Independent and Interactive Effects of Genetic Background and Sex on Tissue Metabolomes of Adipose, Skeletal Muscle, and Liver in Mice
Abstract
1. Introduction
2. Results
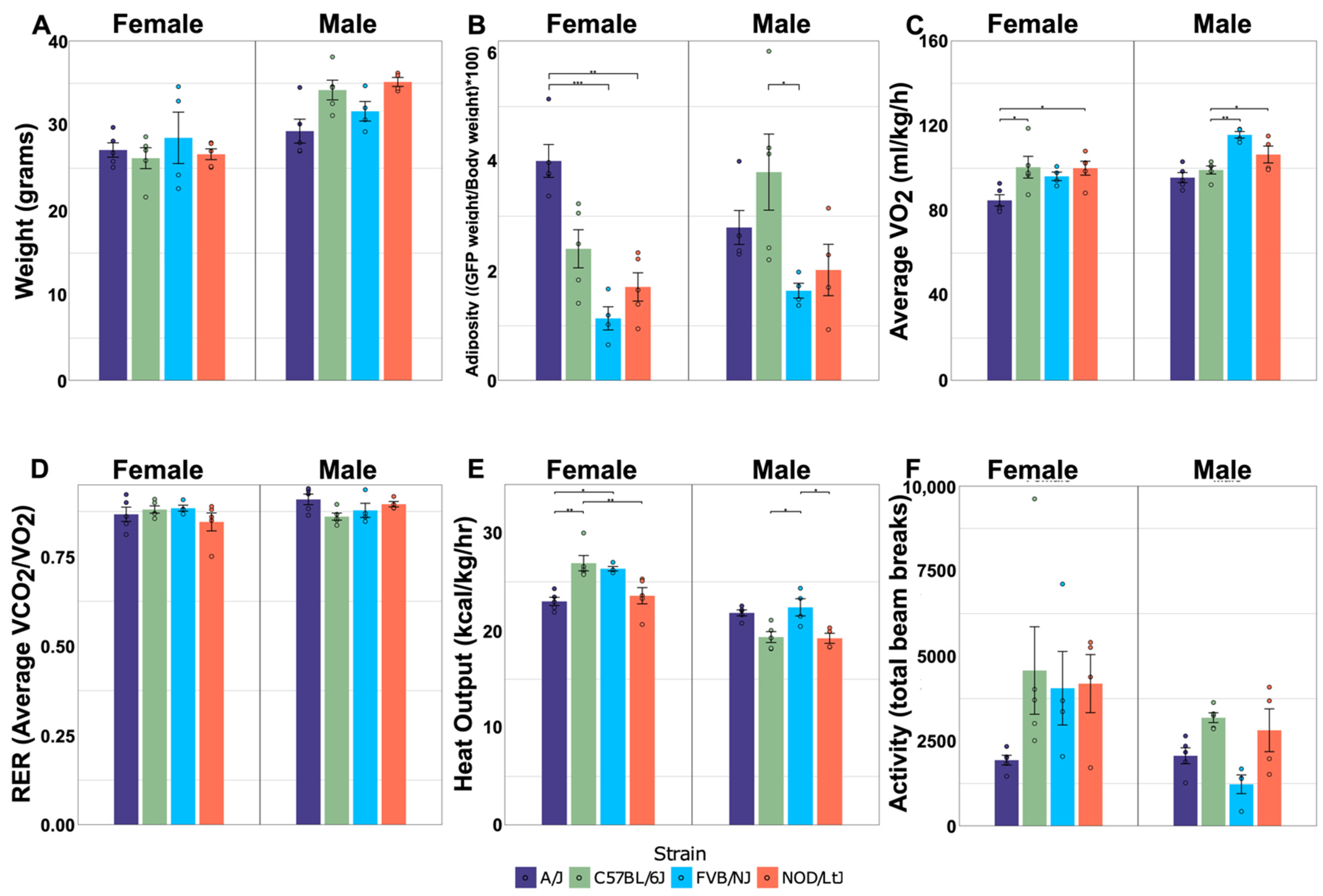
2.1. Physiological Measurements Reveal Differences among Strain and Sex-by-Strain
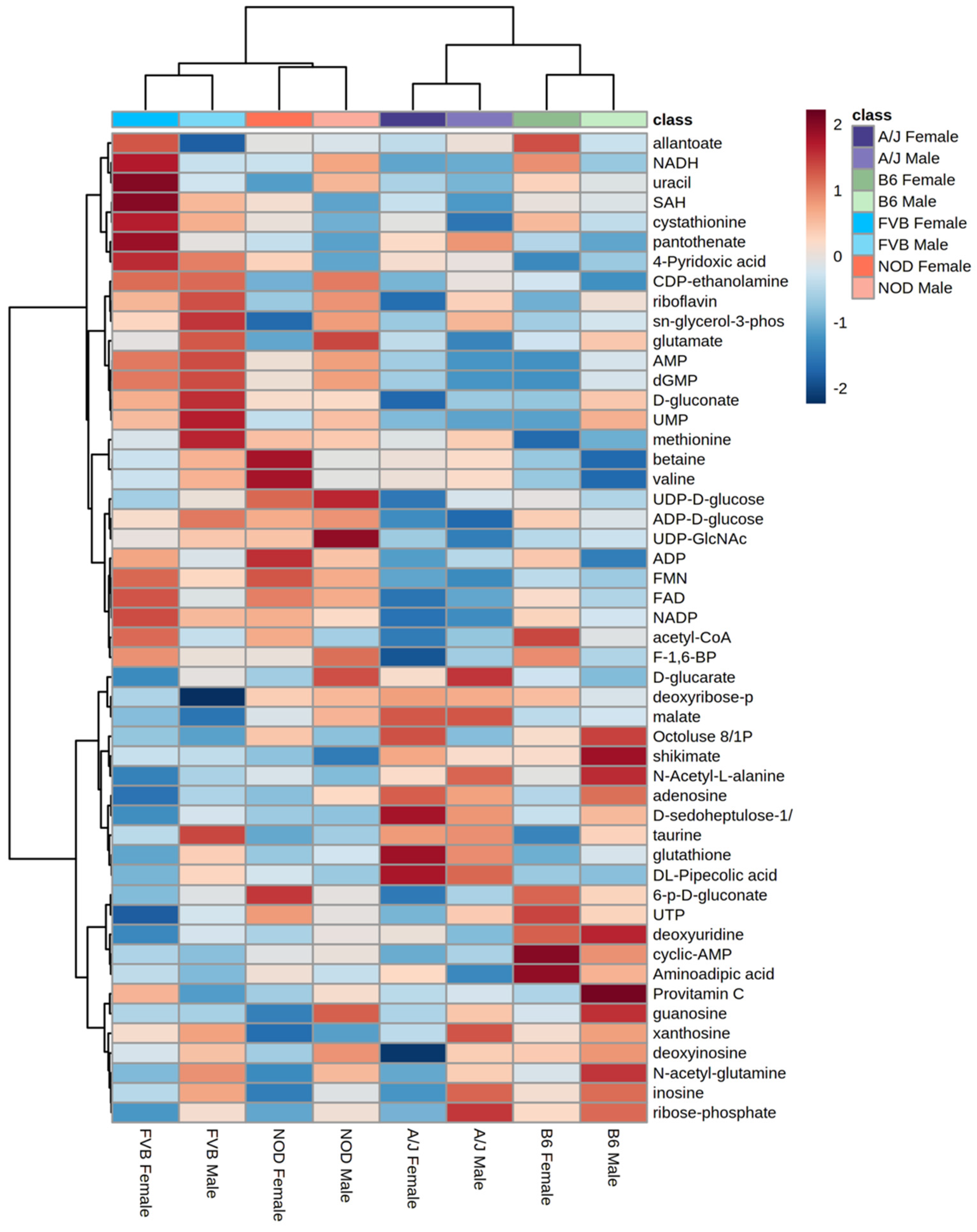
2.2. Significant Effects of Strain, Sex and Their Interactions on Tissue Metabolomes
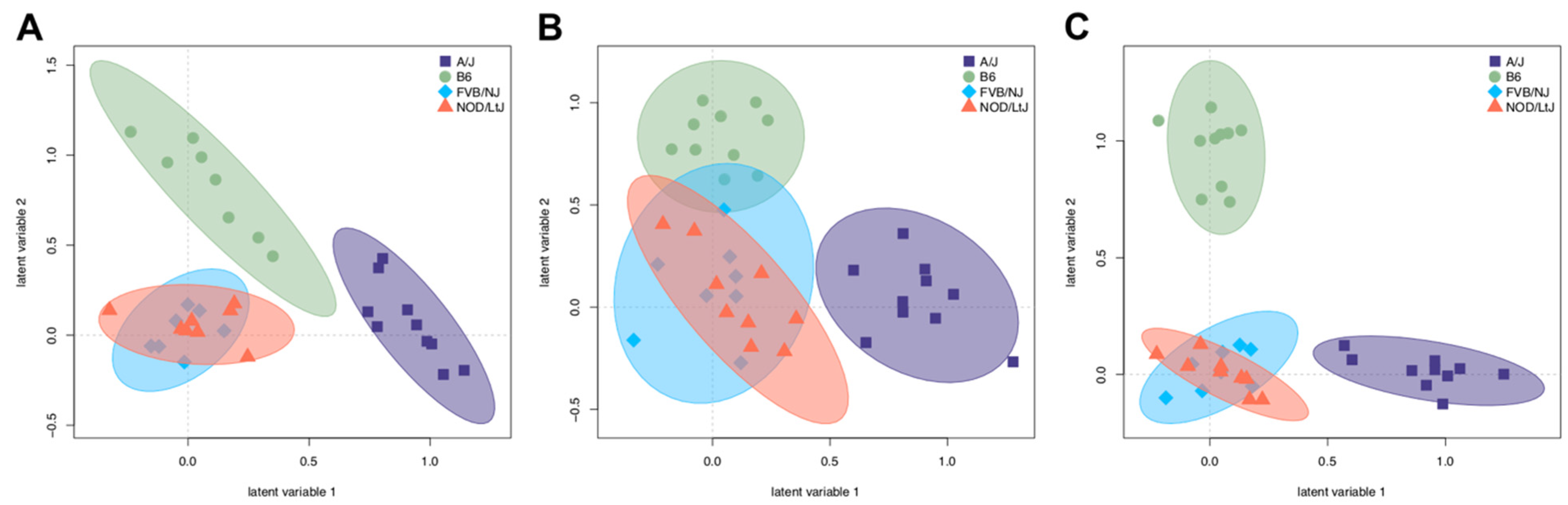
2.3. Partial Least Squares Discriminant Analysis Reveals Strain Is a Discriminant of Metabolites
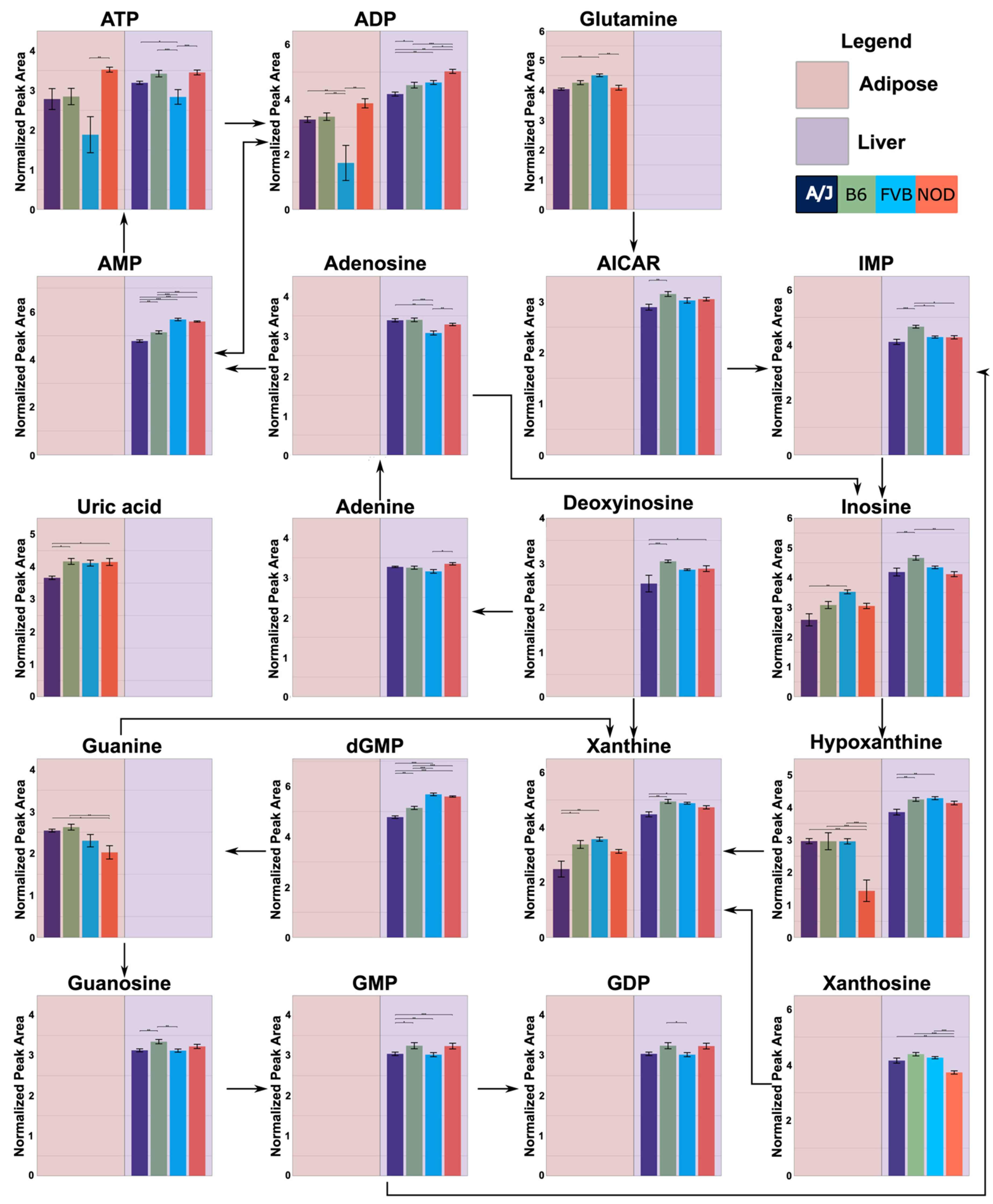
2.4. Functional Annotation of Strain Effects
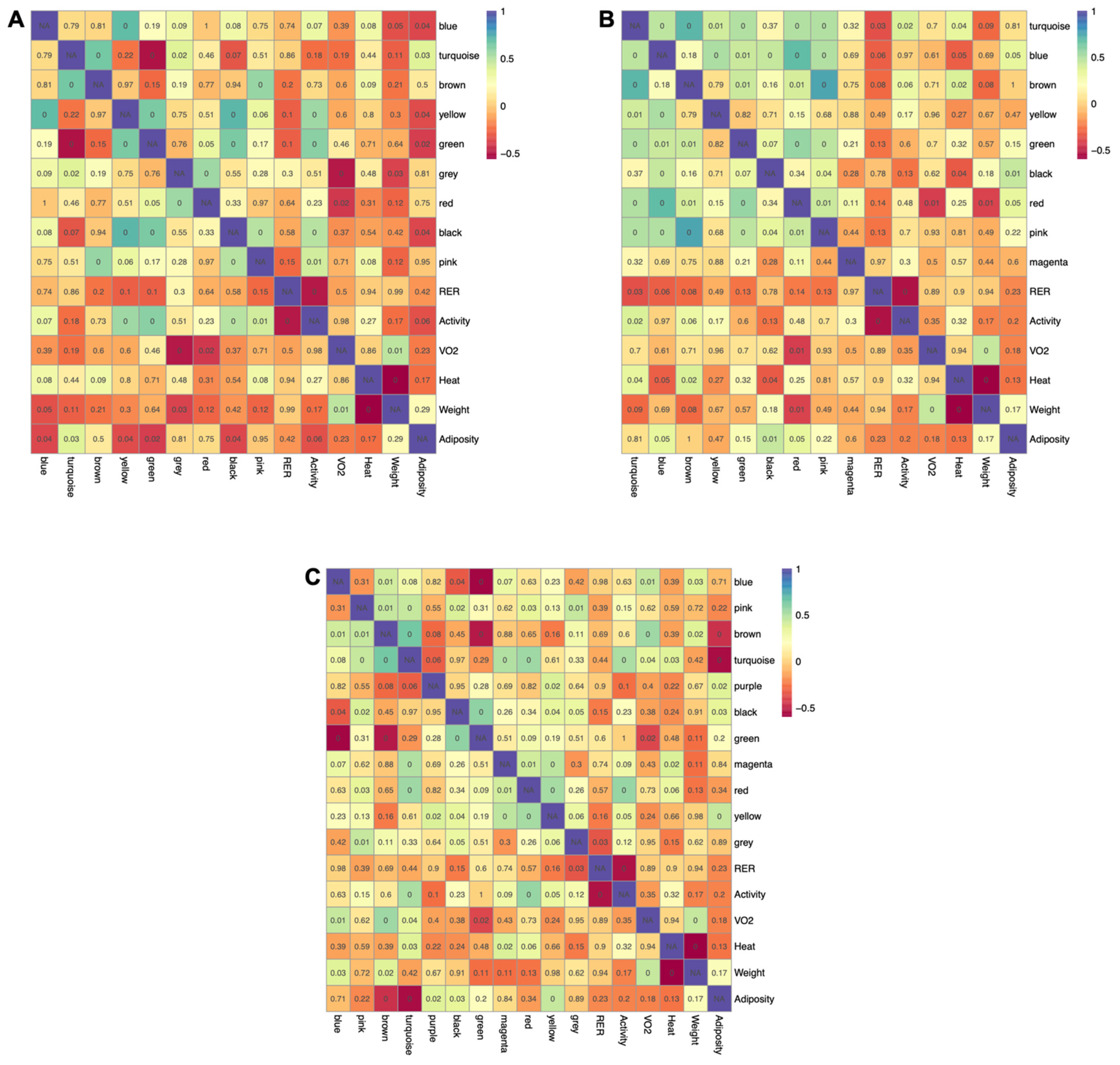
2.5. Connecting Metabolic Profiles to Traits
3. Discussion
4. Materials and Methods
4.1. Animals and Diets
4.2. Metabolite Extraction from Tissues
4.3. Liquid Chromatography Mass Spectrometry
4.4. Generation of 13C-Labeled E. coli
4.5. Metabolomics Data Processing
4.6. Use of 13C-Labelled E. coli Cellular Extracts as an Internal Standard
4.7. Statistical Analysis
internal standard coefficient (mean internal standard − internal standard)
4.8. Partial Least Squares Discriminant Analysis
4.9. Correlation Analysis
4.10. Functional Pathway Analysis
