Role of Polyamines and Hypusine in β Cells and Diabetes Pathogenesis
Abstract
1. Introduction
2. Polyamines in Diabetes Pathogenesis
2.1. Biosynthesis and Regulation of Polyamines
2.2. Polyamines in β-Cell Function and Diabetes Pathogenesis
2.2.1. Polyamines in β-Cell Replication
2.2.2. Polyamines in Glucose Homeostasis
2.2.3. Mechanistic Insight in the Role of Polyamines in Diabetes
2.2.4. Polyamines in Obesity and T2D
2.2.5. Polyamines in T1D
2.2.6. Effects of Polyamine Supplementation
3. Hypusine and eIF5A Pathway in β-Cell Function and Diabetes Pathogenesis
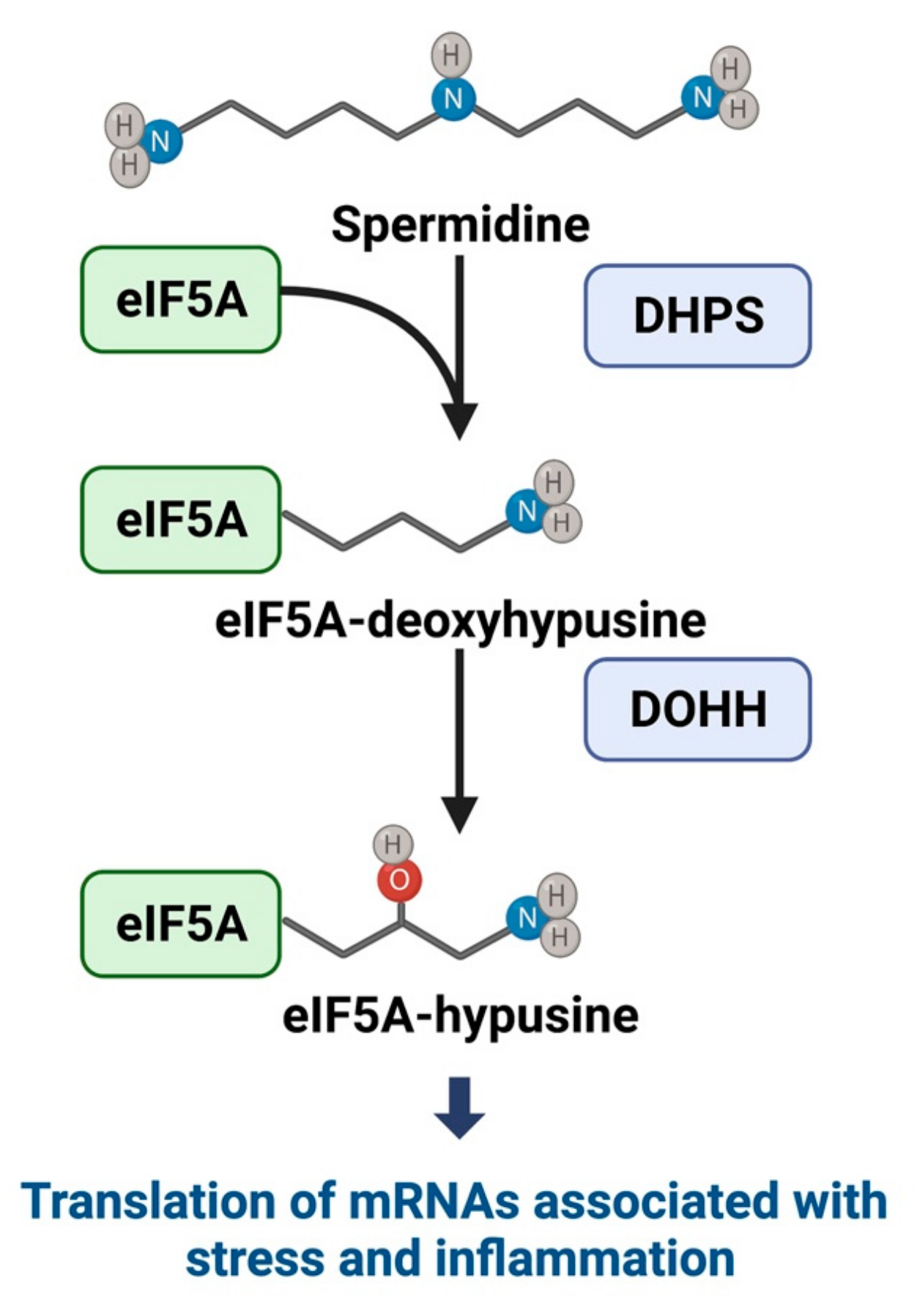
3.1. Hypusine and eIF5A Mechanistic Pathways
3.2. eIF5Ahyp in Diabetes and the β Cell
3.3. Involvement of eIF5A in Islet Inflammation and Diabetic Immune Response
3.4. Future Considerations
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cerf, M.E. Beta Cell Dysfunction and Insulin Resistance. Front. Endocrinol. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Sims, E.K.; Carr, A.L.J.; Oram, R.A.; DiMeglio, L.A.; Evans-Molina, C. 100 Years of Insulin: Celebrating the Past, Present and Future of Diabetes Therapy. Nat. Med. 2021, 27, 1154–1164. [Google Scholar] [CrossRef]
- Saisho, Y. β-Cell Dysfunction: Its Critical Role in Prevention and Management of Type 2 Diabetes. World J. Diabetes 2015, 6, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, J.S.; Nunes, S.; Rolo, A.P.; Reis, F.; Palmeira, C.M. Therapeutic Options Targeting Oxidative Stress, Mitochondrial Dysfunction and Inflammation to Hinder the Progression of Vascular Complications of Diabetes. Front. Physiol. 2019, 9, 1857. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Morón, E.; Abad-Jiménez, Z.; Martínez de Marañón, A.; Iannantuoni, F.; Escribano-López, I.; López-Domènech, S.; Salom, C.; Jover, A.; Mora, V.; Roldan, I.; et al. Relationship between Oxidative Stress, ER Stress, and Inflammation in Type 2 Diabetes: The Battle Continues. J. Clin. Med. 2019, 8, 1385. [Google Scholar] [CrossRef]
- Demirtas, L.; Guclu, A.; Erdur, F.M.; Akbas, E.M.; Ozcicek, A.; Onk, D.; Turkmen, K. Apoptosis, Autophagy & Endoplasmic Reticulum Stress in Diabetes Mellitus. Ind. J. Med. Res. 2016, 144, 515–524. [Google Scholar] [CrossRef]
- Kulkarni, A.; Nadler, J.L.; Mirmira, R.G.; Casimiro, I. Regulation of Tissue Inflammation by 12-Lipoxygenases. Biomolecules 2021, 11, 717. [Google Scholar] [CrossRef]
- Khin, P.-P.; Lee, J.-H.; Jun, H.-S. A Brief Review of the Mechanisms of β-Cell Dedifferentiation in Type 2 Diabetes. Nutrients 2021, 13, 1593. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, C. Targeting β-Cell Dedifferentiation and Transdifferentiation: Opportunities and Challenges. Endocr. Connect. 2021, 10, R213–R228. [Google Scholar] [CrossRef]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in Aging and Disease. Aging 2011, 3, 716–732. [Google Scholar] [CrossRef]
- Schuller, A.P.; Wu, C.C.-C.; Dever, T.E.; Buskirk, A.R.; Green, R. EIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205.e5. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Alepuz, P. EIF5A Facilitates Translation Termination Globally and Promotes the Elongation of Many Non Polyproline-Specific Tripeptide Sequences. Nucl. Acids Res. 2017, 45, 7326–7338. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.A.; Padgett, L.R.; Fine, J.A.; Chopra, G.; Mastracci, T.L. Targeting Polyamine Biosynthesis to Stimulate Beta Cell Regeneration in Zebrafish. Islets 2020, 12, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, E.M.; Yamada, K.; Piñeros, A.R.; Wu, W.; Syed, F.; Orr, K.S.; Anderson-Baucum, E.; Mastracci, T.L.; Maier, B.; Mosley, A.L.; et al. Hypusine Biosynthesis in β Cells Links Polyamine Metabolism to Facultative Cellular Proliferation to Maintain Glucose Homeostasis. Sci. Signal. 2019, 12, eaax0715. [Google Scholar] [CrossRef]
- Cerrada-Gimenez, M.; Tusa, M.; Casellas, A.; Pirinen, E.; Moya, M.; Bosch, F.; Alhonen, L. Altered Glucose-Stimulated Insulin Secretion in a Mouse Line with Activated Polyamine Catabolism. Transgen. Res. 2012, 21, 843–853. [Google Scholar] [CrossRef]
- Maier, B.; Ogihara, T.; Trace, A.P.; Tersey, S.A.; Robbins, R.D.; Chakrabarti, S.K.; Nunemaker, C.S.; Stull, N.D.; Taylor, C.A.; Thompson, J.E.; et al. The Unique Hypusine Modification of EIF5A Promotes Islet Beta Cell Inflammation and Dysfunction in Mice. J. Clin. Investig. 2010, 120, 2156–2170. [Google Scholar] [CrossRef]
- Tersey, S.A.; Colvin, S.C.; Maier, B.; Mirmira, R.G. Protective Effects of Polyamine Depletion in Mouse Models of Type 1 Diabetes: Implications for Therapy. Amino Acids 2014, 46, 633–642. [Google Scholar] [CrossRef]
