Salivary Metabolomics for Oral Cancer Detection: A Narrative Review
Abstract
:1. Introduction
2. Applications for Cancer Biomarker Discovery
2.1. Diagnostic Markers for OC
2.2. Biomarkers That Discriminate between OC and Other Diseases
2.3. Other Biomarkers Identified Using Salivary Metabolomics
2.4. Biomarkers for Other Cancers
2.5. Salivary Metabolomics for Oral Cavity Diseases
3. Technical Challenges in Salivary Metabolomic Studies
3.1. Metabolite Measurement Technologies
3.2. Discrimination Methods
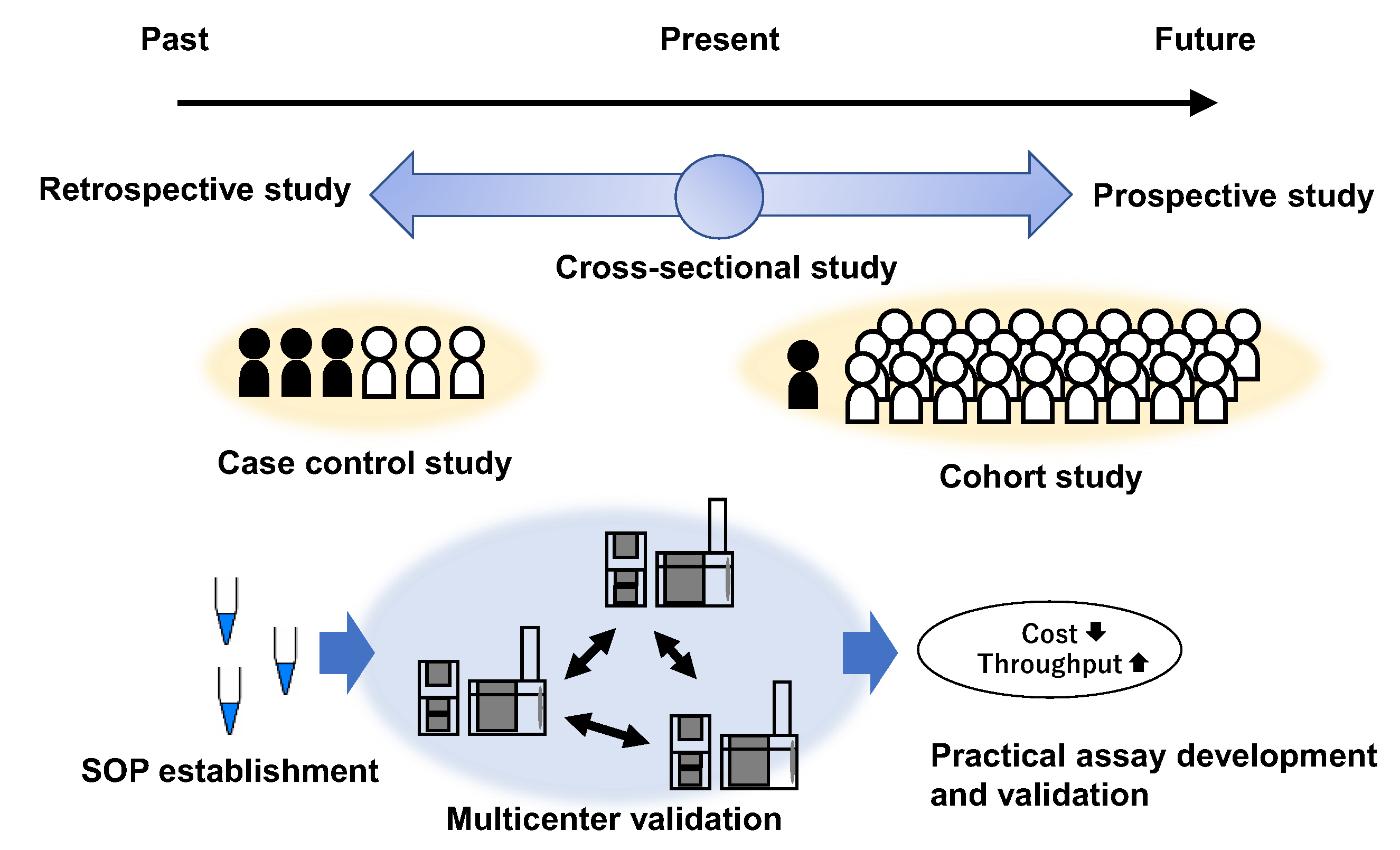
3.3. Standard Operating Protocols (SoP)
4. Discussion
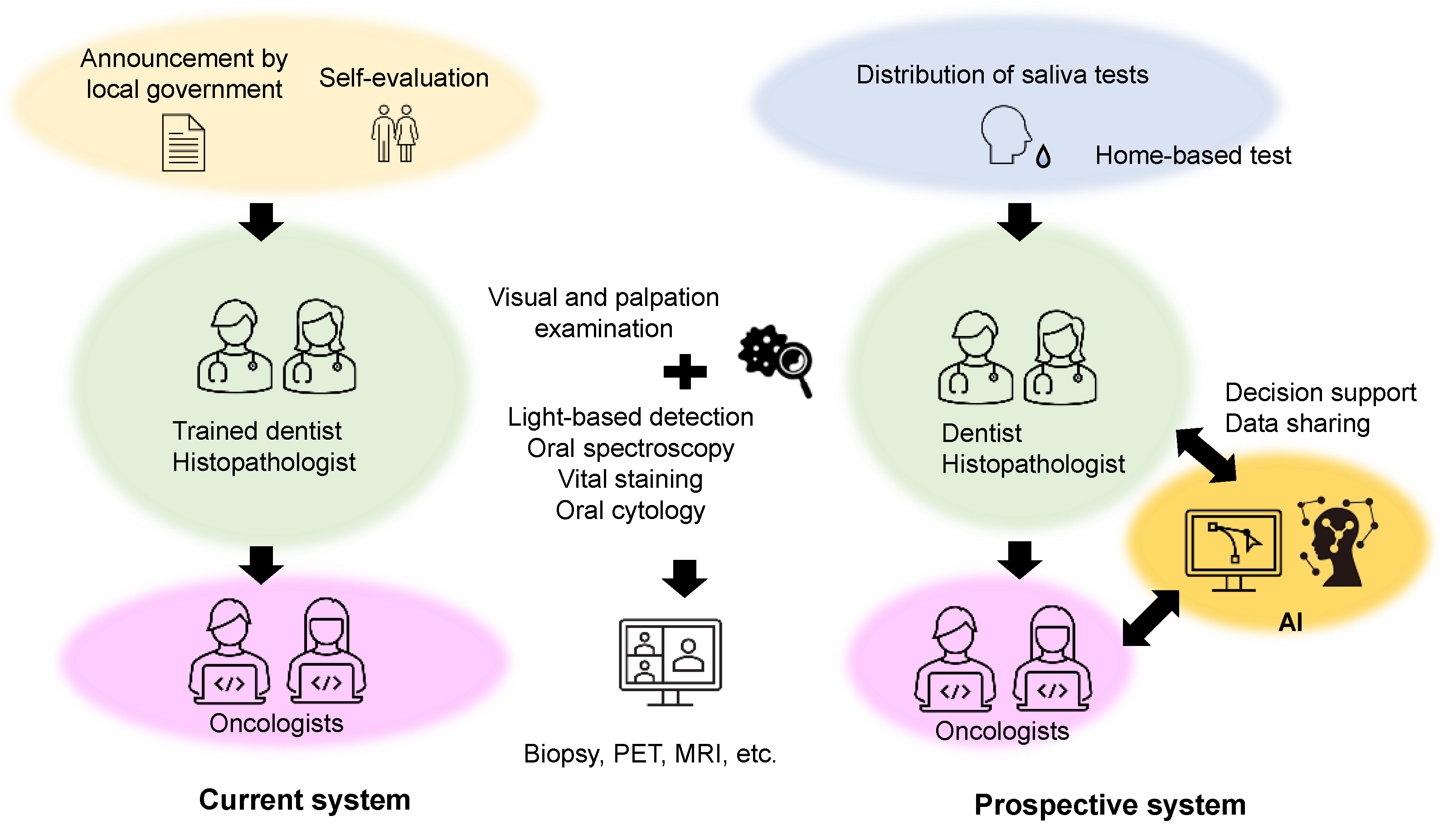
4.1. Current OC Screening Strategies
4.2. Futuer Prospects for Salivary Metabolomics
4.3. Future Perspectives on Cancer Screening Using Salivary Metabolomics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Xu, Q.G.; Chen, X.M.; Fan, M.W. Human Papillomavirus as an Independent Predictor in Oral Squamous Cell Cancer. Int. J. Oral. Sci. 2009, 1, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asthana, S.; Labani, S.; Kailash, U.; Sinha, D.N.; Mehrotra, R. Association of Smokeless Tobacco Use and Oral Cancer: A Systematic Global Review and Meta-Analysis. Nicotine Tob. Res. 2019, 21, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Filho, A.; Bray, F. Global Patterns and Trends in Cancers of the Lip, Tongue and Mouth. Oral Oncol. 2020, 102, 104551. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Xie, L.; Shang, Z.J. Burden of Oral Cancer on the 10 Most Populous Countries from 1990 to 2019: Estimates from the Global Burden of Disease Study 2019. Int. J. Env.. Res. Public Health 2022, 19, 875. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Global Epidemiology of Oral and Oropharyngeal Cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef]
- Sciubba, J.J. Oral Cancer. The Importance of Early Diagnosis and Treatment. Am. J. Clin. Derm.. 2001, 2, 239–251. [Google Scholar] [CrossRef]
- Borse, V.; Konwar, A.N.; Buragohain, P. Oral Cancer Diagnosis and Perspectives in India. Sens. Int. 2020, 1, 100046. [Google Scholar] [CrossRef]
- Akram, M.; Siddiqui, S.A.; Karimi, A.M. Patient Related Factors Associated with Delayed Reporting in Oral Cavity and Oropharyngeal Cancer. Int. J. Prev. Med. 2014, 5, 915–919. [Google Scholar]
- Mummudi, N.; Agarwal, J.P.; Chatterjee, S.; Mallick, I.; Ghosh-Laskar, S. Oral Cavity Cancer in the Indian Subcontinent - Challenges and Opportunities. Clin. Oncol. 2019, 31, 520–528. [Google Scholar] [CrossRef]
- Kawakita, D.; Oze, I.; Iwasaki, S.; Matsuda, T.; Matsuo, K.; Ito, H. Trends in the Incidence of Head and Neck Cancer by Subsite between 1993 and 2015 in Japan. Cancer Med. 2022, 11, 1553–1560. [Google Scholar] [CrossRef]
- Englander, H.; Jeffay, A.; Fuller, J.; Chauncey, H. Glucose Concentrations in Blood Plasma and Parotid Saliva of Individuals with and without Diabetes Mellitus. J. Dent. Res. 1963, 42, 1246–1246. [Google Scholar] [CrossRef]
- Kelsay, J.L.; Behall, K.M.; Holden, J.M.; Crutchfield, H.C. Pyruvate and Lactate in Human Blood and Saliva in Response to Different Carbohydrates. J. Nutr. 1972, 102, 661–666. [Google Scholar] [CrossRef]
- Gardner, A.; Carpenter, G.; So, P.W. Salivary Metabolomics: From Diagnostic Biomarker Discovery to Investigating Biological Function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef] [Green Version]
