Multi-Strain and -Species Investigation of Volatile Metabolites Emitted from Planktonic and Biofilm Candida Cultures
Abstract
1. Introduction
2. Results
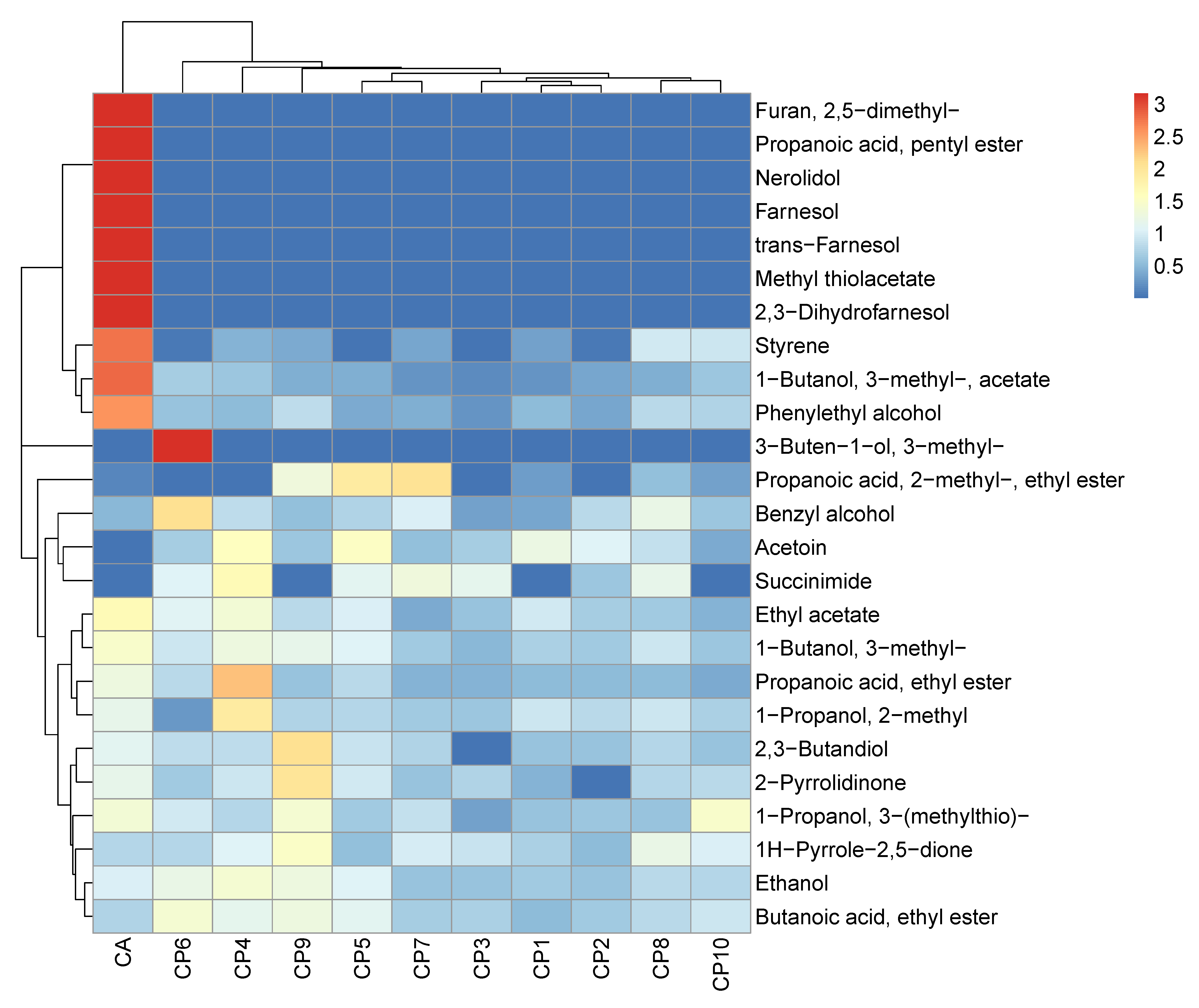
2.1. Discriminative Volatilomics of Planktonic Candida spp. at the Species- and Strain-Level
2.2. Chemical Composition of Planktonic Candida Volatilomes
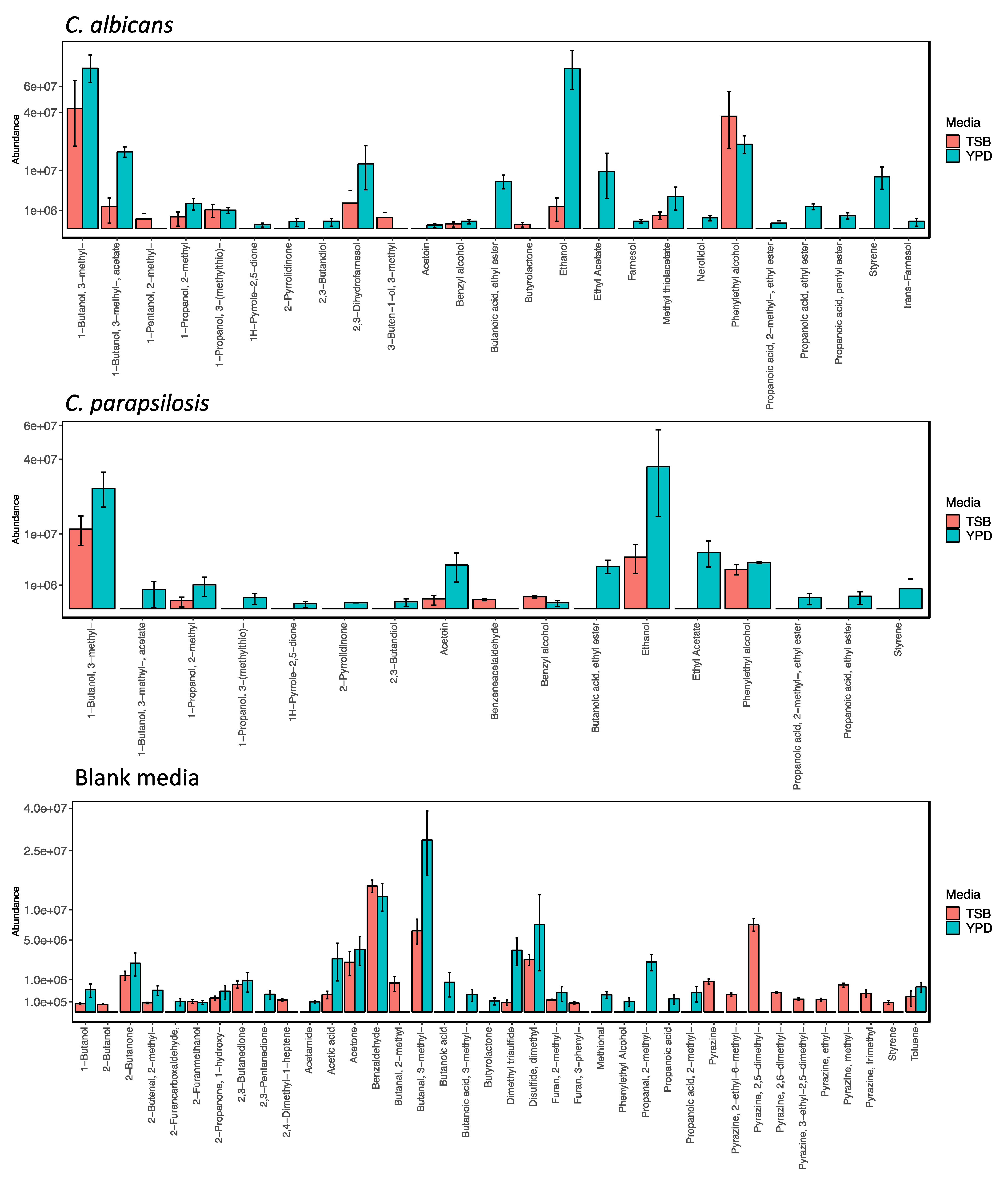
2.3. Media-Dependent Influences on Planktonic Candida Volatilomes
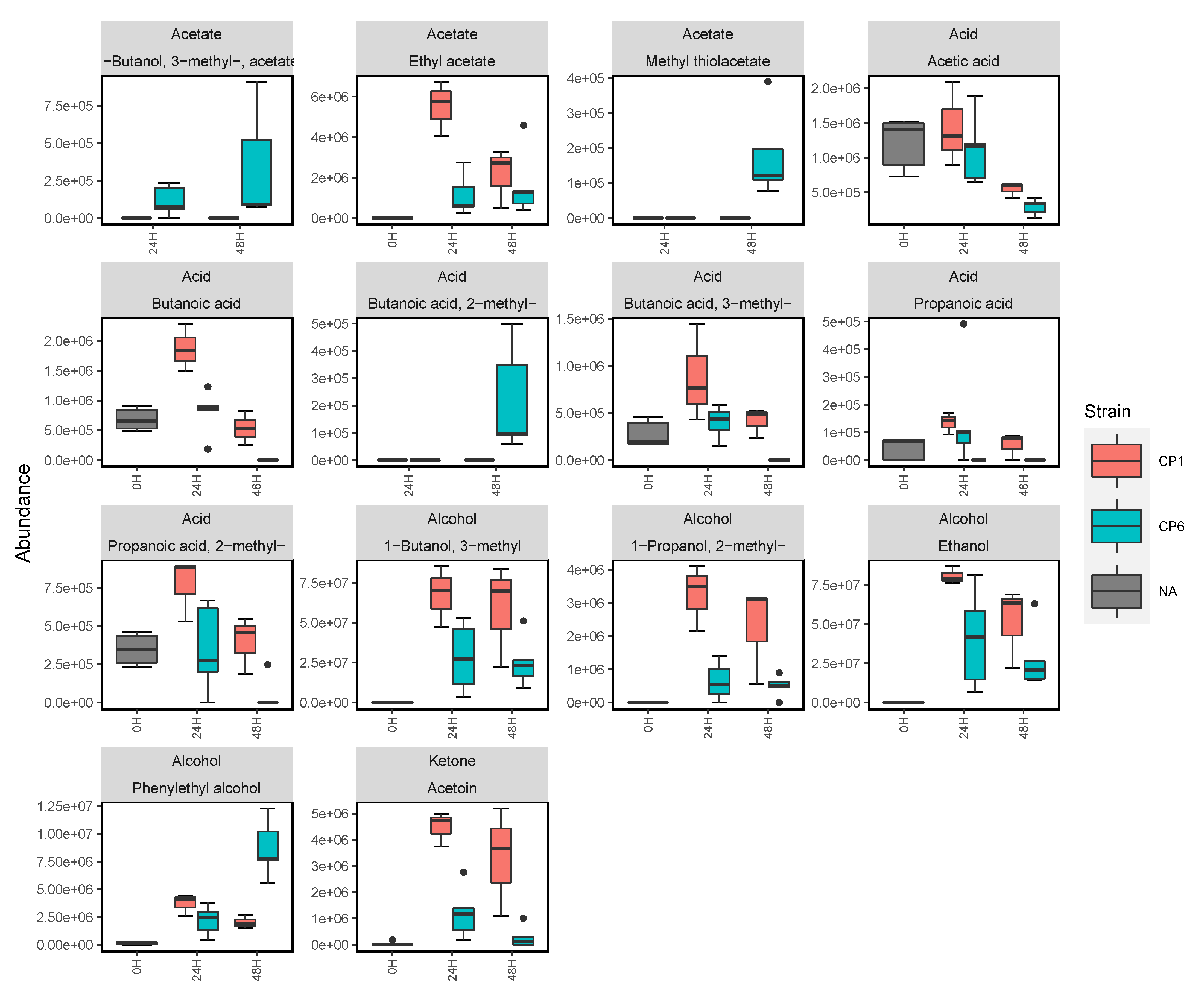
2.4. Biofilm-Dependent Influences on Candida Volatilomes
3. Discussion
4. Methods
4.1. Growth of Candida Planktonic Samples
4.2. Growth of Candida Biofilm Samples
4.3. Crystal Violet Staining of Biofilm Samples
4.4. HS-SPME Sampling
4.5. Gas Chromatography–Mass Spectrometry
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trofa, D.; Gácser, A.; Nosanchuk, J. Candida parapsilosis, an Emerging Fungal Pathogen. Clin. Microbiol. Rev. 2008, 21, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Holland, L.; Butler, G.; Gacser, A.; Bliss, J. Candida parapsilosis Is a Significant Neonatal Pathogen. Pediatr. Infect. Dis. J. 2013, 32, e206–e216. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, S.; Butler, G. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 2005, 151, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef]
- Martins, M.; Henriques, M.; Azeredo, J.; Rocha, S.; Coimbra, M.; Oliveira, R. Morphogenesis Control in Candida albicans and Candida dubliniensis through Signaling Molecules Produced by Planktonic and Biofilm Cells. Eukaryot. Cell 2007, 6, 2429–2436. [Google Scholar] [CrossRef]
- Weisskopf, L.; Schulz, S.; Garbeva, P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 2021, 19, 391–404. [Google Scholar] [CrossRef]
- Schulz, S.; Dickschat, J. Bacterial Volatiles: The Smell of Small Organisms. ChemInform 2007, 24, 814–842. [Google Scholar] [CrossRef]
- Schulz, S.; Schlawis, C.; Koteska, D.; Harig, T.; Biwer, P. Chapter 3: Structural Diversity of Bacterial Volatiles. In Bacterial Volatile Compounds as Mediators of Airborne Interactions, 1st ed.; Ryu, C.M., Weisskopf, L., Piechulla, B., Eds.; Springer: Singapore, 2020; pp. 93–116. [Google Scholar]
