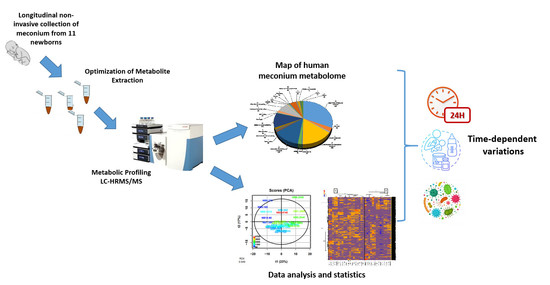
The Human Meconium Metabolome and Its Evolution during the First Days of Life
Abstract
:1. Introduction
2. Results
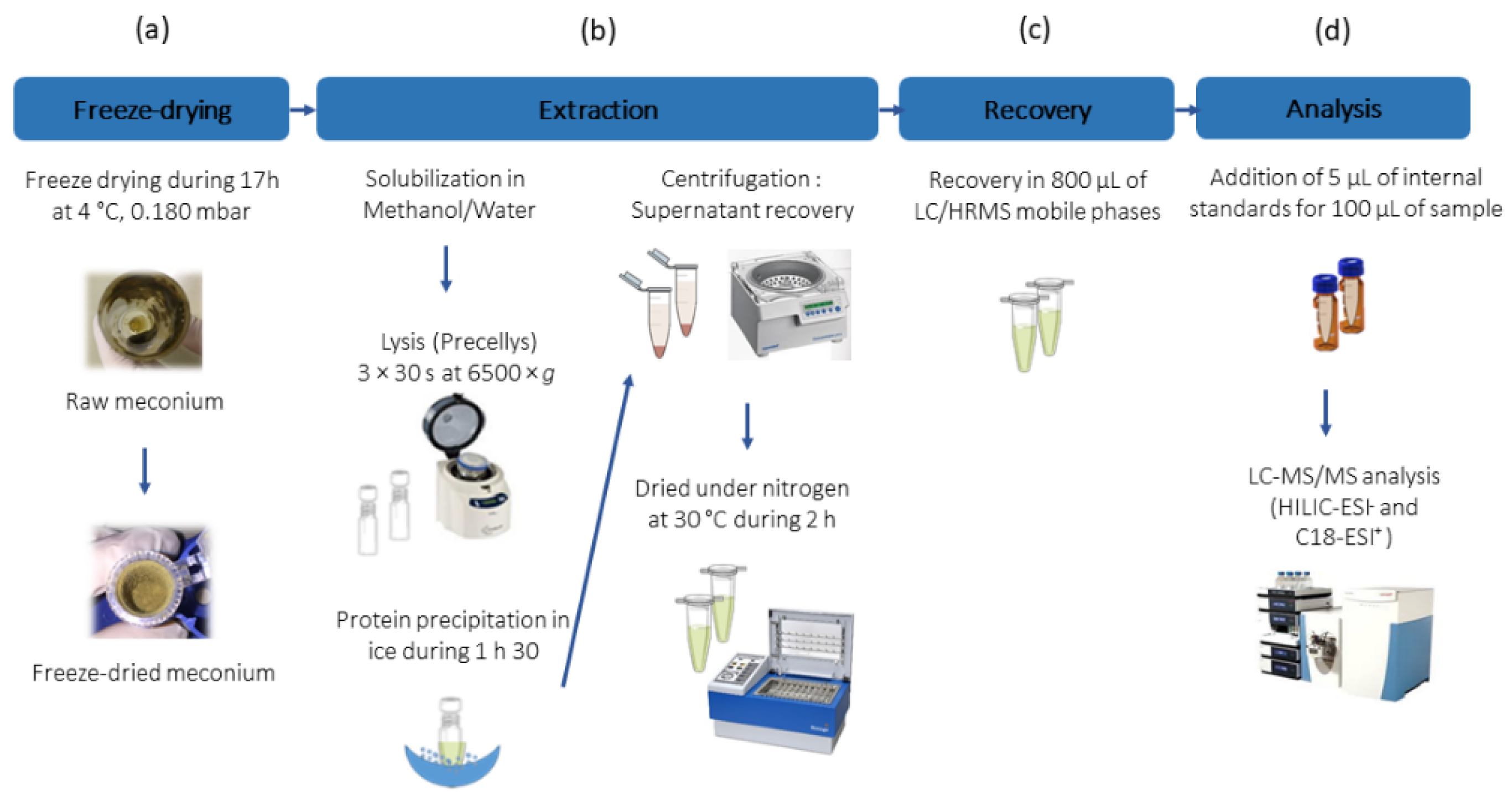
2.1. Optimization of the Workflow for Metabolome Analysis of Meconium
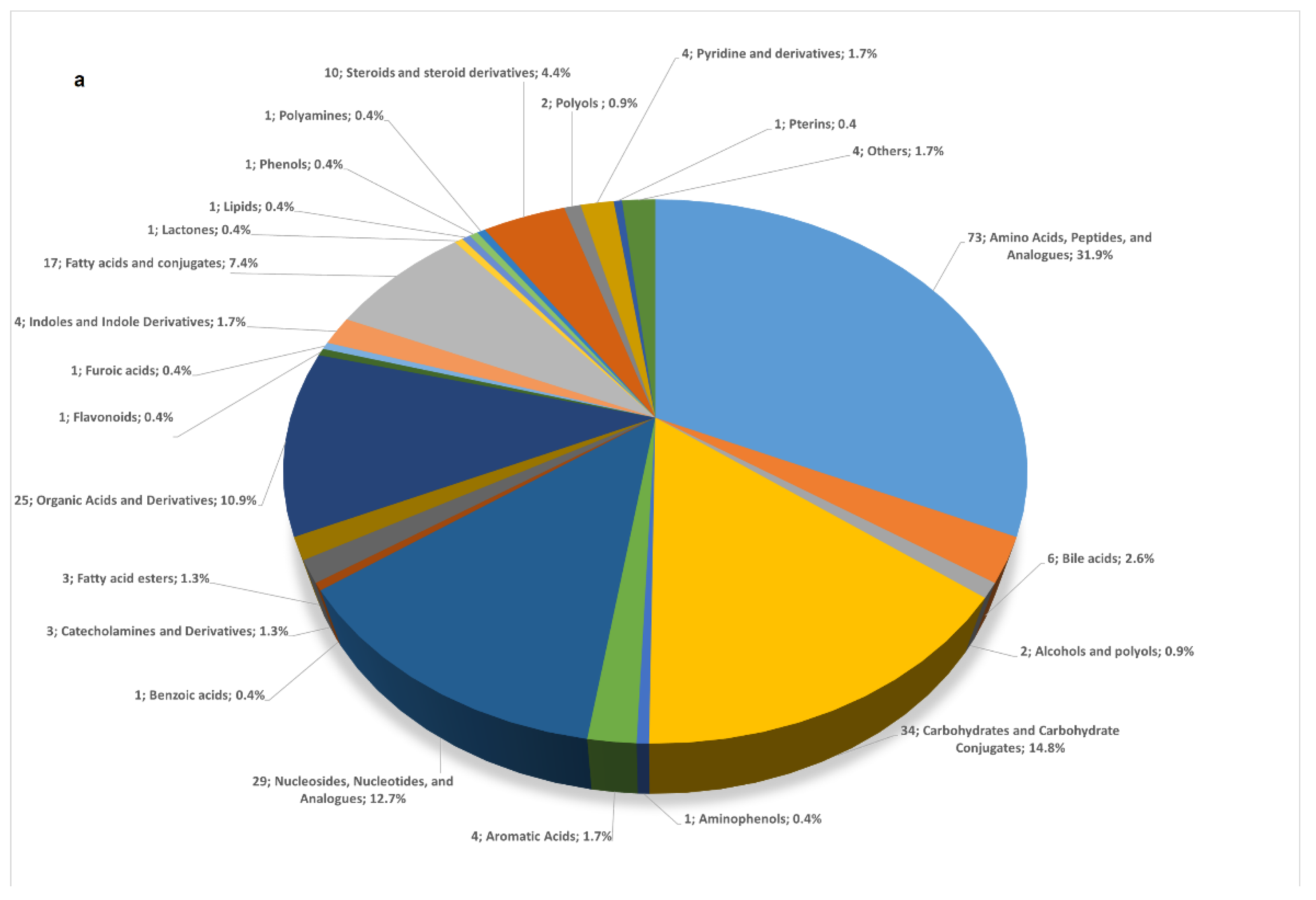
2.2. Characterization of the Human Meconium Metabolome
2.2.1. Comprehensive View of the Meconium Metabolome
2.2.2. A Map of the Human Meconium Metabolome
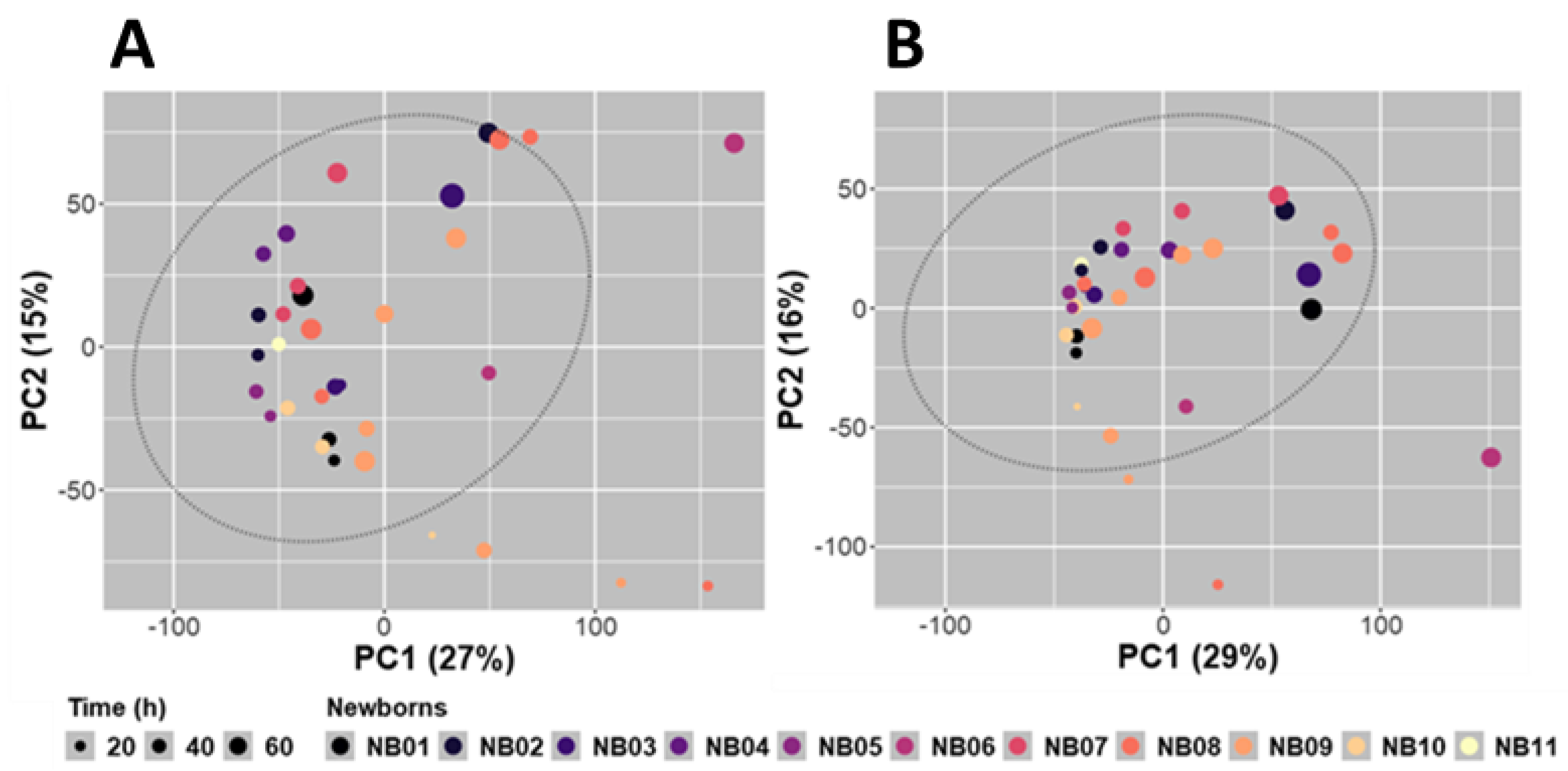
2.3. Analysis of the Whole Dataset Evidenced Rapid Evolution of the Meconium Metabolome during the First Days of Life
2.3.1. Preliminary Analysis of the Whole Dataset through Non-Supervised PCA and Hierarchical Clustering
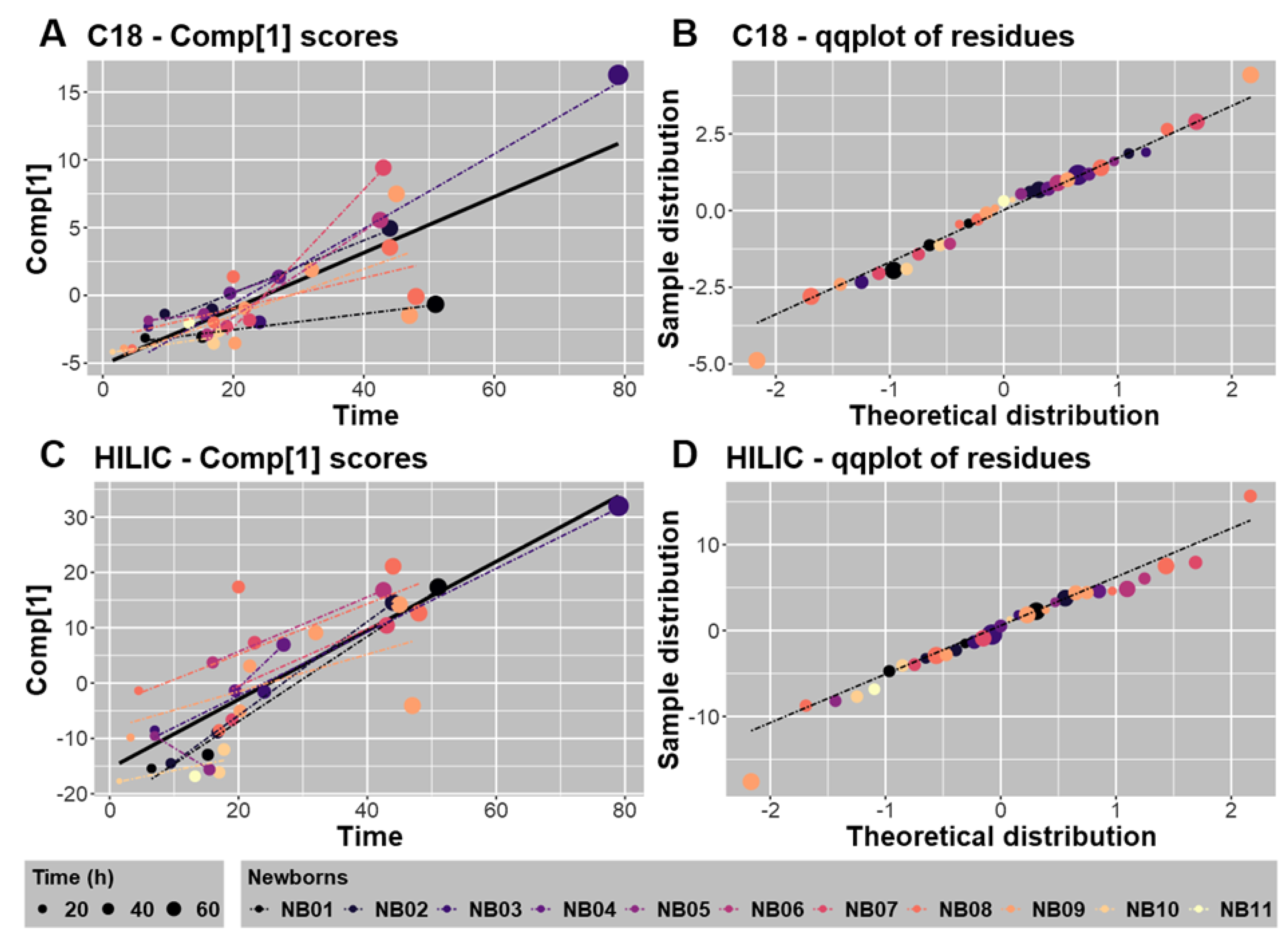
2.3.2. Canonical and Regression Analyses Confirmed a Major Impact of Time on the Meconium Metabolome
2.3.3. Modeling of the Whole Datasets and Metabolic Changes Associated with Time-Dependent Variation
2.4. Identification of Metabolites Showing Time-Dependent Variations: Complementarity of Non-Supervised and Supervised Analyses
3. Discussion
4. Material and Methods
4.1. Study Subjects
4.2. Meconium Sample Preparation
4.3. Metabolic Profiling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyer-Monath, M.; Chatellier, C.; Rouget, F.; Morel, I.; Lestremau, F. Development of a Multi-Residue Method in a Fetal Matrix: Analysis of Meconium. Anal. Bioanal. Chem. 2014, 406, 7785–7797. [Google Scholar] [CrossRef] [PubMed]