- Khurshid, Z.; Zohaib, S.; Najeeb, S.; Zafar, M.S.; Slowey, P.D.; Almas, K. Human Saliva Collection Devices for Proteomics: An Update. Int. J. Mol. Sci. 2016, 17, 846. [Google Scholar] [CrossRef] [Green Version]
- Wallner-Liebmann, S.; Tenori, L.; Mazzoleni, A.; Dieber-Rotheneder, M.; Konrad, M.; Hofmann, P.; Luchinat, C.; Turano, P.; Zatloukal, K. Individual Human Metabolic Phenotype Analyzed by 1H NMR of Saliva Samples. J. Proteome Res. 2016, 15, 1787–1793. [Google Scholar] [CrossRef]
- Dalanon, J.; Matsuka, Y. Decreased Global Interest in Oral Cancer During the COVID-19 Pandemic. Asian Pac. J. Cancer Prev. 2021, 22, 2117–2124. [Google Scholar] [CrossRef]
- Sahajpal, N.S.; Mondal, A.K.; Ananth, S.; Njau, A.; Jones, K.; Ahluwalia, P.; Oza, E.; Ross, T.M.; Kota, V.; Kothandaraman, A.; et al. Clinical Validation of a Multiplex Pcr-Based Detection Assay Using Saliva or Nasopharyngeal Samples for SARS-COV-2, Influenza a and B. Sci. Rep. 2022, 12, 3480. [Google Scholar] [CrossRef]
- Khalid, M.F.; Selvam, K.; Jeffry, A.J.N.; Salmi, M.F.; Najib, M.A.; Norhayati, M.N.; Aziah, I. Performance of Rapid Antigen Tests for COVID-19 Diagnosis: A Systematic Review and Meta-Analysis. Diagnostics 2022, 12, 110. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The Early Diagnosis and Monitoring of Squamous Cell Carcinoma Via Saliva Metabolomics. Sci. Rep. 2014, 4, 6802. [Google Scholar] [CrossRef]
- Ohshima, M.; Sugahara, K.; Kasahara, K.; Katakura, A. Metabolomic Analysis of the Saliva of Japanese Patients with Oral Squamous Cell Carcinoma. Oncol. Rep. 2017, 37, 2727–2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Sá Alves, M.; de Sá Rodrigues, N.; Bandeira, C.M.; Chagas, J.F.S.; Pascoal, M.B.N.; Nepomuceno, G.; da Silva Martinho, H.; Alves, M.G.O.; Mendes, M.A.; Dias, M.; et al. Identification of Possible Salivary Metabolic Biomarkers and Altered Metabolic Pathways in South American Patients Diagnosed with Oral Squamous Cell Carcinoma. Metabolites 2021, 11, 650. [Google Scholar] [CrossRef]
- Song, X.; Yang, X.; Narayanan, R.; Shankar, V.; Ethiraj, S.; Wang, X.; Duan, N.; Ni, Y.H.; Hu, Q.; Zare, R.N. Oral Squamous Cell Carcinoma Diagnosed from Saliva Metabolic Profiling. Proc. Natl. Acad. Sci. Usa 2020, 117, 16167–16173. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Xie, G.; Zhou, Z.; Shi, P.; Qiu, Y.; Zheng, X.; Chen, T.; Su, M.; Zhao, A.; Jia, W. Salivary Metabolite Signatures of Oral Cancer and Leukoplakia. Int. J. Cancer 2011, 129, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Sugimoto, M.; Edamatsu, K.; Sugano, A.; Kitabatake, K.; Iino, M. Discrimination of Oral Squamous Cell Carcinoma from Oral Lichen Planus by Salivary Metabolomics. Oral. Dis. 2020, 26, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, G.; Ramani, P.; Patankar, S.; Vijayaraghavan, R. Evaluation of Salivary Metabolomics in Oral Leukoplakia and Oral Squamous Cell Carcinoma. J. Oral Pathol. Med. 2019, 48, 299–306. [Google Scholar] [CrossRef]
- Ishikawa, S.; Hiraka, T.; Kirii, K.; Sugimoto, M.; Shimamoto, H.; Sugano, A.; Kitabatake, K.; Toyoguchi, Y.; Kanoto, M.; Nemoto, K.; et al. Relationship between Standard Uptake Values of Positron Emission Tomography/Computed Tomography and Salivary Metabolites in Oral Cancer: A Pilot Study. J. Clin. Med. 2020, 9, 3958. [Google Scholar] [CrossRef]