- Rees, C.; Nordick, K.; Franchina, F.; Lewis, A.; Hirsch, E.; Hill, J. Volatile metabolic diversity of Klebsiella pneumoniae in nutrient-replete conditions. Metabolomics 2017, 13, 18. [Google Scholar] [CrossRef]
- Jenkins, C.; Bean, H. Influence of media on the differentiation of Staphylococcus spp. by volatile compounds. J. Breath Res. 2019, 14, 016007. [Google Scholar] [CrossRef]
- Fitzgerald, S.; Holland, L.; Morrin, A. An Investigation of Stability and Species and Strain-Level Specificity in Bacterial Volatilomes. Front. Microbiol. 2021, 12, 693075. [Google Scholar] [CrossRef]
- Boots, A.W.; Smolinska, A.; van Berkel1, J.J.B.N.; Fijten, R.R.R.; Stobberingh, E.E.; Boumans, M.L.L.; Moonen, E.J.; Wouters, E.F.M.; Dallinga, J.W.; van Schooten, F.J. Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography–mass spectrometry. J. Breath Res. 2014, 8, 027106. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, S.; Duffy, E.; Holland, L.; Morrin, A. Multi-strain volatile profiling of pathogenic and commensal cutaneous bacteria. Sci. Rep. 2020, 10, 17971. [Google Scholar] [CrossRef]
- Insam, H.; Seewald, M. Volatile organic compounds (VOCs) in soils. Biol. Fertil. Soils 2010, 46, 199–213. [Google Scholar] [CrossRef]
- Inamdar, A.; Morath, S.; Bennett, J. Fungal Volatile Organic Compounds: More Than Just a Funky Smell? Annu. Rev. Microbiol. 2020, 74, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Cordovez, V.; de Boer, W.; Raaijmakers, J.; Garbeva, P. Volatile affairs in microbial interactions. ISME J. 2015, 9, 2329–2335. [Google Scholar] [CrossRef] [PubMed]
- Duffy, E.; Morrin, A. Endogenous and microbial volatile organic compounds in cutaneous health and disease. TrAC Trends Anal. Chem. 2019, 111, 163–172. [Google Scholar] [CrossRef]
- Ryu, C.; Weisskopf, L.; Piechulla, B. Bacterial Volatile Compounds as Mediators of Airborne Interactions; Springer: Singapore, 2020. [Google Scholar]
- Costa, C.; Bezerra, A.; Almeida, A.; Rocha, S. Candida Species (Volatile) Metabotyping through Advanced Comprehensive Two-Dimensional Gas Chromatography. Microorganisms 2020, 8, 1911. [Google Scholar] [CrossRef] [PubMed]