4.11. Weighted Gene Co-Expression Network Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silventoinen, K.; Jelenkovic, A.; Sund, R.; Hur, Y.-M.; Yokoyama, Y.; Honda, C.; Hjelmborg, J.; Möller, S.; Ooki, S.; Aaltonen, S.; et al. Genetic and environmental effects on body mass index from infancy to the onset of adulthood: An individual-based pooled analysis of 45 twin cohorts participating in the COllaborative project of Development of Anthropometrical measures in Twins (CODATwins) study. Am. J. Clin. Nutr. 2016, 104, 371–379. [Google Scholar] [CrossRef]
- Kastenmüller, G.; Raffler, J.; Gieger, C.; Suhre, K. Genetics of human metabolism: An update. Hum. Mol. Genet. 2015, 24, R93–R101. [Google Scholar] [CrossRef]
- Hagenbeek, F.A.; Pool, R.; van Dongen, J.; Draisma, H.H.M.; jan Hottenga, J.; Willemsen, G.; Abdellaoui, A.; Fedko, I.O.; den Braber, A.; Visser, P.J.; et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat Commun. 2020, 11, 39, Erratum in Nat. Commun. 2020, 11, 1702. [Google Scholar] [CrossRef]
- Long, T.; Hicks, M.; Yu, H.-C.; Biggs, W.H.; Kirkness, E.F.; Menni, C.; Zierer, J.; Small, K.S.; Mangino, M.; Messier, H.; et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017, 49, 568–578. [Google Scholar] [CrossRef]
