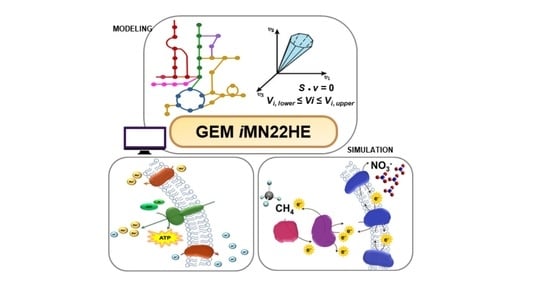
A Genome-Scale Metabolic Model of Methanoperedens nitroreducens: Assessing Bioenergetics and Thermodynamic Feasibility
Abstract
:1. Introduction
2. Results and Discussion
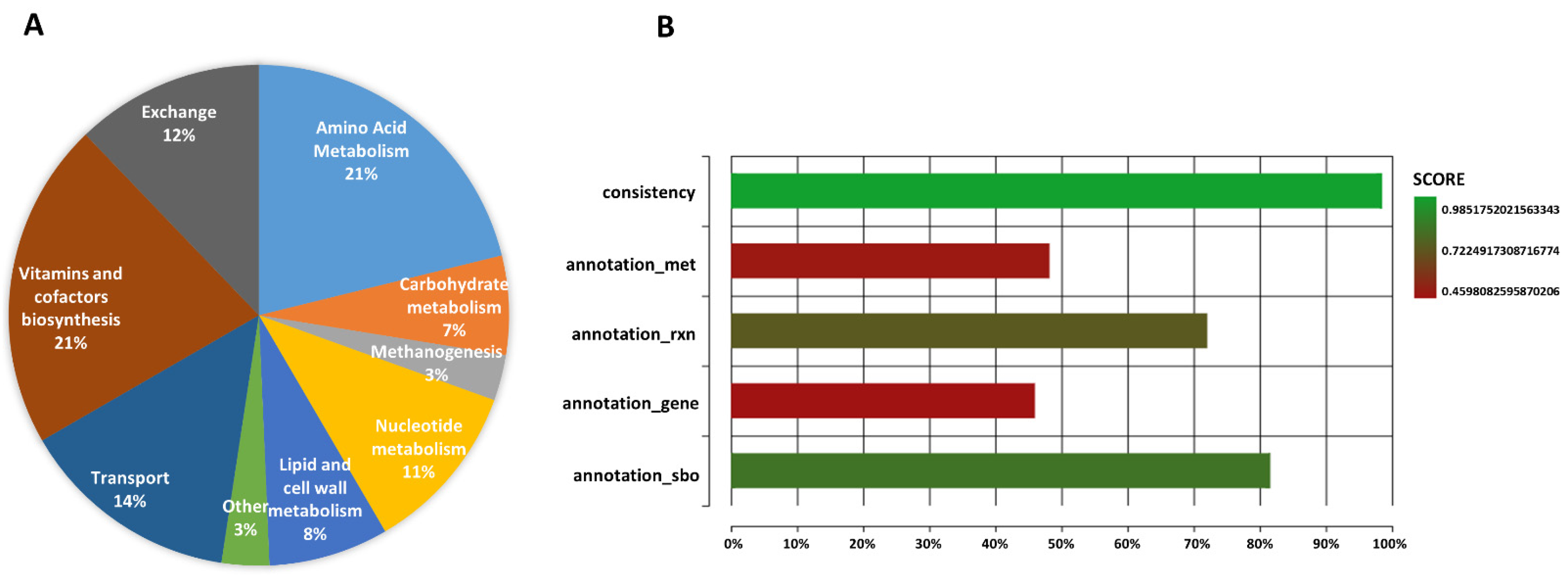
2.1. General Properties and Model Validation of iMN22HE
2.2. Comparison of iMN22HE with Other Relative Models
2.3. Model Prediction of Electron Confurcation Essentiality in Reverse Methanogenesis
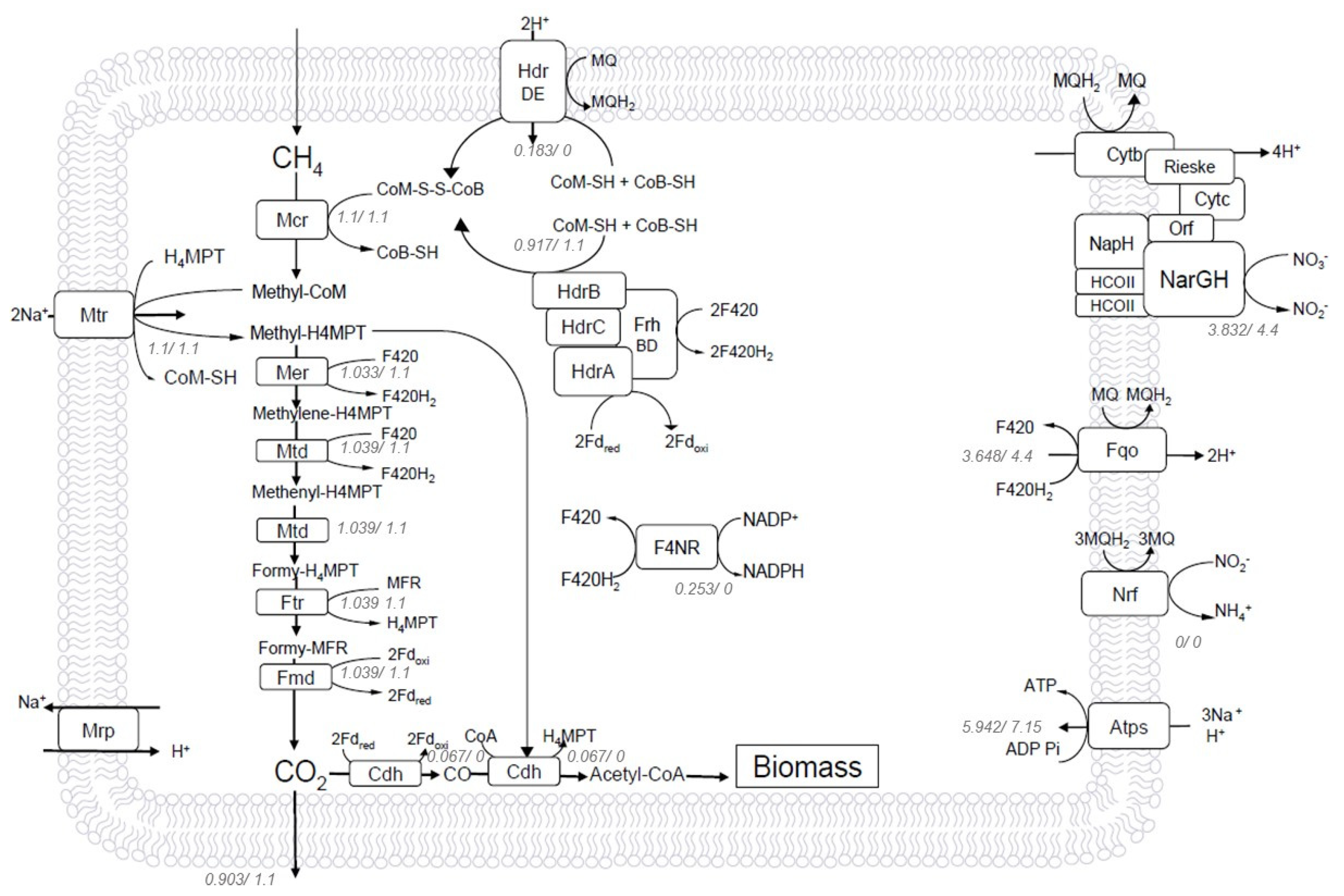
2.4. Bioenergetics Analysis of Steady-State Reverse Methanogenesis Using Flux Balance Analysis (FBA)
2.5. Electron Transfer during Nitrate-Driven Methane Oxidation in M. nitroredencens
2.6. Thermodynamic Feasibility in Endergonic Methane Oxidation
3. Materials and Methods
3.1. Metabolic Model Reconstruction
3.2. Model Simulation with Flux Balance Analysis
3.3. Thermodynamic Calculations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciais, P.; Sabine, C.; Bala, G.; Bopp, L.; Brovkin, V.; House, J.I. Carbon and Other Biogeochemical Cycles. In Climate Change 2013; Edenhofer, O., Pichs-Madruga, R., Sokona, Y., Farahani, E., Kadner, S., Seyboth, K., Adler, A., Baum, I., Brunner, S., Eickemeier, P., et al., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 465–570. [Google Scholar]
- Conrad, R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 2009, 1, 23765881. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, S.; Bousquet, P.; Ciais, P. Three decades of global methane sources and sinks. Nat. Geosci. 2013, 6, 813–823. [Google Scholar] [CrossRef]
- Kurth, J.M.; Camp, H.O.D.; Welte, C.U. Several ways one goal-methanogenesis from unconventional substrates. Appl. Microbiol. Biotechnol. 2020, 104, 6839–6854. [Google Scholar] [CrossRef] [PubMed]
- Niemann, H.; Lösekann, T.; Beer, D.D. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 2006, 443, 854–858. [Google Scholar] [CrossRef]
- Reeburgh, W.S. Oceanic methane biogeochemistry. Chem. Rev. 2007, 107, 486–513. [Google Scholar] [CrossRef] [Green Version]
- Hinrichs, K.U.; Hayes, J.M.; Sylva, S.P.; Brewer, P.G.; Delong, E.F. Methane-consuming archaebacteria in marine sediments. Nature 1999, 398, 802–805. [Google Scholar] [CrossRef]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623–626. [Google Scholar] [CrossRef]