- Berton, T.; Mayhoub, F.; Chardon, K.; Duca, R.-C.; Lestremau, F.; Bach, V.; Tack, K. Development of an Analytical Strategy Based on LC–MS/MS for the Measurement of Different Classes of Pesticides and Theirs Metabolites in Meconium: Application and Characterisation of Foetal Exposure in France. Environ. Res. 2014, 132, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Unsworth, J.; Vause, S. Meconium in Labour. Obstet. Gynaecol. Reprod. Med. 2010, 20, 289–294. [Google Scholar] [CrossRef]
- Ostrea, E.M. Testing for Exposure to Illicit Drugs and Other Agents in the Neonate: A Review of Laboratory Methods and the Role of Meconium Analysis. Curr. Probl. Pediatr. 1999, 29, 41–56. [Google Scholar] [CrossRef]
- Ostrea, E.M. Meconium Analysis to Assess Fetal Exposure to Nicotine by Active and Passive Maternal Smoking. J. Pediatr. 1994, 124, 471–476. [Google Scholar] [CrossRef]
- Ostrea, E.M.; Bielawski, D.M.; Posecion, N.C.; Corrion, M.; Villanueva-Uy, E.; Bernardo, R.C.; Jin, Y.; Janisse, J.J.; Ager, J.W. Combined Analysis of Prenatal (Maternal Hair and Blood) and Neonatal (Infant Hair, Cord Blood and Meconium) Matrices to Detect Fetal Exposure to Environmental Pesticides. Environ. Res. 2009, 109, 116–122. [Google Scholar] [CrossRef] [Green Version]
- Bearer, C.F.; Lee, S.; Salvator, A.E.; Minnes, S.; Swick, A.; Yamashita, T.; Singer, L.T. Ethyl Linoleate in Meconium: A Biomarker for Prenatal Ethanol Exposure. Alcohol. Clin. Exp. Res. 1999, 23, 487–493. [Google Scholar] [CrossRef]