- Chu, X.; Jaeger, M.; Beumer, J.; Bakker, O.B.; Aguirre-Gamboa, R.; Oosting, M.; Smeekens, S.P.; Moorlag, S.; Mourits, V.P.; Koeken, V.A.C.M.; et al. Integration of metabolomics, genomics, and immune phenotypes reveals the causal roles of metabolites in disease. Genome Biol. 2021, 22, 198. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. Opinion: Understanding ‘global’ systems biology: Metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2003, 2, 668–676. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
- Rossmeisl, M.; Rim, J.S.; Koza, R.A.; Kozak, L.P. Variation in Type 2 Diabetes-Related Traits in Mouse Strains Susceptible to Diet-Induced Obesity. Diabetes 2003, 52, 1958–1966. [Google Scholar] [CrossRef]
- Surwit, R.S.; Feinglos, M.N.; Rodin, J.; Sutherland, A.; Petro, A.E.; Opara, E.C.; Rebuffe-Scrive, M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism 1995, 44, 645–651. [Google Scholar]
- West, D.B.; Boozer, C.N.; Moody, D.L.; Atkinson, R.L. Dietary obesity in nine inbred mouse strains. Am. J. Physiol. Integr. Comp. Physiol. 1992, 262, R1025–R1032. [Google Scholar] [CrossRef]
- Haluzik, M.; Colombo, C.; Gavrilova, O.; Chua, S.; Wolf, N.; Chen, M.; Stannard, B.; Dietz, K.; Le Roith, D.; Reitman, M. Genetic Background (C57BL/6J Versus FVB/N) Strongly Influences the Severity of Diabetes and Insulin Resistance in ob/ob Mice. Endocrinology 2004, 145, 3258–3264. [Google Scholar] [CrossRef]