- Bjelakovic, G.; Beninati, S.; Bjelakovic, B.; Sokolovic, D.; Jevtovic, T.; Stojanovic, I.; Rossi, S.; Tabolacci, C.; Kocić, G.; Pavlovic, D.; et al. Does Polyamine Oxidase Activity Influence the Oxidative Metabolism of Children Who Suffer of Diabetes Mellitus? Mol. Cell Biochem. 2010, 341, 79–85. [Google Scholar] [CrossRef]
- Seghieri, G.; Gironi, A.; Niccolai, M.; Mammini, P.; Alviggi, L.; De Giorgio, L.A.; Caselli, P.; Bartolomei, G. Serum Spermidine Oxidase Activity in Patients with Insulin-Dependent Diabetes Mellitus and Microvascular Complications. Acta Diabetol. Lat. 1990, 27, 303–308. [Google Scholar] [CrossRef]
- Fernandez-Garcia, J.C.; Delpino-Rius, A.; Samarra, I.; Castellano-Castillo, D.; Muñoz-Garach, A.; Bernal-Lopez, M.R.; Queipo-Ortuño, M.I.; Cardona, F.; Ramos-Molina, B.; Tinahones, F.J. Type 2 Diabetes Is Associated with a Different Pattern of Serum Polyamines: A Case–Control Study from the PREDIMED-Plus Trial. J. Clin. Med. 2019, 8, 71. [Google Scholar] [CrossRef]
- Robbins, R.D.; Tersey, S.A.; Ogihara, T.; Gupta, D.; Farb, T.B.; Ficorilli, J.; Bokvist, K.; Maier, B.; Mirmira, R.G. Inhibition of Deoxyhypusine Synthase Enhances Islet β Cell Function and Survival in the Setting of Endoplasmic Reticulum Stress and Type 2 Diabetes. J. Biol. Chem. 2010, 285, 39943–39952. [Google Scholar] [CrossRef] [PubMed]
- Fernández, Á.F.; Bárcena, C.; Martínez-García, G.G.; Tamargo-Gómez, I.; Suárez, M.F.; Pietrocola, F.; Castoldi, F.; Esteban, L.; Sierra-Filardi, E.; Boya, P.; et al. Autophagy Couteracts Weight Gain, Lipotoxicity and Pancreatic β-Cell Death upon Hypercaloric pro-Diabetic Regimens. Cell Death Dis. 2017, 8, e2970. [Google Scholar] [CrossRef] [PubMed]
- Sadasivan, S.K.; Vasamsetti, B.; Singh, J.; Marikunte, V.V.; Oommen, A.M.; Jagannath, M.R.; Pralhada Rao, R. Exogenous Administration of Spermine Improves Glucose Utilization and Decreases Bodyweight in Mice. Eur. J. Pharm. 2014, 729, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Anderson-Baucum, E.; Piñeros, A.R.; Kulkarni, A.; Webb-Robertson, B.-J.; Maier, B.; Anderson, R.M.; Wu, W.; Tersey, S.A.; Mastracci, T.L.; Casimiro, I.; et al. Deoxyhypusine Synthase Promotes a Pro-Inflammatory Macrophage Phenotype. Cell Metab. 2021, 33, 1883–1893.e7. [Google Scholar] [CrossRef]
- Karacay, C.; Prietl, B.; Harer, C.; Ehall, B.; Haudum, C.W.; Bounab, K.; Franz, J.; Eisenberg, T.; Madeo, F.; Kolb, D.; et al. The Effect of Spermidine on Autoimmunity and Beta Cell Function in NOD Mice. Sci. Rep. 2022, 12, 4502. [Google Scholar] [CrossRef]
- Imam, S.; Prathibha, R.; Dar, P.; Almotah, K.; Al-Khudhair, A.; Hasan, S.A.-M.; Salim, N.; Jilani, T.N.; Mirmira, R.G.; Jaume, J.C. EIF5A Inhibition Influences T Cell Dynamics in the Pancreatic Microenvironment of the Humanized Mouse Model of Type 1 Diabetes. Sci. Rep. 2019, 9, 1533. [Google Scholar] [CrossRef]
- Colvin, S.C.; Maier, B.; Morris, D.L.; Tersey, S.A.; Mirmira, R.G. Deoxyhypusine Synthase Promotes Differentiation and Proliferation of T Helper Type 1 (Th1) Cells in Autoimmune Diabetes. J. Biol. Chem. 2013, 288, 36226–36235. [Google Scholar] [CrossRef]
- van Dam, L.; Korolev, N.; Nordenskiöld, L. Polyamine–Nucleic Acid Interactions and the Effects on Structure in Oriented DNA Fibers. Nucl. Acids Res. 2002, 30, 419–428. [Google Scholar] [CrossRef]
- Podestà, A.; Indrieri, M.; Brogioli, D.; Manning, G.S.; Milani, P.; Guerra, R.; Finzi, L.; Dunlap, D. Positively Charged Surfaces Increase the Flexibility of DNA. Biophys. J. 2005, 89, 2558–2563. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hall, J. Endogenous Polyamine Function—The RNA Perspective. Nucl. Acids Res. 2014, 42, 11275–11290. [Google Scholar] [CrossRef] [PubMed]
- Brooks, W.H. Increased Polyamines Alter Chromatin and Stabilize Autoantigens in Autoimmune Diseases. Front. Immunol. 2013, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Perepelytsya, S.; Uličný, J.; Laaksonen, A.; Mocci, F. Pattern Preferences of DNA Nucleotide Motifs by Polyamines Putrescine2+, Spermidine3+ and Spermine4+. Nucl. Acids Res. 2019, 47, 6084–6097. [Google Scholar] [CrossRef] [PubMed]
- Gerner, E.W.; Russell, D.H. The Relationship between Polyamine Accumulation and DNA Replication in Synchronized Chinese Hamster Ovary Cells after Heat Shock. Cancer Res. 1977, 37, 482–489. [Google Scholar] [PubMed]
- Gallo, C.J.; Koza, R.A.; Herbst, E.J. Polyamines and HeLa-Cell DNA Replication. Biochem. J. 1986, 238, 37–42. [Google Scholar] [CrossRef]
- Tkachenko, A.G.; Nesterova, L.Y. Polyamines as Modulators of Gene Expression under Oxidative Stress in Escherichia coli. Biochemistry 2003, 68, 850–856. [Google Scholar] [CrossRef]
- Sakamoto, A.; Terui, Y.; Uemura, T.; Igarashi, K.; Kashiwagi, K. Polyamines Regulate Gene Expression by Stimulating Translation of Histone Acetyltransferase MRNAs. J. Biol. Chem. 2020, 295, 8736–8745. [Google Scholar] [CrossRef]
