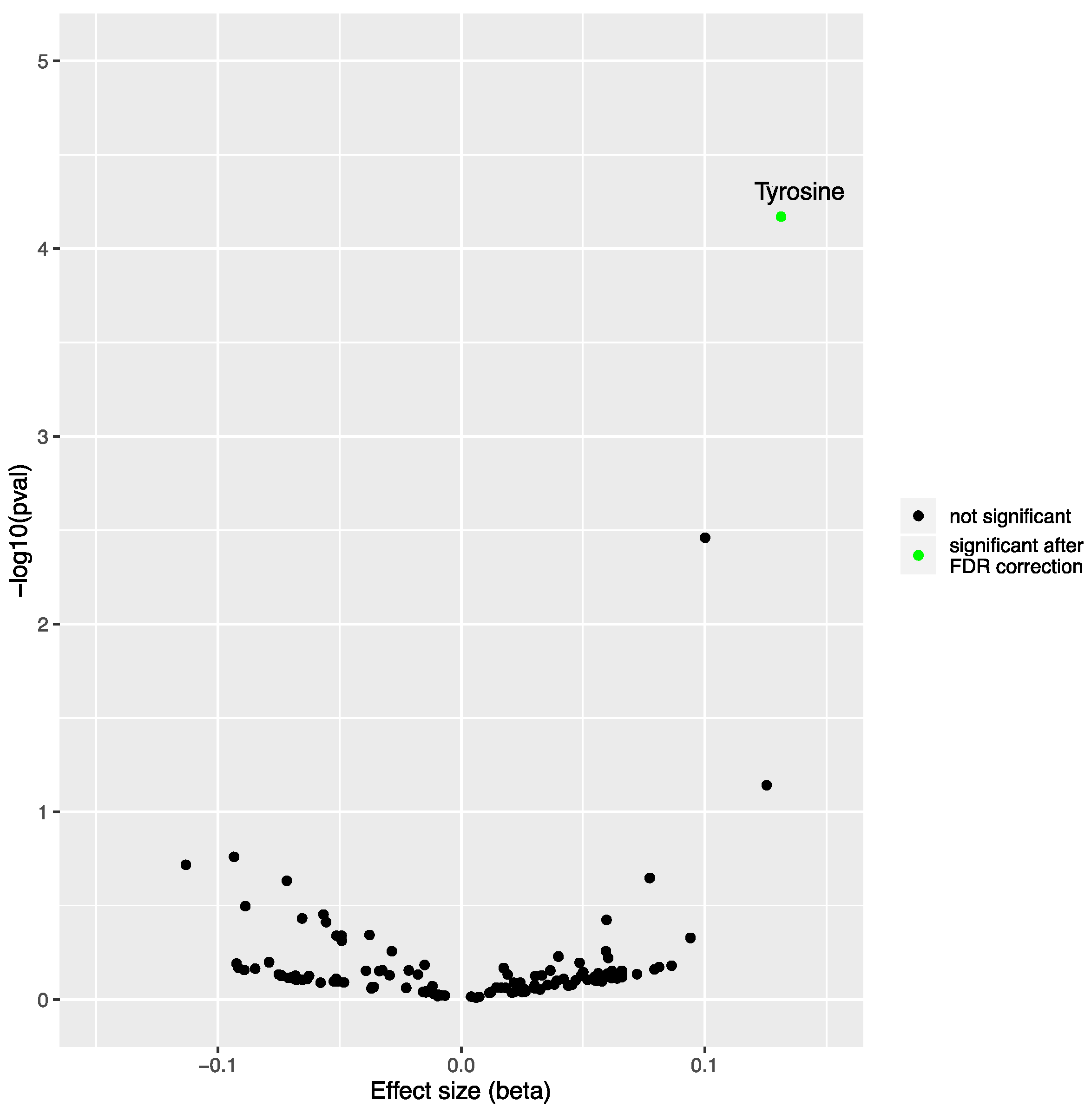
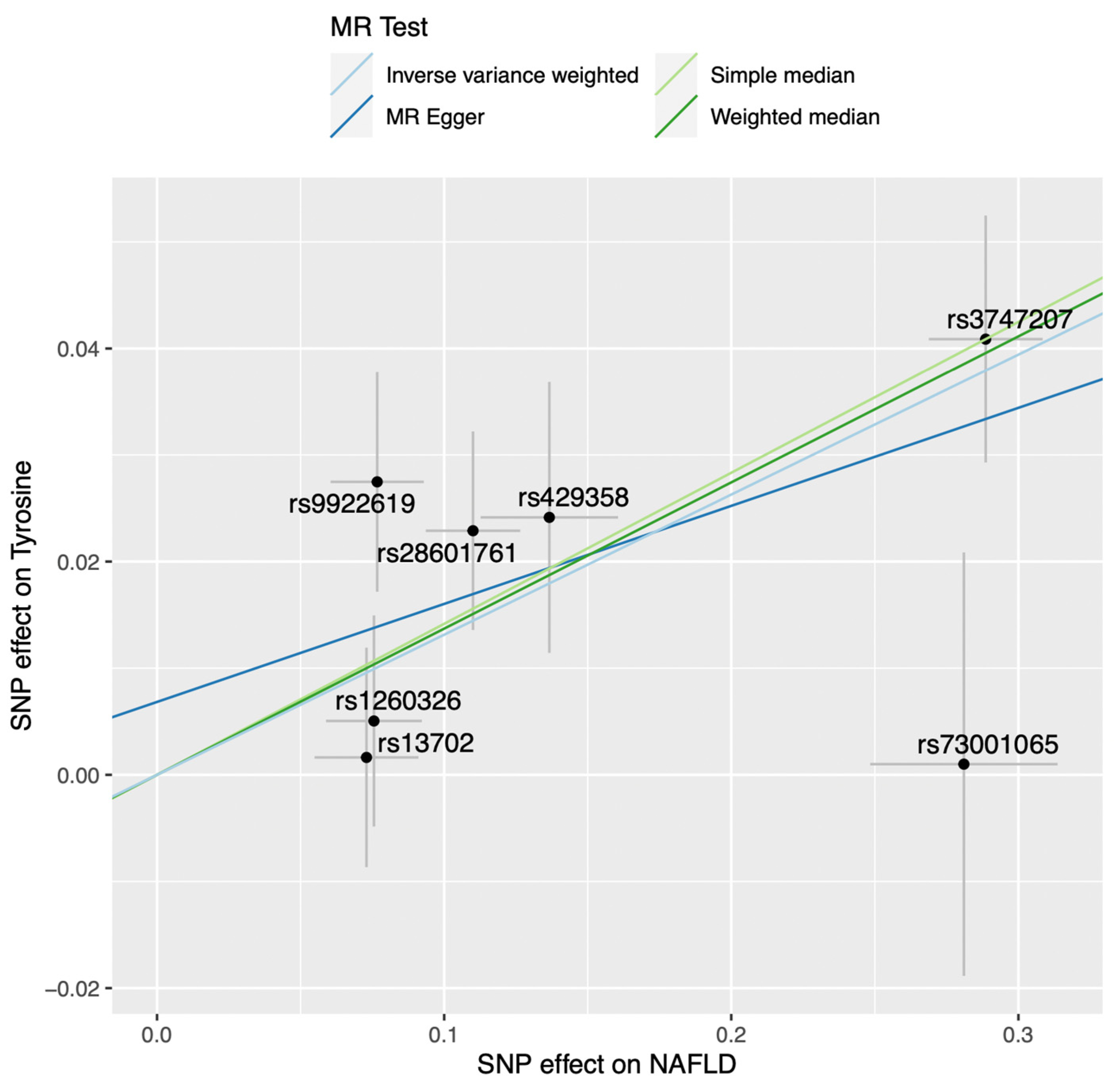

| N SNPs | Inverse-Variance Weighted | Simple Median | Weighted Median | MR-Egger | MR PRESSO | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p-Value | Beta | SE | p-Value | Beta | SE | p-Value | Intercept | p-Value Intercept | Outlier Test p-Value | |
| 7 | 0.131 | 0.033 | 6.75 × 10−5 | 0.142 | 0.045 | 0.002 | 0.137 | 0.038 | 2.82 × 10−4 | 0.010 | 0.515 | 0.334 |
| 12 | 0.104 | 0.027 | 1.47 × 10−4 | 0.042 | 0.040 | 0.297 | 0.103 | 0.036 | 3.88 × 10−3 | −0.002 | 0.759 | 0.293 |
| Tyrosine | Odds Ratio (95% CI) for NAFLD Presence in the Estonian Biobank * | p-Value |
|---|---|---|
| Model 1 | 1.29 (1.18–1.42) | 2.09 × 10−8 |
| Model 2 | 1.23 (1.12–1.36) | 2.19 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gobeil, É.; Maltais-Payette, I.; Taba, N.; Brière, F.; Ghodsian, N.; Abner, E.; Bourgault, J.; Gagnon, E.; Manikpurage, H.D.; Couture, C.; et al. Mendelian Randomization Analysis Identifies Blood Tyrosine Levels as a Biomarker of Non-Alcoholic Fatty Liver Disease. Metabolites 2022, 12, 440. https://doi.org/10.3390/metabo12050440