- Madsen, R.; Banday, V.; Moritz, T.; Trygg, J.; Lejon, K. Altered Metabolic Signature in Pre-Diabetic NOD Mice. PLoS ONE 2012, 7, e35445. [Google Scholar] [CrossRef]
- Bouwknecht, J.; Paylor, R. Behavioral and physiological mouse assays for anxiety: A survey in nine mouse strains. Behav. Brain Res. 2002, 136, 489–501. [Google Scholar] [CrossRef]
- Hennings, H.; Glick, A.B.; Lowry, D.T.; Krsmanovic, L.S.; Sly, L.M.; Yuspa, S.H. FVB/N mice: An inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis 1993, 14, 2353–2358. [Google Scholar] [CrossRef]
- Goios, A.; Pereira, L.; Bogue, M.; Macaulay, V.; Amorim, A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007, 17, 293–298. [Google Scholar] [CrossRef]
- Gavaghan, C.L.; Holmes, E.; Lenz, E.; Wilson, I.D.; Nicholson, J.K. An NMR-based metabonomic approach to investigate the biochemical consequences of genetic strain differences: Application to the C57BL10J and Alpk:ApfCD mouse. FEBS Lett. 2000, 484, 169–174. [Google Scholar] [CrossRef]
- Lacruz, M.E.; Kluttig, A.; Tiller, D.; Medenwald, D.; Giegling, I.; Rujescu, D.; Prehn, C.; Adamski, J.; Greiser, K.H.; Kastenmüller, G. Instability of personal human metabotype is linked to all-cause mortality. Sci. Rep. 2018, 8, 9810. [Google Scholar] [CrossRef]