- Lamy, S.; Hennart, B.; Houivet, E.; Dulaurent, S.; Delavenne, H.; Benichou, J.; Allorge, D.; Marret, S.; Thibaut, F. Assessment of Tobacco, Alcohol and Cannabinoid Metabolites in 645 Meconium Samples of Newborns Compared to Maternal Self-Reports. J. Psychiatr. Res. 2017, 90, 86–93. [Google Scholar] [CrossRef]
- Ristimaa, J.; Gergov, M.; Pelander, A.; Halmesmäki, E.; Ojanperä, I. Broad-Spectrum Drug Screening of Meconium by Liquid Chromatography with Tandem Mass Spectrometry and Time-of-Flight Mass Spectrometry. Anal. Bioanal. Chem. 2010, 398, 925–935. [Google Scholar] [CrossRef]
- Naritaka, N.; Suzuki, M.; Sato, H.; Takei, H.; Murai, T.; Kurosawa, T.; Iida, T.; Nittono, H.; Shimizu, T. Profile of Bile Acids in Fetal Gallbladder and Meconium Using Liquid Chromatography-Tandem Mass Spectrometry. Clin. Chim. Acta 2015, 446, 76–81. [Google Scholar] [CrossRef]
- Peng, S.; Zhang, J.; Liu, L.; Zhang, X.; Huang, Q.; Alamdar, A.; Tian, M.; Shen, H. Newborn Meconium and Urinary Metabolome Response to Maternal Gestational Diabetes Mellitus: A Preliminary Case–Control Study. J. Proteome Res. 2015, 14, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Qin, Y.; Chen, M.; Zhang, Y.; Wang, X.; Dong, T.; Chen, G.; Sun, X.; Lu, T.; White, R.A.; et al. Gestational Diabetes Mellitus Is Associated with the Neonatal Gut Microbiota and Metabolome. BMC Med. 2021, 19, 120. [Google Scholar] [CrossRef] [PubMed]
- Righetti, C.; Peroni, D.G.; Pietrobelli, A.; Zancanaro, C. Proton Nuclear Magnetic Resonance Analysis of Meconium Composition in Newborns. J. Pediatric Gastroenterol. Nutr. 2003, 36, 498–501. [Google Scholar] [CrossRef]