- López-Ramos, J.; Bautista, E.; Gutiérrez-Escobedo, G.; Mancilla-Montelongo, G.; Castaño, I.; González-Chávez, M.; De Las Peñas, A. Analysis of Volatile Molecules Present in the Secretome of the Fungal Pathogen Candida glabrata. Molecules 2021, 26, 3881. [Google Scholar] [CrossRef]
- Perl, T.; Jünger, M.; Vautz, W.; Nolte, J.; Kuhns, M.; Borg-von Zepelin, M.; Quintel, M. Detection of characteristic metabolites of Aspergillus fumigatus and Candida species using ion mobility spectrometry—Metabolic profiling by volatile organic compounds. Mycoses 2011, 54, e828–e837. [Google Scholar] [CrossRef]
- Slade, E.; Thorn, R.; Young, A.; Reynolds, D. Real-time detection of volatile metabolites enabling species-level discrimination of bacterial biofilms associated with wound infection. J. Appl. Microbiol. 2019, 111, 163–172. [Google Scholar] [CrossRef]
- Bhargava, P.; Collins, J. Boosting Bacterial Metabolism to Combat Antibiotic Resistance. Cell Metab. 2015, 21, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Serrazanetti, D.; Ndagijimana, M.; Sado-Kamdem, S.; Corsetti, A.; Vogel, R.; Ehrmann, M.; Guerzoni, M. Acid Stress-Mediated Metabolic Shift in Lactobacillus sanfranciscensis LSCE1. Appl. Environ. Microbiol. 2011, 77, 2656–2666. [Google Scholar] [CrossRef] [PubMed]
- Mitri, S.; Koubaa, M.; Maroun, R.; Rossignol, T.; Nicaud, J.; Louka, N. Bioproduction of 2-Phenylethanol through Yeast Fermentation on Synthetic Media and on Agro-Industrial Waste and By-Products: A Review. Foods 2022, 11, 109. [Google Scholar] [CrossRef]
- McNerney, R.; Mallard, K.; Okolo, P.; Turner, C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol. Lett. 2012, 328, 150–156. [Google Scholar] [CrossRef]
- Saerens, S.; Delvaux, F.; Verstrepen, K.; Thevelein, J. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microb. Biotechnol. 2010, 3, 165–177. [Google Scholar] [CrossRef]
- Wolfe, A. The Acetate Switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef]
- Guo, W.; Sheng, J.; Feng, X. 13C-Metabolic Flux Analysis: An Accurate Approach to Demystify Microbial Metabolism for Biochemical Production. Bioengineering 2015, 3, 3. [Google Scholar] [CrossRef]