- Matthews, H.R. Polyamines, Chromatin Structure and Transcription. Bioessays 1993, 15, 561–566. [Google Scholar] [CrossRef]
- Xiao, L.; Rao, J.N.; Zou, T.; Liu, L.; Marasa, B.S.; Chen, J.; Turner, D.J.; Zhou, H.; Gorospe, M.; Wang, J.-Y. Polyamines Regulate the Stability of Activating Transcription Factor-2 MRNA through RNA-Binding Protein HuR in Intestinal Epithelial Cells. Mol. Biol. Cell 2007, 18, 4579–4590. [Google Scholar] [CrossRef]
- Huang, S.C.; Panagiotidis, C.A.; Canellakis, E.S. Transcriptional Effects of Polyamines on Ribosomal Proteins and on Polyamine-Synthesizing Enzymes in Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 3464–3468. [Google Scholar] [CrossRef]
- Igarashi, K.; Kashiwagi, K. Effects of Polyamines on Protein Synthesis and Growth of Escherichia coli. J. Biol. Chem. 2018, 293, 18702–18709. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Shin, B.-S.; Loughran, G.; Tzani, I.; Young-Baird, S.K.; Cao, C.; Atkins, J.F.; Dever, T.E. Polyamine Control of Translation Elongation Regulates Start Site Selection on the Antizyme Inhibitor MRNA via Ribosome Queuing. Mol. Cell 2018, 70, 254–264.e6. [Google Scholar] [CrossRef] [PubMed]
- Perez-Leal, O.; Barrero, C.A.; Clarkson, A.B.; Casero, R.A.; Merali, S. Polyamine-Regulated Translation of Spermidine/Spermine-N1-Acetyltransferase. Mol. Cell. Biol. 2012, 32, 1453–1467. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Thomas, T.J. Polyamines in Cell Growth and Cell Death: Molecular Mechanisms and Therapeutic Applications. Cell Mol. Life Sci. 2001, 58, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Cherry, S.; Minocha, R.; Majumdar, R.; Thangavel, P.; Long, S.; Minocha, S.C. The Response of High and Low Polyamine-Producing Cell Lines to Aluminum and Calcium Stress. Plant Physiol. Biochem. 2010, 48, 612–620. [Google Scholar] [CrossRef]
- Kahana, C. Regulation of Cellular Polyamine Levels and Cellular Proliferation by Antizyme and Antizyme Inhibitor. Essays Biochem. 2009, 46, 47–61. [Google Scholar] [CrossRef]
- Whelly, S.M. Role of Polyamine in the Regulation of RNA Synthesis in Uterine Nucleoli. J. Steroid. Biochem. Mol. Biol. 1991, 39, 161–167. [Google Scholar] [CrossRef]
- Terui, Y.; Tabei, Y.; Akiyama, M.; Higashi, K.; Tomitori, H.; Yamamoto, K.; Ishihama, A.; Igarashi, K.; Kashiwagi, K. Ribosome Modulation Factor, an Important Protein for Cell Viability Encoded by the Polyamine Modulon. J. Biol. Chem. 2010, 285, 28698–28707. [Google Scholar] [CrossRef]
- Rato, C.; Amirova, S.R.; Bates, D.G.; Stansfield, I.; Wallace, H.M. Translational Recoding as a Feedback Controller: Systems Approaches Reveal Polyamine-Specific Effects on the Antizyme Ribosomal Frameshift. Nucl. Acids Res. 2011, 39, 4587–4597. [Google Scholar] [CrossRef]
- McMurry, L.M.; Algranati, I.D. Effect of Polyamines on Translation Fidelity in Vivo. Eur. J. Biochem. 1986, 155, 383–390. [Google Scholar] [CrossRef]
- Landau, G.; Bercovich, Z.; Park, M.H.; Kahana, C. The Role of Polyamines in Supporting Growth of Mammalian Cells Is Mediated through Their Requirement for Translation Initiation and Elongation. J. Biol. Chem. 2010, 285, 12474–12481. [Google Scholar] [CrossRef]
- Park, M.H.; Wolff, E.C. Hypusine, a Polyamine-Derived Amino Acid Critical for Eukaryotic Translation. J. Biol. Chem. 2018, 293, 18710–18718. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Esparza, N.C.; Latorre-Moratalla, M.L.; Comas-Basté, O.; Toro-Funes, N.; Veciana-Nogués, M.T.; Vidal-Carou, M.C. Polyamines in Food. Front. Nutr. 2019, 6, 108. [Google Scholar] [CrossRef] [PubMed]
- Tofalo, R.; Cocchi, S.; Suzzi, G. Polyamines and Gut Microbiota. Front Nutr. 2019, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Seiler, N. Polyamine Metabolism. Digestion 1990, 46, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Morris, S.M. Arginine Metabolism: Nitric Oxide and Beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.K.; Brooks, H.B.; Myers, D.P.; Phillips, M.A. Ornithine Decarboxylase Promotes Catalysis by Binding the Carboxylate in a Buried Pocket Containing Phenylalanine 397. Biochemistry 2003, 42, 2933–2940. [Google Scholar] [CrossRef] [PubMed]
- Tabor, C.W.; Tabor, H. Polyamines. Annu. Rev. Biochem. 1984, 53, 749–790. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Chen, S.-F.; Hsieh, J.-Y.; Chou, F.; Wang, Y.-H.; Lin, W.-T.; Lee, P.-Y.; Yu, Y.-J.; Lin, L.-Y.; Lin, T.-S.; et al. Structural Basis of Antizyme-Mediated Regulation of Polyamine Homeostasis. Proc. Natl. Acad. Sci. USA 2015, 112, 11229–11234. [Google Scholar] [CrossRef]
- Pegg, A.E. S-Adenosylmethionine Decarboxylase. Essays Biochem. 2009, 46, 25–45. [Google Scholar] [CrossRef]