- Jensen, R.G.; Ferris, A.M.; Lammi-Keefe, C.J.; Henderson, R.A. Lipids of Bovine and Human Milks: A Comparison. J. Dairy Sci. 1990, 73, 223–240. [Google Scholar] [CrossRef]
- Del Chierico, F.; Vernocchi, P.; Petrucca, A.; Paci, P.; Fuentes, S.; Praticò, G.; Capuani, G.; Masotti, A.; Reddel, S.; Russo, A.; et al. Phylogenetic and Metabolic Tracking of Gut Microbiota during Perinatal Development. PLoS ONE 2015, 10, e0137347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bittinger, K.; Zhao, C.; Li, Y.; Ford, E.; Friedman, E.S.; Ni, J.; Kulkarni, C.V.; Cai, J.; Tian, Y.; Liu, Q.; et al. Bacterial Colonization Reprograms the Neonatal Gut Metabolome. Nat. Microbiol. 2020, 5, 838–847. [Google Scholar] [CrossRef]
- Wandro, S.; Osborne, S.; Enriquez, C.; Bixby, C.; Arrieta, A.; Whiteson, K. The Microbiome and Metabolome of Preterm Infant Stool Are Personalized and Not Driven by Health Outcomes, Including Necrotizing Enterocolitis and Late-Onset Sepsis. mSphere 2018, 3, e00104-18. [Google Scholar] [CrossRef] [Green Version]
- Petersen, C.; Dai, D.L.Y.; Boutin, R.C.T.; Sbihi, H.; Sears, M.R.; Moraes, T.J.; Becker, A.B.; Azad, M.B.; Mandhane, P.J.; Subbarao, P.; et al. A Rich Meconium Metabolome in Human Infants Is Associated with Early-Life Gut Microbiota Composition and Reduced Allergic Sensitization. Cell Rep. Med. 2021, 2, 100260. [Google Scholar] [CrossRef]