Gobeil É, Maltais-Payette I, Taba N, Brière F, Ghodsian N, Abner E, Bourgault J, Gagnon E, Manikpurage HD, Couture C, et al. Mendelian Randomization Analysis Identifies Blood Tyrosine Levels as a Biomarker of Non-Alcoholic Fatty Liver Disease. Metabolites. 2022; 12(5):440. https://doi.org/10.3390/metabo12050440
Chicago/Turabian StyleGobeil, Émilie, Ina Maltais-Payette, Nele Taba, Francis Brière, Nooshin Ghodsian, Erik Abner, Jérôme Bourgault, Eloi Gagnon, Hasanga D. Manikpurage, Christian Couture, and et al. 2022. "Mendelian Randomization Analysis Identifies Blood Tyrosine Levels as a Biomarker of Non-Alcoholic Fatty Liver Disease" Metabolites 12, no. 5: 440. https://doi.org/10.3390/metabo12050440
APA StyleGobeil, É., Maltais-Payette, I., Taba, N., Brière, F., Ghodsian, N., Abner, E., Bourgault, J., Gagnon, E., Manikpurage, H. D., Couture, C., Mitchell, P. L., Mathieu, P., Julien, F., Corbeil, J., Vohl, M.-C., Thériault, S., Esko, T., Tchernof, A., & Arsenault, B. J. (2022). Mendelian Randomization Analysis Identifies Blood Tyrosine Levels as a Biomarker of Non-Alcoholic Fatty Liver Disease. Metabolites, 12(5), 440. https://doi.org/10.3390/metabo12050440