- Long, C.; Antoniewicz, M. High-resolution 13C metabolic flux analysis. Nat. Protoc. 2019, 14, 2856–2877. [Google Scholar] [CrossRef]
- Rodrigues, C.; Černáková, L. Farnesol and Tyrosol: Secondary Metabolites with a Crucial quorum-sensing Role in Candida Biofilm Development. Genes 2020, 11, 444. [Google Scholar] [CrossRef]
- Lindsay, A.; Deveau, A.; Piispanen, A.; Hogan, D. Farnesol and Cyclic AMP Signaling Effects on the Hypha-to-Yeast Transition in Candida albicans. Eukaryot. Cell 2012, 11, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Hornby, J.; Jensen, E.; Lisec, A.; Tasto, J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K. Quorum Sensing in the Dimorphic Fungus Candida albicans Is Mediated by Farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed]
- Jabra-Rizk, M.; Meiller, T.; James, C.; Shirtliff, M. Effect of Farnesol on Staphylococcus aureus Biofilm Formation and Antimicrobial Susceptibility. Antimicrob. Agents Chemother. 2006, 50, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Liang, R.; Hicks, J.; Barrish, J.; Versalovic, J. Farnesol Decreases Biofilms of Staphylococcus epidermidis and Exhibits Synergy With Nafcillin and Vancomycin. Pediatr. Res. 2011, 70, 578–583. [Google Scholar] [CrossRef]
- Strobel, G.; Sears, J.; Dirkse, E.; Markworth, C. Volatile antimicrobials from Muscodor albus, a novel endophytic fungus. Microbiology 2001, 147, 2943–2950. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.; Sohr, R.; Schulz, B.; Fleischhacker, M.; Ruhnke, M. Secretion of E,E-Farnesol and Biofilm Formation in Eight Different Candida Species. Antimicrob. Agents Chemother. 2009, 53, 848. [Google Scholar] [CrossRef]
- Ezra, D.; Hess, W.; Strobel, G. New endophytic isolates of Muscodor albus, a volatile-antibiotic-producing fungus. Microbiology 2004, 150, 4023–4031. [Google Scholar] [CrossRef]
- Mitchell, A.; Strobel, G.; Moore, E.; Robison, R.; Sears, J. Volatile antimicrobials from Muscodor crispans, a novel endophytic fungus. Microbiology 2010, 156, 270–277. [Google Scholar] [CrossRef]