- Casero, R.A.; Pegg, A.E. Polyamine Catabolism and Disease. Biochem. J. 2009, 421, 323–338. [Google Scholar] [CrossRef]
- Vujcic, S.; Diegelman, P.; Bacchi, C.J.; Kramer, D.L.; Porter, C.W. Identification and Characterization of a Novel Flavin-Containing Spermine Oxidase of Mammalian Cell Origin. Biochem. J. 2002, 367, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Sessa, A.; Perin, A. Diamine Oxidase in Relation to Diamine and Polyamine Metabolism. Agents Act. 1994, 43, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.L.; Judd, G.G.; Leyser, A.; Choe, C. Osmotic Stress Induces Variation in Cellular Levels of Ornithine Decarboxylase-Antizyme. Biochem. J. 1998, 329, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.M.; Viar, M.J.; Patel, T.B.; Johnson, L.R. Interaction of Asparagine and EGF in the Regulation of Ornithine Decarboxylase in IEC-6 Cells. Am. J. Physiol. 1999, 276, G773–G780. [Google Scholar] [CrossRef]
- Rinehart, C.A.; Viceps-Madore, D.; Fong, W.F.; Ortiz, J.G.; Canellakis, E.S. The Effect of Transport System A and N Amino Acids and of Nerve and Epidermal Growth Factors on the Induction of Ornithine Decarboxylase Activity. J. Cell Physiol. 1985, 123, 435–441. [Google Scholar] [CrossRef]
- Ramos-Molina, B.; Lambertos, A.; Peñafiel, R. Antizyme Inhibitors in Polyamine Metabolism and Beyond: Physiopathological Implications. Med. Sci. 2018, 6, 89. [Google Scholar] [CrossRef]
- Hill, J.R.; Morris, D.R. Cell-Specific Translational Regulation of S-Adenosylmethionine Decarboxylase MRNA. Dependence on Translation and Coding Capacity of the Cis-Acting Upstream Open Reading Frame. J. Biol. Chem. 1993, 268, 726–731. [Google Scholar] [CrossRef]
- Casero, R.A.; Pegg, A.E. Spermidine/Spermine N1-Acetyltransferase--the Turning Point in Polyamine Metabolism. FASEB J. 1993, 7, 653–661. [Google Scholar] [CrossRef]
- Pegg, A.E. Spermidine/Spermine-N(1)-Acetyltransferase: A Key Metabolic Regulator. Am. J. Physiol. Endocr. Metab. 2008, 294, E995–E1010. [Google Scholar] [CrossRef]
- Abdulhussein, A.A.; Wallace, H.M. Polyamines and Membrane Transporters. Amino Acids 2014, 46, 655–660. [Google Scholar] [CrossRef]
- Uemura, T.; Gerner, E.W. Polyamine Transport Systems in Mammalian Cells and Tissues. Methods Mol. Biol. 2011, 720, 339–348. [Google Scholar] [CrossRef]
- Hougaard, D.M.; Larsson, L.I. Localization and Possible Function of Polyamines in Protein and Peptide Secreting Cells. Med. Biol. 1986, 64, 89–94. [Google Scholar] [PubMed]
- Hougaard, D.M.; Nielsen, J.H.; Larsson, L.I. Localization and Biosynthesis of Polyamines in Insulin-Producing Cells. Biochem. J. 1986, 238, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Mastracci, T.L.; Robertson, M.A.; Mirmira, R.G.; Anderson, R.M. Polyamine Biosynthesis Is Critical for Growth and Differentiation of the Pancreas. Sci. Rep. 2015, 5, 13269. [Google Scholar] [CrossRef] [PubMed]
- Sjöholm, A. Role of Polyamines in the Regulation of Proliferation and Hormone Production by Insulin-Secreting Cells. Am. J. Physiol. 1993, 264, C501–C518. [Google Scholar] [CrossRef]
- Sjöholm, A.; Arkhammar, P.; Berggren, P.O.; Andersson, A. Polyamines in Pancreatic Islets of Obese-Hyperglycemic (Ob/Ob) Mice of Different Ages. Am. J. Physiol. Cell Physiol. 2001, 280, C317–C323. [Google Scholar] [CrossRef]
- Yang, B.; Covington, B.A.; Chen, W. In Vivo Generation and Regeneration of β Cells in Zebrafish. Cell Regen. 2020, 9, 9. [Google Scholar] [CrossRef]
- Moro, E.; Gnügge, L.; Braghetta, P.; Bortolussi, M.; Argenton, F. Analysis of Beta Cell Proliferation Dynamics in Zebrafish. Dev. Biol. 2009, 332, 299–308. [Google Scholar] [CrossRef]
- Kulkarni, A.A.; Conteh, A.M.; Sorrell, C.A.; Mirmira, A.; Tersey, S.A.; Mirmira, R.G.; Linnemann, A.K.; Anderson, R.M. An In Vivo Zebrafish Model for Interrogating ROS-Mediated Pancreatic β-Cell Injury, Response, and Prevention. Oxid. Med. Cell Longev. 2018, 2018, 1324739. [Google Scholar] [CrossRef]
- Hernandez-Perez, M.; Kulkarni, A.; Samala, N.; Sorrell, C.; El, K.; Haider, I.; Mukhtar Aleem, A.; Holman, T.R.; Rai, G.; Tersey, S.A.; et al. A 12-Lipoxygenase-Gpr31 Signaling Axis Is Required for Pancreatic Organogenesis in the Zebrafish. FASEB J. 2020, 34, 14850–14862. [Google Scholar] [CrossRef]