- Boudah, S.; Olivier, M.-F.; Aros-Calt, S.; Oliveira, L.; Fenaille, F.; Tabet, J.-C.; Junot, C. Annotation of the Human Serum Metabolome by Coupling Three Liquid Chromatography Methods to High-Resolution Mass Spectrometry. J. Chromatogr. B 2014, 966, 34–47. [Google Scholar] [CrossRef]
- Roux, A.; Lison, D.; Junot, C.; Heilier, J.-F. Applications of Liquid Chromatography Coupled to Mass Spectrometry-Based Metabolomics in Clinical Chemistry and Toxicology: A Review. Clin. Biochem. 2011, 44, 119–135. [Google Scholar] [CrossRef] [Green Version]
- Aros-Calt, S.; Muller, B.H.; Boudah, S.; Ducruix, C.; Gervasi, G.; Junot, C.; Fenaille, F. Annotation of the Staphylococcus Aureus Metabolome Using Liquid Chromatography Coupled to High-Resolution Mass Spectrometry and Application to the Study of Methicillin Resistance. J. Proteome Res. 2015, 14, 4863–4875. [Google Scholar] [CrossRef] [PubMed]
- Gratton, J.; Phetcharaburanin, J.; Mullish, B.H.; Williams, H.R.T.; Thursz, M.; Nicholson, J.K.; Holmes, E.; Marchesi, J.R.; Li, J.V. Optimized Sample Handling Strategy for Metabolic Profiling of Human Feces. Anal. Chem. 2016, 88, 4661–4668. [Google Scholar] [CrossRef] [PubMed]
- Karu, N.; Deng, L.; Slae, M.; Guo, A.C.; Sajed, T.; Huynh, H.; Wine, E.; Wishart, D.S. A Review on Human Fecal Metabolomics: Methods, Applications and the Human Fecal Metabolome Database. Anal. Chim. Acta 2018, 1030, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Tabone, M.; Bressa, C.; García-Merino, J.A.; Moreno-Pérez, D.; Van, E.C.; Castelli, F.A.; Fenaille, F.; Larrosa, M. The Effect of Acute Moderate-Intensity Exercise on the Serum and Fecal Metabolomes and the Gut Microbiota of Cross-Country Endurance Athletes. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Sindelar, M.; Patti, G.J. Chemical Discovery in the Era of Metabolomics. J. Am. Chem. Soc. 2020, 142, 9097–9105. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Brydges, C.; Fiehn, O.; Mayberg, H.; Kaddurah-Daouk, R. Indoxyl sulfate, a gut microbiome-derived uremic toxin, is associated with psychic anxiety and its functional magnetic resonance imaging-based neurologic signature. Sci. Rep. 2021, 11, 21011. Available online: https://www.nature.com/articles/s41598-021-99845-1 (accessed on 20 December 2021). [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards More Transparent and Integrative Metabolomics Analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Aristizabal-Henao, J.J.; Lemas, D.J.; Griffin, E.K.; Costa, K.A.; Camacho, C.; Bowden, J.A. Metabolomic Profiling of Biological Reference Materials Using a Multiplatform High-Resolution Mass Spectrometric Approach. J. Am. Soc. Mass Spectrom. 2021, 32, 2481–2489. [Google Scholar] [CrossRef]
- Tabone, M.; García-Merino, J.A.; Bressa, C.; Rocha Guzman, N.E.; Herrera Rocha, K.; Chu Van, E.; Castelli, F.A.; Fenaille, F.; Larrosa, M. Chronic Consumption of Cocoa Rich in Procyanidins Has a Marginal Impact on Gut Microbiota and on Serum and Fecal Metabolomes in Male Endurance Athletes. J. Agric. Food Chem. 2022, 70(6), 1878–1889. [Google Scholar] [CrossRef] [PubMed]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-F.; Jacob, D.; et