- Yatsuoka, W.; Ueno, T.; Miyano, K.; Uezono, Y.; Enomoto, A.; Kaneko, M.; Ota, S.; Soga, T.; Sugimoto, M.; Ushijima, T. Metabolomic Profiling Reveals Salivary Hypotaurine as a Potential Early Detection Marker for Medication-Related Osteonecrosis of the Jaw. PLoS ONE 2019, 14, e0220712. [Google Scholar] [CrossRef] [Green Version]
- Yatsuoka, W.; Ueno, T.; Miyano, K.; Enomoto, A.; Ota, S.; Sugimoto, M.; Uezono, Y. Time-Course of Salivary Metabolomic Profiles During Radiation Therapy for Head and Neck Cancer. J. Clin. Med. 2021, 10, 2631. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary Electrophoresis Mass Spectrometry-Based Saliva Metabolomics Identified Oral, Breast and Pancreatic Cancer-Specific Profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Cavaco, C.; Pereira, J.A.M.; Taunk, K.; Taware, R.; Rapole, S.; Nagarajaram, H.; Câmara, J.S. Screening of Salivary Volatiles for Putative Breast Cancer Discrimination: An Exploratory Study Involving Geographically Distant Populations. Anal. Bioanal. Chem. 2018, 410, 4459–4468. [Google Scholar] [CrossRef]
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; De Luca Canto, G.; Acevedo, A.C.; Guerra, E.N. Salivary Biomarkers in the Diagnosis of Breast Cancer: A Review. Crit. Rev. Oncol. Hematol. 2017, 110, 62–73. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, Z.; Huang, Y.; Duan, Y.; Wang, X. Investigation of Salivary Free Amino Acid Profile for Early Diagnosis of Breast Cancer with Ultra Performance Liquid Chromatography-Mass Spectrometry. Clin. Chim. Acta 2015, 447, 23–31. [Google Scholar] [CrossRef]
- Zhong, L.; Cheng, F.; Lu, X.; Duan, Y.; Wang, X. Untargeted Saliva Metabonomics Study of Breast Cancer Based on Ultra Performance Liquid Chromatography Coupled to Mass Spectrometry with Hilic and Rplc Separations. Talanta 2016, 158, 351–360. [Google Scholar] [CrossRef]
- Xavier Assad, D.; Acevedo, A.C.; Cançado Porto Mascarenhas, E.; Costa Normando, A.G.; Pichon, V.; Chardin, H.; Neves Silva Guerra, E.; Combes, A. Using an Untargeted Metabolomics Approach to Identify Salivary Metabolites in Women with Breast Cancer. Metabolites 2020, 10, 506. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Solomatin, D.V.; Kosenok, V.K. Metabolic Features of Saliva in Breast Cancer Patients. Metabolites 2022, 12, 166. [Google Scholar] [CrossRef]
- Takayama, T.; Tsutsui, H.; Shimizu, I.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Inoue, K.; Todoroki, K.; Min, J.Z.; Mizuno, H.; et al. Diagnostic Approach to Breast Cancer Patients Based on Target Metabolomics in Saliva by Liquid Chromatography with Tandem Mass Spectrometry. Clin. Chim. Acta 2016, 452, 18–26. [Google Scholar] [CrossRef]
- Tsutsui, H.; Mochizuki, T.; Inoue, K.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Todoroki, K.; Min, J.Z.; Toyo’oka, T. High-Throughput Lc-Ms/Ms Based Simultaneous Determination of Polyamines Including N-Acetylated Forms in Human Saliva and the Diagnostic Approach to Breast Cancer Patients. Anal. Chem. 2013, 85, 11835–11842. [Google Scholar] [CrossRef]