- Welsh, N.; Sjöholm, A. Polyamines and Insulin Production in Isolated Mouse Pancreatic Islets. Biochem. J. 1988, 252, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Welsh, N. A Role for Polyamines in Glucose-Stimulated Insulin-Gene Expression. Biochem. J. 1990, 271, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Pirinen, E.; Kuulasmaa, T.; Pietilä, M.; Heikkinen, S.; Tusa, M.; Itkonen, P.; Boman, S.; Skommer, J.; Virkamäki, A.; Hohtola, E.; et al. Enhanced Polyamine Catabolism Alters Homeostatic Control of White Adipose Tissue Mass, Energy Expenditure, and Glucose Metabolism. Mol. Cell. Biol. 2007, 27, 4953–4967. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Zhang, L.; Cao, Y.; Gao, W.; Zhao, C.; Fang, Y.; Zahedi, K.; Soleimani, M.; Lu, X.; Fang, Z.; et al. Spermidine/Spermine N1-Acetyltransferase-Mediated Polyamine Catabolism Regulates Beige Adipocyte Biogenesis. Metabolism 2018, 85, 298–304. [Google Scholar] [CrossRef]
- Niiranen, K.; Keinänen, T.A.; Pirinen, E.; Heikkinen, S.; Tusa, M.; Fatrai, S.; Suppola, S.; Pietilä, M.; Uimari, A.; Laakso, M.; et al. Mice with Targeted Disruption of Spermidine/Spermine N1-Acetyltransferase Gene Maintain Nearly Normal Tissue Polyamine Homeostasis but Show Signs of Insulin Resistance upon Aging. J. Cell. Mol. Med. 2006, 10, 933–945. [Google Scholar] [CrossRef]
- Puleston, D.J.; Baixauli, F.; Sanin, D.E.; Edwards-Hicks, J.; Villa, M.; Kabat, A.M.; Kamiński, M.M.; Stanckzak, M.; Weiss, H.J.; Grzes, K.M.; et al. Polyamine Metabolism Is a Central Determinant of Helper T Cell Lineage Fidelity. Cell 2021, 184, 4186–4202.e20. [Google Scholar] [CrossRef]
- Tsalamandris, S.; Antonopoulos, A.S.; Oikonomou, E.; Papamikroulis, G.-A.; Vogiatzi, G.; Papaioannou, S.; Deftereos, S.; Tousoulis, D. The Role of Inflammation in Diabetes: Current Concepts and Future Perspectives. Eur. Cardiol. 2019, 14, 50–59. [Google Scholar] [CrossRef]
- Piñeros, A.; Kulkarni, A.; Gao, H.; Orr, K.; Glenn, L.; Huang, F.; Liu, Y.; Gannon, M.; Syed, F.; Wu, W.; et al. Proinflammatory Signaling in Islet β Cells Propagates Invasion of Pathogenic Immune Cells in Autoimmune Diabetes; Social Science Research Network: Rochester, NY, USA, 2021. [Google Scholar]
- Marro, B.S.; Legrain, S.; Ware, B.C.; Oldstone, M.B.A. Macrophage IFN-I Signaling Promotes Autoreactive T Cell Infiltration into Islets in Type 1 Diabetes Model. JCI Insight 2019, 4, e125067. [Google Scholar] [CrossRef]
- Kulkarni, A.; Pineros, A.R.; Walsh, M.A.; Casimiro, I.; Ibrahim, S.; Hernandez-Perez, M.; Orr, K.S.; Glenn, L.; Nadler, J.L.; Morris, M.A.; et al. 12-Lipoxygenase Governs the Innate Immune Pathogenesis of Islet Inflammation and Autoimmune Diabetes. JCI Insight 2021, 6, e147812. [Google Scholar] [CrossRef]
- Davanso, M.R.; Crisma, A.R.; Braga, T.T.; Masi, L.N.; do Amaral, C.L.; Leal, V.N.C.; de Lima, D.S.; Patente, T.A.; Barbuto, J.A.; Corrêa-Giannella, M.L.; et al. Macrophage Inflammatory State in Type 1 Diabetes: Triggered by NLRP3/INOS Pathway and Attenuated by Docosahexaenoic Acid (DHA). Clin. Sci. 2020, 22, CS20201348. [Google Scholar] [CrossRef]
- Mensah-Brown, E.; Shahin, A.; Parekh, K.; Hakim, A.A.; Shamisi, M.A.; Hsu, D.K.; Lukic, M.L. Functional Capacity of Macrophages Determines the Induction of Type 1 Diabetes. Ann. N. Y. Acad. Sci. 2006, 1084, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Tesch, G.H. Role of Macrophages in Complications of Type 2 Diabetes. Clin. Exp. Pharm. Physiol. 2007, 34, 1016–1019. [Google Scholar] [CrossRef] [PubMed]
- Roep, B.O. The Role of T-Cells in the Pathogenesis of Type 1 Diabetes: From Cause to Cure. Diabetologia 2003, 46, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; Shameli, A.; Santamaria, P. CD8+ T Cells in Type 1 Diabetes. Adv. Immunol. 2008, 100, 79–124. [Google Scholar] [CrossRef]
- Kent, S.C.; Mannering, S.I.; Michels, A.W.; Babon, J.A.B. Deciphering the Pathogenesis of Human Type 1 Diabetes (T1D) by Interrogating T Cells from the Scene of the Crime. Curr. Diab. Rep. 2017, 17, 95. [Google Scholar] [CrossRef]
- Hull, C.M.; Peakman, M.; Tree, T.I.M. Regulatory T Cell Dysfunction in Type 1 Diabetes: What’s Broken and How Can We Fix It? Diabetologia 2017, 60, 1839–1850. [Google Scholar] [CrossRef]
- Xia, C.; Rao, X.; Zhong, J. Role of T Lymphocytes in Type 2 Diabetes and Diabetes-Associated Inflammation. J. Diabetes Res. 2017, 2017, 6494795. [Google Scholar] [CrossRef]
- Lau, E.Y.M.; Carroll, E.C.; Callender, L.A.; Hood, G.A.; Berryman, V.; Pattrick, M.; Finer, S.; Hitman, G.A.; Ackland, G.L.; Henson, S.M. Type 2 Diabetes Is Associated with the Accumulation of Senescent T Cells. Clin. Exp. Immunol. 2019, 197, 205–213. [Google Scholar] [CrossRef]
- Latour, Y.L.; Gobert, A.P.; Wilson, K.T. The Role of Polyamines in the Regulation of Macrophage Polarization and Function. Amino Acids 2020, 52, 151–160. [Google Scholar] [CrossRef]
- Wang, Z. Polyamines Instruct T-Cell Differentiation. Nat. Cell Biol. 2021, 23, 811. [Google Scholar] [CrossRef]
- Sandler, S.; Bendtzen, K.; Eizirik, D.L.; Sjöholm, A.; Welsh, N. Decreased Cell Replication and Polyamine Content in Insulin-Producing Cells after Exposure to Human Interleukin 1 Beta. Immunol. Lett. 1989, 22, 267–272. [Google Scholar] [CrossRef]