- Yousri, N.A.; Kastenmüller, G.; Gieger, C.; Shin, S.-Y.; Erte, I.; Menni, C.; Peters, A.; Meisinger, C.; Mohney, R.P.; Illig, T.; et al. Long term conservation of human metabolic phenotypes and link to heritability. Metabolomics 2014, 10, 1005–1017. [Google Scholar] [CrossRef]
- Clayton, T.A.; Lindon, J.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.-P.; Le Net, J.-L.; Baker, D.; Walley, R.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef]
- Rezzi, S.; Martin, F.-P.J.; Kochhar, S. Defining Personal Nutrition and Metabolic Health Through Metabonomics. Ernst Scher. Found Symp. Proc. 2007, 4, 251–264. [Google Scholar] [CrossRef]
- Riedl, A.; Wawro, N.; Gieger, C.; Meisinger, C.; Peters, A.; Rathmann, W.; Koenig, W.; Strauch, K.; Quante, A.S.; Thorand, B.; et al. Modifying effect of metabotype on diet–diabetes associations. Eur. J. Nutr. 2019, 59, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Assfalg, M.; Bertini, I.; Colangiuli, D.; Luchinat, C.; Schäfer, H.; Schütz, B.; Spraul, M. Evidence of different metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Adav, S.S.; Wang, Y. Metabolomics Signatures of Aging: Recent Advances. Aging Dis. 2021, 12, 646–661. [Google Scholar] [CrossRef] [PubMed]