- Stinson, M.; Ezra, D.; Hess, W.; Sears, J.; Strobel, G. An endophytic Gliocladium sp. of Eucryphia cordifolia producing selective volatile antimicrobial compounds. Plant Sci. 2003, 165, 913–922. [Google Scholar] [CrossRef]
- Singh, S.; Strobel, G.; Knighton, B.; Geary, B.; Sears, J.; Ezra, D. An Endophytic Phomopsis sp. Possessing Bioactivity and Fuel Potential with its Volatile Organic Compounds. Microb. Ecol. 2011, 61, 729–739. [Google Scholar] [CrossRef]
- Hou, Q.; Keren-Paz, A.; Korenblum, E.; Oved, R.; Malitsky, S.; Kolodkin-Gal, I. Weaponizing volatiles to inhibit competitor biofilms from a distance. NPJ Biofilms Microbiomes 2021, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Slade, E.; Thorn, R.; Young, A.; Reynolds, D. An in vitro collagen perfusion wound biofilm model; with applications for antimicrobial studies and microbial metabolomics. BMC Microbiol. 2019, 19, 310. [Google Scholar] [CrossRef] [PubMed]
- Frade, J.; Arthington-Skaggs, B. Effect of serum and surface characteristics on Candida albicans biofilm formation. Mycoses 2010, 54, e154–e162. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Fujarte, I.; López-Romero, E.; Reyna-López, G.; Martínez-Gámez, M.; Vega-González, A.; Cuéllar-Cruz, M. Influence of Culture Media on Biofilm Formation by Candida Species and Response of Sessile Cells to Antifungals and Oxidative Stress. BioMed Res. Int. 2015, 2015, 783639. [Google Scholar] [CrossRef]
- Chen, Y.; Gozzi, K.; Yan, F.; Chai, Y. Acetic Acid Acts as a Volatile Signal to Stimulate Bacterial Biofilm Formation. MBio 2015, 6, e00392-15. [Google Scholar] [CrossRef]
- Xiao, Z.; Xu, P. Acetoin Metabolism in Bacteria. Crit. Rev. Microbiol. 2007, 33, 127–140. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Shang, Q.; Jiang, Y.; Cao, Y.; Chai, Y. Time Course Analysis of Candida albicans Metabolites during Biofilm Development. J. Proteome Res. 2012, 12, 2375–2385. [Google Scholar] [CrossRef]
- Lei, X.; Deng, B.; Ruan, C.; Deng, L.; Zeng, K. Phenylethanol as a quorum sensing molecule to promote biofilm formation of the antagonistic yeast Debaryomyces nepalensis for the control of black spot rot on jujube. Postharvest Biol. Technol. 2022, 185, 111788. [Google Scholar] [CrossRef]