- Smismans, A.; Eizirik, D.L.; Pipeleers, D.G. Interleukin-1beta Induces Ornithine Decarboxylase Activity in Insulin-Producing Cells. Cytokine 2000, 12, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Mizuno, I.; Kojima, Y.; Ishikawa, Y.; Sodeno, M.; Asakura, Y.; Samejima, K.; Oka, T. Spermidine Regulates Insulin Synthesis and Cytoplasmic Ca2+ in Mouse Beta-TC6 Insulinoma Cells. Cell Struct. Funct. 2009, 34, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Lenzen, S.; Rustenbeck, I. Effects of IP3, Spermine, and Mg2+ on Regulation of Ca2+ Transport by Endoplasmic Reticulum and Mitochondria in Permeabilized Pancreatic Islets. Diabetes 1991, 40, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.; Johnson, J.D.; Arvan, P.; Han, J.; Kaufman, R.J. Therapeutic Opportunities for Pancreatic β-Cell ER Stress in Diabetes Mellitus. Nat. Rev. Endocrinol. 2021, 17, 455–467. [Google Scholar] [CrossRef]
- Codoñer-Franch, P.; Tavárez-Alonso, S.; Murria-Estal, R.; Herrera-Martín, G.; Alonso-Iglesias, E. Polyamines Are Increased in Obese Children and Are Related to Markers of Oxidative/Nitrosative Stress and Angiogenesis. J. Clin. Endocrinol. Metab. 2011, 96, 2821–2825. [Google Scholar] [CrossRef] [PubMed]
- Marselli, L.; Bosi, E.; De Luca, C.; Del Guerra, S.; Tesi, M.; Suleiman, M.; Marchetti, P. Arginase 2 and Polyamines in Human Pancreatic Beta Cells: Possible Role in the Pathogenesis of Type 2 Diabetes. Int. J. Mol. Sci. 2021, 22, 12099. [Google Scholar] [CrossRef]
- Hesterberg, R.S.; Cleveland, J.L.; Epling-Burnette, P.K. Role of Polyamines in Immune Cell Functions. Med. Sci. 2018, 6, 22. [Google Scholar] [CrossRef]
- Gao, M.; Zhao, W.; Li, C.; Xie, X.; Li, M.; Bi, Y.; Fang, F.; Du, Y.; Liu, X. Spermidine Ameliorates Non-Alcoholic Fatty Liver Disease through Regulating Lipid Metabolism via AMPK. Biochem. Biophys. Res. Commun. 2018, 505, 93–98. [Google Scholar] [CrossRef]
- Méndez, J.D.; Balderas, F.L. Inhibition by L-Arginine and Spermidine of Hemoglobin Glycation and Lipid Peroxidation in Rats with Induced Diabetes. Biomed. Pharm. 2006, 60, 26–31. [Google Scholar] [CrossRef]
- Méndez, J.D.; Hernández, R.D.H. L-Arginine and Polyamine Administration Protect Beta-Cells against Alloxan Diabetogenic Effect in Sprague-Dawley Rats. Biomed. Pharm. 2005, 59, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Jafarnejad, A.; Bathaie, S.Z.; Nakhjavani, M.; Hassan, M.Z. Effect of Spermine on Lipid Profile and HDL Functionality in the Streptozotocin-Induced Diabetic Rat Model. Life Sci. 2008, 82, 301–307. [Google Scholar] [CrossRef]
- Fetterman, J.L.; Holbrook, M.; Flint, N.; Feng, B.; Bretón-Romero, R.; Linder, E.A.; Berk, B.D.; Duess, M.-A.; Farb, M.G.; Gokce, N.; et al. Restoration of Autophagy in Endothelial Cells from Patients with Diabetes Mellitus Improves Nitric Oxide Signaling. Atherosclerosis 2016, 247, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, E.; Shin, B.-S.; Woolstenhulme, C.J.; Kim, J.-R.; Saini, P.; Buskirk, A.R.; Dever, T.E. EIF5A Promotes Translation of Polyproline Motifs. Mol. Cell 2013, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Wolff, E.C.; Park, M.H. Assay of Deoxyhypusine Hydroxylase Activity. Methods Mol. Biol. 2011, 720, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Clement, P.M.J.; Johansson, H.E.; Wolff, E.C.; Park, M.H. Differential Expression of EIF5A-1 and EIF5A-2 in Human Cancer Cells. FEBS J. 2006, 273, 1102–1114. [Google Scholar] [CrossRef]
- Benne, R.; Hershey, J.W. The Mechanism of Action of Protein Synthesis Initiation Factors from Rabbit Reticulocytes. J. Biol. Chem. 1978, 253, 3078–3087. [Google Scholar] [CrossRef]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-Containing Protein EIF5A Promotes Translation Elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef]
- Park, M.H.; Wolff, E.C.; Smit-McBride, Z.; Hershey, J.W.; Folk, J.E. Comparison of the Activities of Variant Forms of EIF-4D. The Requirement for Hypusine or Deoxyhypusine. J. Biol. Chem. 1991, 266, 7988–7994. [Google Scholar] [CrossRef]
- Park, M.H.; Nishimura, K.; Zanelli, C.F.; Valentini, S.R. Functional Significance of EIF5A and Its Hypusine Modification in Eukaryotes. Amino Acids 2010, 38, 491–500. [Google Scholar] [CrossRef]
- Mastracci, T.L.; Colvin, S.C.; Padgett, L.R.; Mirmira, R.G. Hypusinated EIF5A Is Expressed in the Pancreas and Spleen of Individuals with Type 1 and Type 2 Diabetes. PLoS ONE 2020, 15, e0230627. [Google Scholar] [CrossRef] [PubMed]