- Murata, T.; Yanagisawa, T.; Kurihara, T.; Kaneko, M.; Ota, S.; Enomoto, A.; Tomita, M.; Sugimoto, M.; Sunamura, M.; Hayashida, T.; et al. Salivary Metabolomics with Alternative Decision Tree-Based Machine Learning Methods for Breast Cancer Discrimination. Breast Cancer Res. Treat. 2019, 177, 591–601. [Google Scholar] [CrossRef]
- Gerner, E.W.; Meyskens, F.L., Jr. Polyamines and Cancer: Old Molecules, New Understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef] [Green Version]
- Soda, K. The Mechanisms by Which Polyamines Accelerate Tumor Spread. J. Exp. Clin. Cancer Res. 2011, 30, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeFelice, B.C.; Fiehn, O. Rapid Lc-Ms/Ms Quantification of Cancer Related Acetylated Polyamines in Human Biofluids. Talanta 2019, 196, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Liu, C.; Guo, M.; Cheng, H.; Lu, Y.; Jin, K.; Liu, L.; Long, J.; Xu, J.; Lu, R.; et al. Potential Biomarkers in Lewis Negative Patients with Pancreatic Cancer. Ann. Surg. 2017, 265, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Asai, Y.; Itoi, T.; Sugimoto, M.; Sofuni, A.; Tsuchiya, T.; Tanaka, R.; Tonozuka, R.; Honjo, M.; Mukai, S.; Fujita, M.; et al. Elevated Polyamines in Saliva of Pancreatic Cancer. Cancers 2018, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Löser, C.; Fölsch, U.R.; Paprotny, C.; Creutzfeldt, W. Polyamine Concentrations in Pancreatic Tissue, Serum, and Urine of Patients with Pancreatic Cancer. Pancreas 1990, 5, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Nakkina, S.P.; Gitto, S.B.; Pandey, V.; Parikh, J.G.; Geerts, D.; Maurer, H.C.; Olive, K.P.; Phanstiel, O.t.; Altomare, D.A. Differential Expression of Polyamine Pathways in Human Pancreatic Tumor Progression and Effects of Polyamine Blockade on Tumor Microenvironment. Cancers 2021, 13, 6391. [Google Scholar] [CrossRef] [PubMed]
- Nissinen, S.I.; Venäläinen, M.; Kumpulainen, P.; Roine, A.; Häkkinen, M.R.; Vepsäläinen, J.; Oksala, N.; Rantanen, T. Discrimination between Pancreatic Cancer, Pancreatitis and Healthy Controls Using Urinary Polyamine Panel. Cancer Control 2021, 28, 10732748211039762. [Google Scholar] [CrossRef]
- Baima, G.; Iaderosa, G.; Citterio, F.; Grossi, S.; Romano, F.; Berta, G.N.; Buduneli, N.; Aimetti, M. Salivary Metabolomics for the Diagnosis of Periodontal Diseases: A Systematic Review with Methodological Quality Assessment. Metabolomics 2021, 17, 1. [Google Scholar] [CrossRef]
- Barnes, V.M.; Ciancio, S.G.; Shibly, O.; Xu, T.; Devizio, W.; Trivedi, H.M.; Guo, L.; Jönsson, T.J. Metabolomics Reveals Elevated Macromolecular Degradation in Periodontal Disease. J. Dent. Res. 2011, 90, 1293–1297. [Google Scholar] [CrossRef]
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; Devizio, W.; Trivedi, H.M.; Jönsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global Metabolomic Analysis of Human Saliva and Plasma from Healthy and Diabetic Subjects, with and without Periodontal Disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef]
- Monton, M.R.; Soga, T. Metabolome Analysis by Capillary Electrophoresis-Mass Spectrometry. J. Chromatogr. A 2007, 1168, 237–246. [Google Scholar] [CrossRef]
- Ranjan, R.; Sinha, N. Nuclear Magnetic Resonance (NMR)-Based Metabolomics for Cancer Research. NMR Biomed. 2019, 32, e3916. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef]