- Burlikowska, K.; Stryjak, I.; Bogusiewicz, J.; Kupcewicz, B.; Jaroch, K.; Bojko, B. Comparison of Metabolomic Profiles of Organs in Mice of Different Strains Based on SPME-LC-HRMS. Metabolites 2020, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Wells, A.; Barrington, W.T.; Dearth, S.; May, A.; Threadgill, D.W.; Campagna, S.R.; Voy, B.H. Tissue Level Diet and Sex-by-Diet Interactions Reveal Unique Metabolite and Clustering Profiles Using Untargeted Liquid Chromatography-Mass Spectrometry on Adipose, Skeletal Muscle, and Liver Tissue in C57BL6/J Mice. J. Proteome Res. 2018, 17, 1077–1090. [Google Scholar] [CrossRef]
- Lu, W.; Clasquin, M.F.; Melamud, E.; Amador-Noguez, D.; Caudy, A.A.; Rabinowitz, J.D. Metabolomic Analysis via Reversed-Phase Ion-Pairing Liquid Chromatography Coupled to a Stand Alone Orbitrap Mass Spectrometer. Anal. Chem. 2010, 82, 3212–3221. [Google Scholar] [CrossRef]
- Bennett, B.D.; Kimball, E.H.; Gao, M.; Osterhout, R.; Van Dien, S.J.; Rabinowitz, J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5, 593–599. [Google Scholar] [CrossRef]
- Wu, L.; Mashego, M.R.; van Dam, J.C.; Proell, A.M.; Vinke, J.L.; Ras, C.; van Winden, W.A.; van Gulik, W.M.; Heijnen, J.J. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 2005, 336, 164–171. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic Analysis and Visualization Engine for LC−MS Data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef]
- Clasquin, M.F.; Melamud, E.; Rabinowitz, J.D.; Clasquin, M.F.; Melamud, E.; Rabinowitz, J.D. LC-MS Data Processing with MAVEN: A Metabolomic Analysis and Visualization Engine. In Current Protocols in Bioinformatics, Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 14.11.1–14.11.23. ISBN 978-0-471-25095-1. [Google Scholar]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2008, 37, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- GmbH, M.S. XLConnect: Excel Connector for R. 2015. Available online: https://cran.r-project.org/web/packages/XLConnect/index.html (accessed on 22 February 2022).
- Hastie, T.; Tibshirani, R.; Narasimhan, B.; Chu, G. Impute: Imputation for Microarray Data, 2016.
- Nyamundanda, G.; Brennan, L.; Gormley, I.C. Probabilistic principal component analysis for metabolomic data. BMC Bioinform. 2010, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. Available online: http://www.jstor.org/stable/2346101 (accessed on 21 March 2022). [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Sanchez, G. DiscriMiner: Tools of the Trade for Discriminant Analysis, 2013.
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef]
- John Fox, S.W. An {R} Companion to Applied Regression, 2nd ed.; Sage: Thousand Oaks, CA, USA, 2011. [Google Scholar]
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous, 2015.