al. Workflow4Metabolomics: A Collaborative Research Infrastructure for Computational Metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunn, A.; Korpela, M. An Introduction to DplR; Washington University: Seattle, WA, USA, 2020. [Google Scholar]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLOS Comput. Biol. 2017, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. arXiv 2014, arXiv:1406.5823. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bekhti, N.; Castelli, F.; Paris, A.; Guillon, B.; Junot, C.; Moiron, C.; Fenaille, F.; Adel-Patient, K. The Human Meconium Metabolome and Its Evolution during the First Days of Life. Metabolites 2022, 12, 414. https://doi.org/10.3390/metabo12050414
Bekhti N, Castelli F, Paris A, Guillon B, Junot C, Moiron C, Fenaille F, Adel-Patient K. The Human Meconium Metabolome and Its Evolution during the First Days of Life. Metabolites. 2022; 12(5):414. https://doi.org/10.3390/metabo12050414
Chicago/Turabian StyleBekhti, Nihel, Florence Castelli, Alain Paris, Blanche Guillon, Christophe Junot, Clémence Moiron, François Fenaille, and Karine Adel-Patient. 2022. "The Human Meconium Metabolome and Its Evolution during the First Days of Life" Metabolites 12, no. 5: 414. https://doi.org/10.3390/metabo12050414
APA StyleBekhti, N., Castelli, F., Paris, A., Guillon, B., Junot, C., Moiron, C., Fenaille, F., & Adel-Patient, K. (2022). The Human Meconium Metabolome and Its Evolution during the First Days of Life. Metabolites, 12(5), 414. https://doi.org/10.3390/metabo12050414