- Timmers, P.H.; Widjaja-Greefkes, H.C.; Ramiro-Garcia, J.; Plugge, J.; Stams, C.M.; Stams, A. Growth and activity of ANME clades with different sulfate and sulfide concentrations in the presence of methane. Front. Microbiol. 2015, 6, 988. [Google Scholar] [CrossRef] [Green Version]
- Hinrichs, K.U.; Boetius, A. The Anaerobic Oxidation of Methane: New Insights in Microbial Ecology and Biogeochemistry; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Knittel, K.; Lösekann, T.; Boetius, A.; Kort, R.; Amann, R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 2005, 71, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Hoehler, T.M.; Alperin, M.J.; Albert, D.B.; Martens, C.S. Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methanogen-sulfate reducer consortium. Glob. Biogeochem. Cycles 1994, 8, 451–463. [Google Scholar] [CrossRef]
- Orphan, V.J.; Hinrichs, K.U.; Ussler, W.P.C.K., III. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 2001, 67, 1922–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knittel, K.; Boetius, A. Anaerobic oxidation of methane: Progress with an unknown process. Annu. Rev. Microbiol. 2009, 63, 311–334. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Blumenberg, M.; Michaelis, W.; Siegert, M.; Krüger, M.; Seifert, R. Methanogenic capabilities of ANME-archaea deduced from (13) C-labelling approaches. Environ. Microbiol. 2013, 15, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.G.; Alperin, M.J.; Teske, A. Environmental evidence for net methane production and oxidation in putative Anaerobic MEthanotrophic (ANME) archaea. Environ. Microbiol. 2011, 13, 2548–2564. [Google Scholar] [CrossRef] [PubMed]
- Amos, R.T.; Bekins, B.A.; Cozzarelli, I.M. Evidence for iron-mediated anaerobic methane oxidation in a crude oil-contaminated aquifer. Geobiology 2012, 10, 506–517. [Google Scholar] [CrossRef]
- Sivan, O.; Antler, G.; Turchyn, A.V.; Marlow, J.J.; Orphan, V.J. Iron oxides stimulate sulfate-driven anaerobic methane oxidation in seeps. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4147. [Google Scholar] [CrossRef] [Green Version]
- Ettwig, K.F.; Zhu, B.; Speth, D.; Keltjens, J.T.; Jetten, M.; Kartal, B. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. USA 2016, 113, 12792–12796. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Leu, A.O.; Xie, G.J. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 2018, 12, 1929–1939. [Google Scholar] [CrossRef]