- Padgett, L.R.; Robertson, M.A.; Anderson-Baucum, E.K.; Connors, C.T.; Wu, W.; Mirmira, R.G.; Mastracci, T.L. Deoxyhypusine Synthase, an Essential Enzyme for Hypusine Biosynthesis, Is Required for Proper Exocrine Pancreas Development. FASEB J. 2021, 35, e21473. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Kurihara, S.; Takahashi, D.; Ohashi, W.; Nakamura, Y.; Kimura, S.; Onuki, M.; Kume, A.; Sasazawa, Y.; Furusawa, Y.; et al. Symbiotic Polyamine Metabolism Regulates Epithelial Proliferation and Macrophage Differentiation in the Colon. Nat. Commun. 2021, 12, 2105. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Murozumi, K.; Shirahata, A.; Park, M.H.; Kashiwagi, K.; Igarashi, K. Independent Roles of EIF5A and Polyamines in Cell Proliferation. Biochem. J. 2005, 385, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Saisho, Y.; Butler, A.E.; Manesso, E.; Elashoff, D.; Rizza, R.A.; Butler, P.C. β-Cell Mass and Turnover in Humans: Effects of Obesity and Aging. Diabetes Care 2013, 36, 111–117. [Google Scholar] [CrossRef]
- Mosser, R.E.; Maulis, M.F.; Moullé, V.S.; Dunn, J.C.; Carboneau, B.A.; Arasi, K.; Pappan, K.; Poitout, V.; Gannon, M. High-Fat Diet-Induced β-Cell Proliferation Occurs Prior to Insulin Resistance in C57Bl/6J Male Mice. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E573–E582. [Google Scholar] [CrossRef]
- Stamateris, R.E.; Sharma, R.B.; Hollern, D.A.; Alonso, L.C. Adaptive β-Cell Proliferation Increases Early in High-Fat Feeding in Mice, Concurrent with Metabolic Changes, with Induction of Islet Cyclin D2 Expression. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E149–E159. [Google Scholar] [CrossRef]
- Keenan, H.A.; Sun, J.K.; Levine, J.; Doria, A.; Aiello, L.P.; Eisenbarth, G.; Bonner-Weir, S.; King, G.L. Residual Insulin Production and Pancreatic SS-Cell Turnover after 50 Years of Diabetes: Joslin Medalist Study. Diabetes 2010, 59, 2846–2853. [Google Scholar] [CrossRef]
- Sreenan, S.; Pick, A.J.; Levisetti, M.; Baldwin, A.C.; Pugh, W.; Polonsky, K.S. Increased Beta-Cell Proliferation and Reduced Mass before Diabetes Onset in the Nonobese Diabetic Mouse. Diabetes 1999, 48, 989–996. [Google Scholar] [CrossRef]
- Dirice, E.; Kahraman, S.; De Jesus, D.F.; El Ouaamari, A.; Basile, G.; Baker, R.L.; Yigit, B.; Piehowski, P.D.; Kim, M.-J.; Dwyer, A.J.; et al. Increased β-Cell Proliferation before Immune Cell Invasion Prevents Progression of Type 1 Diabetes. Nat. Metab. 2019, 1, 509–518. [Google Scholar] [CrossRef]
- Johannesen, J.; Pie, A.; Pociot, F.; Kristiansen, O.P.; Karlsen, A.E.; Nerup, J.; Danish Study Group of Diabetes in Childhood. The Danish Insulin-dependent Diabetes Mellitus Epidemiology and Genetics Group Linkage of the Human Inducible Nitric Oxide Synthase Gene to Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2001, 86, 2792–2796. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Templin, A.T.; Maier, B.; Nishiki, Y.; Tersey, S.A.; Mirmira, R.G. Deoxyhypusine Synthase Haploinsufficiency Attenuates Acute Cytokine Signaling. Cell Cycle 2011, 10, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Turpaev, K.; Krizhanovskii, C.; Wang, X.; Sargsyan, E.; Bergsten, P.; Welsh, N. The Protein Synthesis Inhibitor Brusatol Normalizes High-Fat Diet-Induced Glucose Intolerance in Male C57BL/6 Mice: Role of Translation Factor EIF5A Hypusination. FASEB J. 2019, 33, 3510–3522. [Google Scholar] [CrossRef] [PubMed]
- Rosorius, O.; Reichart, B.; Krätzer, F.; Heger, P.; Dabauvalle, M.C.; Hauber, J. Nuclear Pore Localization and Nucleocytoplasmic Transport of EIF-5A: Evidence for Direct Interaction with the Export Receptor CRM1. J. Cell Sci. 1999, 112, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.P.; Finley, J.L.; Latour, Y.L.; Asim, M.; Smith, T.M.; Verriere, T.G.; Barry, D.P.; Allaman, M.M.; Delagado, A.G.; Rose, K.L.; et al. Hypusination Orchestrates the Antimicrobial Response of Macrophages. Cell Rep. 2020, 33, 108510. [Google Scholar] [CrossRef]
- de Almeida, O.P.; Toledo, T.R.; Rossi, D.; de Rossetto, D.B.; Watanabe, T.F.; Galvão, F.C.; Medeiros, A.I.; Zanelli, C.F.; Valentini, S.R. Hypusine Modification of the Ribosome-Binding Protein EIF5A, a Target for New Anti-Inflammatory Drugs: Understanding the Action of the Inhibitor GC7 on a Murine Macrophage Cell Line. Curr. Pharm. Des. 2014, 20, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Puleston, D.J.; Buck, M.D.; Klein Geltink, R.I.; Kyle, R.L.; Caputa, G.; O’Sullivan, D.; Cameron, A.M.; Castoldi, A.; Musa, Y.; Kabat, A.M.; et al. Polyamines and EIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 2019, 30, 352–363.e8. [Google Scholar] [CrossRef]
- Oliverio, S.; Corazzari, M.; Sestito, C.; Piredda, L.; Ippolito, G.; Piacentini, M. The Spermidine Analogue GC7 (N1-Guanyl-1,7-Diamineoheptane) Induces Autophagy through a Mechanism Not Involving the Hypusination of EIF5A. Amino Acids 2014, 46, 2767–2776. [Google Scholar] [CrossRef]