- Chauhan, N.; Mohan Karuppayil, S. Dual identities for various alcohols in two different yeasts. Mycology 2021, 12, 25–38. [Google Scholar] [CrossRef]
- Pu, L.; Jingfan, F.; Kai, C.; Chao-an, L.; Yunjiang, C. Phenylethanol promotes adhesion and biofilm formation of the antagonistic yeast Kloeckera apiculate for the control of blue mold on citrus. FEMS Yeast Res. 2014, 14, 536–546. [Google Scholar] [CrossRef]
- Han, T.; Tumanov, S.; Cannon, R.; Villas-Boas, S. Metabolic Response of Candida albicans to Phenylethyl Alcohol under Hyphae-Inducing Conditions. PLoS ONE 2013, 8, e71364. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.; Johnson, A. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Blank, H.; Gajjar, S.; Belyanin, A.; Polymenis, M. Sulfur Metabolism Actively Promotes Initiation of Cell Division in Yeast. PLoS ONE 2009, 4, e8018. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, J.; Zeef, L.; Hoyle, D.; Zhang, N.; Hayes, A.; Gardner, D.; Cornell, M.; Petty, J.; Hakes, L.; Wardleworth, L.; et al. Growth control of the eukaryote cell: A systems biology study in yeast. J. Biol. 2007, 6, 4. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fitzgerald, S.; Furlong, C.; Holland, L.; Morrin, A. Multi-Strain and -Species Investigation of Volatile Metabolites Emitted from Planktonic and Biofilm Candida Cultures. Metabolites 2022, 12, 432. https://doi.org/10.3390/metabo12050432
Fitzgerald S, Furlong C, Holland L, Morrin A. Multi-Strain and -Species Investigation of Volatile Metabolites Emitted from Planktonic and Biofilm Candida Cultures. Metabolites. 2022; 12(5):432. https://doi.org/10.3390/metabo12050432
Chicago/Turabian StyleFitzgerald, Shane, Ciara Furlong, Linda Holland, and Aoife Morrin. 2022. "Multi-Strain and -Species Investigation of Volatile Metabolites Emitted from Planktonic and Biofilm Candida Cultures" Metabolites 12, no. 5: 432. https://doi.org/10.3390/metabo12050432
APA StyleFitzgerald, S., Furlong, C., Holland, L., & Morrin, A. (2022). Multi-Strain and -Species Investigation of Volatile Metabolites Emitted from Planktonic and Biofilm Candida Cultures. Metabolites, 12(5), 432. https://doi.org/10.3390/metabo12050432