- Gardner, A.; Parkes, H.G.; Carpenter, G.H.; So, P.W. Developing and Standardizing a Protocol for Quantitative Proton Nuclear Magnetic Resonance (1H NMR) Spectroscopy of Saliva. J. Proteome Res. 2018, 17, 1521–1531. [Google Scholar]
- Lohavanichbutr, P.; Zhang, Y.; Wang, P.; Gu, H.; Nagana Gowda, G.A.; Djukovic, D.; Buas, M.F.; Raftery, D.; Chen, C. Salivary Metabolite Profiling Distinguishes Patients with Oral Cavity Squamous Cell Carcinoma from Normal Controls. PLoS ONE 2018, 13, e0204249. [Google Scholar] [CrossRef] [Green Version]
- Mikkonen, J.J.W.; Singh, S.P.; Akhi, R.; Salo, T.; Lappalainen, R.; González-Arriagada, W.A.; Ajudarte Lopes, M.; Kullaa, A.M.; Myllymaa, S. Potential Role of Nuclear Magnetic Resonance Spectroscopy to Identify Salivary Metabolite Alterations in Patients with Head and Neck Cancer. Oncol. Lett. 2018, 16, 6795–6800. [Google Scholar] [CrossRef] [Green Version]
- García-Villaescusa, A.; Morales-Tatay, J.M.; Monleón-Salvadó, D.; González-Darder, J.M.; Bellot-Arcis, C.; Montiel-Company, J.M.; Almerich-Silla, J.M. Using NMR in Saliva to Identify Possible Biomarkers of Glioblastoma and Chronic Periodontitis. PLoS ONE 2018, 13, e0188710. [Google Scholar] [CrossRef]
- Gilany, K.; Mohamadkhani, A.; Chashmniam, S.; Shahnazari, P.; Amini, M.; Arjmand, B.; Malekzadeh, R.; Nobakht Motlagh Ghoochani, B.F. Metabolomics Analysis of the Saliva in Patients with Chronic Hepatitis B Using Nuclear Magnetic Resonance: A Pilot Study. Iran J. Basic Med. Sci. 2019, 22, 1044–1049. [Google Scholar]
- Kumari, S.; Goyal, V.; Kumaran, S.S.; Dwivedi, S.; Srivastava, A.; Jagannathan, N. Quantitative Metabolomics of Saliva Using Proton NMR Spectroscopy in Patients with Parkinson’s Disease and Healthy Controls. Neurol. Sci. 2020, 41, 1201–1210. [Google Scholar] [CrossRef]
- Yilmaz, A.; Geddes, T.; Han, B.; Bahado-Singh, R.O.; Wilson, G.D.; Imam, K.; Maddens, M.; Graham, S.F. Diagnostic Biomarkers of Alzheimer’s Disease as Identified in Saliva Using 1H NMR-Based Metabolomics. J. Alzheimer’s Dis. 2017, 58, 355–359. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. Investigation and Identification of Potential Biomarkers in Human Saliva for the Early Diagnosis of Oral Squamous Cell Carcinoma. Clin. Chim. Acta 2014, 427, 79–85. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Sugano, A.; Nakamura, M.; Kaneko, M.; Ota, S.; Hiwatari, K.; Enomoto, A.; Soga, T.; et al. Identification of Salivary Metabolomic Biomarkers for Oral Cancer Screening. Sci. Rep. 2016, 6, 31520. [Google Scholar]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Tu, M.; Sugano, A.; Yamamori, I.; Iba, A.; Yusa, K.; Kaneko, M.; Ota, S.; et al. Effect of Timing of Collection of Salivary Metabolomic Biomarkers on Oral Cancer Detection. Amino Acids 2017, 49, 761–770. [Google Scholar] [CrossRef]
- Hyvärinen, E.; Savolainen, M.; Mikkonen, J.J.W.; Kullaa, A.M. Salivary Metabolomics for Diagnosis and Monitoring Diseases: Challenges and Possibilities. Metabolites 2021, 11, 587. [Google Scholar] [CrossRef]
- Jain, N.S.; Dürr, U.H.; Ramamoorthy, A. Bioanalytical Methods for Metabolomic Profiling: Detection of Head and Neck Cancer, Including Oral Cancer. Chin. Chem. Lett. 2015, 26, 407–415. [Google Scholar] [CrossRef]