- Bengtsson, H. The R.oo package—Object-Oriented Programming with References Using Standard R Code. In Proceedings of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003), Vienna, Austria, 20–22 March 2003. [Google Scholar]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2011, 28, 373–380. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]

| Sex | Strain | Sex-by-Strain | |
|---|---|---|---|
| Weight | <0.001 | 0.284 | 0.052 |
| Adiposity | 0.338 | <0.001 | 0.015 |
| VO2 | 0.001 | <0.001 | 0.023 |
| RER | 0.148 | 0.594 | 0.100 |
| Heat Output | <0.001 | 0.001 | <0.001 |
| Activity | 0.009 | 0.020 | 0.124 |
| Tissue | Factor | Pathway | Hits | p-Value | (i/m)q |
|---|---|---|---|---|---|
| All Tissues | Sex | Purine metabolism | 9 | <0.001 | 0.001 |
| Strain | Purine metabolism | 20 | <0.001 | 0.008 | |
| Pyrimidine metabolism | 15 | <0.001 | 0.008 | ||
| Alanine, aspartate and glutamate metabolism | 8 | <0.001 | 0.008 | ||
| Ascorbate and aldarate metabolism | 5 | <0.001 | 0.008 | ||
| Citrate cycle (TCA cycle) | 6 | 0.002 | 0.008 | ||
| Pantothenate and CoA biosynthesis | 5 | 0.003 | 0.008 | ||
| Aminoacyl-tRNA biosynthesis | 12 | 0.003 | 0.008 | ||
| Adipose | Strain | Purine metabolism | 8 | <0.001 | 0.007 |
| Pyrimidine metabolism | 5 | 0.001 | 0.007 | ||
| Arginine and proline metabolism | 5 | 0.002 | 0.007 | ||
| Aminoacyl-tRNA biosynthesis | 6 | 0.002 | 0.007 | ||
| Muscle | Sex | Aminoacyl-tRNA biosynthesis | 5 | 0.001 | 0.002 |
| Strain | Aminoacyl-tRNA biosynthesis | 6 | 0.001 | 0.004 | |
| Liver | Sex | Purine metabolism | 9 | <0.001 | 0.003 |
| Strain | Purine metabolism | 17 | <0.001 | 0.003 | |
| Pyrimidine metabolism | 13 | <0.001 | 0.003 | ||
| Pantothenate and CoA biosynthesis | 5 | 0.002 | 0.003 | ||
| Sex-by-Strain | Pyrimidine metabolism | 4 | 0.001 | 0.002 |
| SNP Type per Mouse Strain | |||||
|---|---|---|---|---|---|
| Intermediary Gene | Pathway Step | A/J | C57BL/6J | FVB/NJ | NOD/ShiLtJ |
| Enpp1 | ATP → AMP | Cn, Cs | Cn, Cs | U3, Cn, Cs | |
| Ak7 | AMP → ADP | U3, Cn, Cs | |||
| Gdr | Guanine → Xanthine | U3 | U3 | ||
| Nt5c3b | Adenosine → Inosine | Cn, Cs | |||
| Xdh | Xanthine → Uric acid | U3, Cn, Cs | |||
| Guk1 | GMP → GDP | U3, U5, Cn | U3, U5, Cn | U3, U5, Cn | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wells, A.E.; Barrington, W.T.; Dearth, S.; Milind, N.; Carter, G.W.; Threadgill, D.W.; Campagna, S.R.; Voy, B.H. Independent and Interactive Effects of Genetic Background and Sex on Tissue Metabolomes of Adipose, Skeletal Muscle, and Liver in Mice. Metabolites 2022, 12, 337. https://doi.org/10.3390/metabo12040337
Wells AE, Barrington WT, Dearth S, Milind N, Carter GW, Threadgill DW, Campagna SR, Voy BH. Independent and Interactive Effects of Genetic Background and Sex on Tissue Metabolomes of Adipose, Skeletal Muscle, and Liver in Mice. Metabolites. 2022; 12(4):337. https://doi.org/10.3390/metabo12040337
Chicago/Turabian StyleWells, Ann E., William T. Barrington, Stephen Dearth, Nikhil Milind, Gregory W. Carter, David W. Threadgill, Shawn R. Campagna, and Brynn H. Voy. 2022. "Independent and Interactive Effects of Genetic Background and Sex on Tissue Metabolomes of Adipose, Skeletal Muscle, and Liver in Mice" Metabolites 12, no. 4: 337. https://doi.org/10.3390/metabo12040337
APA StyleWells, A. E., Barrington, W. T., Dearth, S., Milind, N., Carter, G. W., Threadgill, D. W., Campagna, S. R., & Voy, B. H. (2022). Independent and Interactive Effects of Genetic Background and Sex on Tissue Metabolomes of Adipose, Skeletal Muscle, and Liver in Mice. Metabolites, 12(4), 337. https://doi.org/10.3390/metabo12040337