- Wang, F.; Sun, F.; Luo, J.; Yue, T.; Chen, L.; Zhou, H.; Zhang, J.; Yang, C.; Luo, X.; Zhou, Q.; et al. Loss of Ubiquitin-Conjugating Enzyme E2 (Ubc9) in Macrophages Exacerbates Multiple Low-Dose Streptozotocin-Induced Diabetes by Attenuating M2 Macrophage Polarization. Cell Death Dis. 2019, 10, 892. [Google Scholar] [CrossRef]
- Calderon, B.; Suri, A.; Unanue, E.R. In CD4+ T-Cell-Induced Diabetes, Macrophages Are the Final Effector Cells That Mediate Islet Beta-Cell Killing: Studies from an Acute Model. Am. J. Pathol. 2006, 169, 2137–2147. [Google Scholar] [CrossRef]
- Tsiavou, A.; Hatziagelaki, E.; Chaidaroglou, A.; Koniavitou, K.; Degiannis, D.; Raptis, S.A. Correlation between Intracellular Interferon-Gamma (IFN-Gamma) Production by CD4+ and CD8+ Lymphocytes and IFN-Gamma Gene Polymorphism in Patients with Type 2 Diabetes Mellitus and Latent Autoimmune Diabetes of Adults (LADA). Cytokine 2005, 31, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Lee, S.B.; Park, J.H.; Park, M.H. Essential Role of EIF5A-1 and Deoxyhypusine Synthase in Mouse Embryonic Development. Amino Acids 2012, 42, 703–710. [Google Scholar] [CrossRef] [PubMed]

| Role | References | Inhibitor | Model | Key Findings |
|---|---|---|---|---|
| β-cell function | Robertson et al., 2020 [13] | DFMO (inhibits ODC) | Zebrafish | DFMO promoted β-cell regeneration after β-cell injury. |
| Levasseur et al., 2019 [14] | - | β cell-specific Dhps KO mice | With HFD, mice with a β-cell knockout of Dhps exhibited impaired glucose tolerance and reduced insulin secretion. | |
| Cerrada-Gimenez et al., 2012 [15] | - | Ssat overexpressing mice | Depletion of spermidine and spermine levels led to impaired glucose-stimulated insulin secretion. | |
| Type 1 diabetes | Maier et al., 2010 [16] | - | STZ-treated mice | siRNA knockdown of Eif5a prevented hyperglycemia and maintained insulin secretory capacity in diabetic mice. |
| Tersey et al., 2014 [17] | DFMO (inhibits ODC) | NOD mice | Inhibition of polyamine biosynthesis significantly delayed T1D incidence, with reduced insulitis. | |
| Bjelakovic et al., 2010 [18] | - | Human patients with T1D | Polyamine oxidase activity was increased in T1D. | |
| Seghieri et al., 1990 [19] | - | Human patients with T1D | Spermidine oxidase activity was significantly lower in individuals with T1D | |
| Obesity and Type 2 diabetes | Fernandez-Garcia 2019 [20] | - | Human patients with T2D | Serum polyamine levels were elevated in T2D subjects and positively correlated with glycosylated Hb and fasting insulin. |
| Robbins et al., 2010 [21] | GC7 (inhibits DHPS) | db/db Mice | Treatment with GC7 resulted in improved glucose tolerance and insulin secretion. | |
| Fernández et al., 2017 [22] | - | HFD-induced obese mice | Spermidine supplementation led to a decrease in body weight, improved glucose tolerance, and enhanced insulin sensitivity. | |
| Sadasivan et al., 2014 [23] | - | HFD-induced obese mice | Exogenous spermine decreased body weight and fasting glucose and improved glucose tolerance in obese mice. | |
| Diabetic immunity | Anderson-Baucum et al., 2021 [24] | - | Myeloid-specific Dhps KO mice | eIF5Ahyp promoted M1 polarization and migration of macrophages in obese mice. |
| Karacay et al., 2022 [25] | - | NOD mice | Spermidine supplementation increased diabetes incidence with an increased proportion of pro-inflammatory T-cells. | |
| Imam et al., 2019 [26] | GC7 (inhibits DHPS) | NOD mice | GC7 treatment reduced pancreatic Th1 cells and increased Treg cells, resulting in overall delay of T1D onset. | |
| Colvin et al., 2013 [27] | GC7 (inhibits DHPS) | NOD mice | Inhibition of DHPS led to an impairment in proliferation and proinflammatory polarization of Th1 immune cells. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulkarni, A.; Anderson, C.M.; Mirmira, R.G.; Tersey, S.A. Role of Polyamines and Hypusine in β Cells and Diabetes Pathogenesis. Metabolites 2022, 12, 344. https://doi.org/10.3390/metabo12040344
Kulkarni A, Anderson CM, Mirmira RG, Tersey SA. Role of Polyamines and Hypusine in β Cells and Diabetes Pathogenesis. Metabolites. 2022; 12(4):344. https://doi.org/10.3390/metabo12040344
Chicago/Turabian StyleKulkarni, Abhishek, Cara M. Anderson, Raghavendra G. Mirmira, and Sarah A. Tersey. 2022. "Role of Polyamines and Hypusine in β Cells and Diabetes Pathogenesis" Metabolites 12, no. 4: 344. https://doi.org/10.3390/metabo12040344
APA StyleKulkarni, A., Anderson, C. M., Mirmira, R. G., & Tersey, S. A. (2022). Role of Polyamines and Hypusine in β Cells and Diabetes Pathogenesis. Metabolites, 12(4), 344. https://doi.org/10.3390/metabo12040344