- Figueira, J.; Gouveia-Figueira, S.; Öhman, C.; Lif Holgerson, P.; Nording, M.L.; Öhman, A. Metabolite Quantification by NMR and Lc-Ms/Ms Reveals Differences between Unstimulated, Stimulated, and Pure Parotid Saliva. J. Pharm. Biomed. Anal. 2017, 140, 295–300. [Google Scholar] [CrossRef]
- Kawanishi, N.; Hoshi, N.; Masahiro, S.; Enomoto, A.; Ota, S.; Kaneko, M.; Soga, T.; Tomita, M.; Kimoto, K. Effects of Inter-Day and Intra-Day Variation on Salivary Metabolomic Profiles. Clin. Chim. Acta 2019, 489, 41–48. [Google Scholar] [CrossRef]
- Na, H.S.; Kim, S.; Kim, S.; Yu, Y.; Kim, S.Y.; Kim, H.J.; Lee, J.Y.; Lee, J.H.; Chung, J. Molecular Subgroup of Periodontitis Revealed by Integrated Analysis of the Microbiome and Metabolome in a Cross-Sectional Observational Study. J. Oral Microbiol. 2021, 13, 1902707. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.J.; Song, Y.; Lee, H.A.; Kim, S.; Chung, J. Metabolic Phenotyping of Saliva to Identify Possible Biomarkers of Periodontitis Using Proton Nuclear Magnetic Resonance. J. Clin. Periodontol. 2021, 48, 1240–1249. [Google Scholar] [CrossRef]
- Sugimoto, M.; Ota, S.; Kaneko, M.; Enomoto, A.; Soga, T. Quantification of Salivary Charged Metabolites Using Capillary Electrophoresis Time-of-Flight-Mass Spectrometry. Bio Protoc. 2020, 10, e3797. [Google Scholar] [CrossRef]
- Sugimoto, M.; Saruta, J.; Matsuki, C.; To, M.; Onuma, H.; Kaneko, M.; Soga, T.; Tomita, M.; Tsukinoki, K. Physiological and Environmental Parameters Associated with Mass Spectrometry-Based Salivary Metabolomic Profiles. Metabolomics 2013, 9, 454–463. [Google Scholar] [CrossRef]
- Okuma, N.; Saita, M.; Hoshi, N.; Soga, T.; Tomita, M.; Sugimoto, M.; Kimoto, K. Effect of Masticatory Stimulation on the Quantity and Quality of Saliva and the Salivary Metabolomic Profile. PLoS ONE 2017, 12, e0183109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomita, A.; Mori, M.; Hiwatari, K.; Yamaguchi, E.; Itoi, T.; Sunamura, M.; Soga, T.; Tomita, M.; Sugimoto, M. Effect of Storage Conditions on Salivary Polyamines Quantified Via Liquid Chromatography-Mass Spectrometry. Sci. Rep. 2018, 8, 12075. [Google Scholar] [CrossRef] [PubMed]
- Aristizabal-Henao, J.J.; Lemas, D.J.; Griffin, E.K.; Costa, K.A.; Camacho, C.; Bowden, J.A. Metabolomic Profiling of Biological Reference Materials Using a Multiplatform High-Resolution Mass Spectrometric Approach. J. Am. Soc. Mass. Spectrom. 2021, 32, 2481–2489. [Google Scholar] [CrossRef]
- Philip, P.M.; Nayak, P.; Philip, S.; Parambil, N.A.; Duraisamy, K.; Balasubramanian, S. Population-Based Cancer Screening through Community Participation: Outcome of a District Wide Oral Cancer Screening Program from Rural Kannur, Kerala, India. South Asian J. Cancer 2018, 7, 244–248. [Google Scholar] [CrossRef]
- Rajaraman, P.; Anderson, B.O.; Basu, P.; Belinson, J.L.; Cruz, A.D.; Dhillon, P.K.; Gupta, P.; Jawahar, T.S.; Joshi, N.; Kailash, U.; et al. Recommendations for Screening and Early Detection of Common Cancers in India. Lancet Oncol. 2015, 16, e352–e361. [Google Scholar] [CrossRef]
- D’Cruz, A.K.; Vaish, R. Risk-Based Oral Cancer Screening - Lessons to Be Learnt. Nat. Rev. Clin. Oncol. 2021, 18, 471–472. [Google Scholar] [CrossRef]
- Kowalski, L.P.; Carvalho, A.L. Influence of Time Delay and Clinical Upstaging in the Prognosis of Head and Neck Cancer. Oral. Oncol. 2001, 37, 94–98. [Google Scholar] [CrossRef]
- Messadi, D.V.; Wilder-Smith, P.; Wolinsky, L. Improving Oral Cancer Survival: The Role of Dental Providers. J. Calif. Dent. Assoc. 2009, 37, 789–798. [Google Scholar]
- Strome, A.; Kossatz, S.; Zanoni, D.K.; Rajadhyaksha, M.; Patel, S.; Reiner, T. Current Practice and Emerging Molecular Imaging Technologies in Oral Cancer Screening. Mol. Imaging 2018, 17, 1536012118808644. [Google Scholar] [CrossRef] [Green Version]
- Teh, M.T.; Ma, H.; Liang, Y.Y.; Solomon, M.C.; Chaurasia, A.; Patil, R.; Tekade, S.A.; Mishra, D.; Qadir, F.; Yeung, J.S.; et al. Molecular Signatures of Tumour and Its Microenvironment for Precise Quantitative Diagnosis of Oral Squamous Cell Carcinoma: An International Multi-Cohort Diagnostic Validation Study. Cancers 2022, 14, 1389. [Google Scholar] [CrossRef]
- Breier, M.; Wahl, S.; Prehn, C.; Fugmann, M.; Ferrari, U.; Weise, M.; Banning, F.; Seissler, J.; Grallert, H.; Adamski, J.; et al. Targeted Metabolomics Identifies Reliable and Stable Metabolites in Human Serum and Plasma Samples. PLoS ONE 2014, 9, e89728. [Google Scholar] [CrossRef]
- González-Domínguez, R.; González-Domínguez, Á.; Sayago, A.; Fernández-Recamales, Á. Recommendations and Best Practices for Standardizing the Pre-Analytical Processing of Blood and Urine Samples in Metabolomics. Metabolites 2020, 10, 229. [Google Scholar] [CrossRef]
- Patil, D.J.; More, C.B. Salivary Metabolomics–a Diagnostic and Biologic Signature for Oral Cancer. J. Oral Maxillofac. Surg. Med. Pathol. 2021, 33, 546–554. [Google Scholar] [CrossRef]
- Urdiales, J.L.; Medina, M.Á.; Sánchez-Jiménez, F. Polyamine Metabolism Revisited. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1015–1019. [Google Scholar] [CrossRef]
- Bachmann, A.S.; Geerts, D. Polyamine Synthesis as a Target of Myc Oncogenes. J. Biol. Chem. 2018, 293, 18757–18769. [Google Scholar] [CrossRef] [Green Version]
- Tanuma, J.; Shisa, H.; Hiai, H.; Higashi, S.; Yamada, Y.; Kamoto, T.; Hirayama, Y.; Matsuuchi, H.; Kitano, M. Quantitative Trait Loci Affecting 4-Nitroquinoline 1-Oxide-Induced Tongue Carcinogenesis in the Rat. Cancer Res. 1998, 58, 1660–1664. [Google Scholar]
- Zhao, L.; Cheng, Z.; Lu, Z.; Jin, J. Nad-Dependent Methylenetetrahydrofolate Dehydrogenase Inhibits Oral Squamous Cell Carcinoma Cell Proliferation and Promotes Apoptosis. Transl. Cancer Res. 2021, 10, 1457–1469. [Google Scholar] [CrossRef]
- Sawczuk, B.; Maciejczyk, M.; Sawczuk-Siemieniuk, M.; Posmyk, R.; Zalewska, A.; Car, H. Salivary Gland Function, Antioxidant Defence and Oxidative Damage in the Saliva of Patients with Breast Cancer: Does the Brca1 Mutation Disturb the Salivary Redox Profile? Cancers 2019, 11, 1501. [Google Scholar] [CrossRef] [Green Version]
- Lau, C.S.; Wong, D.T. Breast Cancer Exosome-Like Microvesicles and Salivary Gland Cells Interplay Alters Salivary Gland Cell-Derived Exosome-Like Microvesicles in Vitro. PLoS ONE 2012, 7, e33037. [Google Scholar] [CrossRef]
- Tse, R.T.; Wong, C.Y.; Chiu, P.K.; Ng, C.F. The Potential Role of Spermine and Its Acetylated Derivative in Human Malignancies. Int. J. Mol. Sci. 2022, 23, 1258. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Katsumata, K.; Kuwabara, H.; Soya, R.; Enomoto, M.; Ishizaki, T.; Tsuchida, A.; Mori, M.; Hiwatari, K.; Soga, T.; et al. Urinary Polyamine Biomarker Panels with Machine-Learning Differentiated Colorectal Cancers, Benign Disease, and Healthy Controls. Int. J. Mol. Sci. 2018, 19, 756. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Katsumata, K.; Udo, R.; Tago, T.; Kasahara, K.; Mazaki, J.; Kuwabara, H.; Kawakita, H.; Enomoto, M.; Ishizaki, T.; et al. Validation of Urinary Charged Metabolite Profiles in Colorectal Cancer Using Capillary Electrophoresis-Mass Spectrometry. Metabolites 2022, 12, 59. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.W.; Kang, W.P.; Huang, B.L.; Qiu, Z.H.; Wei, L.F.; Zhang, B.; Ding, T.Y.; Luo, Y.; Liu, C.T.; Chu, L.Y.; et al. Nomogram Based on Clinical Characteristics and Serological Inflammation Markers to Predict Overall Survival of Oral Tongue Squamous Cell Carcinoma Patient after Surgery. BMC Oral Health 2021, 21, 667. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Shibahara, T.; Takano, M.; Iwamoto, M.; Takaki, T.; Kasahara, K.; Nomura, T.; Takano, N.; Katakura, A. Countermeasure and Opportunistic Screening Systems for Oral Cancer. Oral Oncol. 2021, 112, 105047. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Warnakulasuriya, S.; Lingen, M.W.; Kerr, A.R.; Ogden, G.R.; Glenny, A.M.; Macey, R. Clinical Assessment for the Detection of Oral Cavity Cancer and Potentially Malignant Disorders in Apparently Healthy Adults. Cochrane Database Syst. Rev. 2021, 12, Cd010173. [Google Scholar] [PubMed]
- Ren, R.; Luo, H.; Su, C.; Yao, Y.; Liao, W. Machine Learning in Dental, Oral and Craniofacial Imaging: A Review of Recent Progress. PeerJ 2021, 9, e11451. [Google Scholar] [CrossRef]
- Shah, P.; Roy, N.; Dhandhukia, P. Algorithm Mediated Early Detection of Oral Cancer from Image Analysis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2022, 133, 70–79. [Google Scholar] [CrossRef]


| Analytical Instrument | Discrimination Method or Univariant Statistics | Discrimination Design | Accuracy (AUC or p-Value) a | Ref. |
|---|---|---|---|---|
| RPLC-MS and HILIC-MS | MLR | OSCC from HC | 0.997 (Stage I–II) | 20 |
| 0.971 (Stage III–IV) | ||||
| CE-TOFMS | Wilcoxon rank sum test | OSCC vs. HC | 0.00006 b | 21 |
| GC-MS | Mann–Whitney test with FDR correction | OSCC vs. HC | 3.1755 × 10−16 c | 22 |
| CPSI-MS | Lasso regression model | OSCC from HC | 0.992 | 23 |
| PML from HC | 0.978 | |||
| OSC from PML | 0.917 | |||
| UPLC-QTOFMS | MLR | OSCC from HC | 0.89 | 24 |
| OSFF from OLK | 0.97 | |||
| CE-TOFMS | MLR | OC from OLP | 0.865 | 25 |
| LC-QTOFMS | ANOVA | OC vs. HC | <0.05 d | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panneerselvam, K.; Ishikawa, S.; Krishnan, R.; Sugimoto, M. Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites 2022, 12, 436. https://doi.org/10.3390/metabo12050436
Panneerselvam K, Ishikawa S, Krishnan R, Sugimoto M. Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites. 2022; 12(5):436. https://doi.org/10.3390/metabo12050436
Chicago/Turabian StylePanneerselvam, Karthika, Shigeo Ishikawa, Rajkumar Krishnan, and Masahiro Sugimoto. 2022. "Salivary Metabolomics for Oral Cancer Detection: A Narrative Review" Metabolites 12, no. 5: 436. https://doi.org/10.3390/metabo12050436
APA StylePanneerselvam, K., Ishikawa, S., Krishnan, R., & Sugimoto, M. (2022). Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites, 12(5), 436. https://doi.org/10.3390/metabo12050436