- Li, H.; Yang, Q.; Zhou, H. Niche Differentiation of Sulfate- and Iron-Dependent Anaerobic Methane Oxidation and Methylotrophic Methanogenesis in Deep Sea Methane Seeps. Front. Microbiol. 2020, 11, 1409. [Google Scholar] [CrossRef]
- Raghoebarsing, A.A.; Pol, A.; Pas-Schoonen, K.T.V.D. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 2006, 440, 918–921. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 2013, 501, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Arshad, A.; Speth, D.R.; Graaf, R.M.D.; Camp, H.J.O.D.; Jetten, M.S.; Welte, C.U. A Metagenomics-Based Metabolic Model of Nitrate-Dependent Anaerobic Oxidation of Methane by Methanoperedens-Like Archaea. Front. Microbiol. 2015, 6, 1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Zeng, R.J.; Burow, L.C.; Lant, P.; Keller, J.; Yuan, Z. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ. Microbiol. Rep. 2009, 1, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.W.; Lu, Y.Z.; Fu, L.; Ding, J.; Zeng, R.J. Simultaneous enrichment of denitrifying anaerobic methane-oxidizing microorganisms and anammox bacteria in a hollow-fiber membrane biofilm reactor. Appl. Microbiol. Biotechnol. 2017, 101, 437–446. [Google Scholar] [CrossRef]
- Vaksmaa, A.; Lüke, C.; Alen, T.V. Distribution and activity of the anaerobic methanotrophic community in a nitrogen-fertilized Italian paddy soil. FEMS Microbiol. Ecol. 2016, 92, fiw181. [Google Scholar] [CrossRef]
- Ding, J.; Ding, Z.W.; Fu, L.; Lu, Y.Z.; Cheng, S.H.; Zeng, R.J. New primers for detecting and quantifying denitrifying anaerobic methane oxidation archaea in different ecological niches. Appl. Microbiol. Biotechnol. 2015, 99, 9805–9812. [Google Scholar] [CrossRef]
- Mcglynn, S.E. Energy Metabolism during Anaerobic Methane Oxidation in ANME Archaea. Microbes Environ. 2017, 32, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Scheller, S.; Goenrich, M.; Boecher, R.; Thauer, R.K.; Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 2010, 465, 606–608. [Google Scholar] [CrossRef]
- Meyerdierks, A.; Kube, M.; Kostadinov, I. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 2010, 12, 422–439. [Google Scholar] [CrossRef]
- Wang, F.P.; Zhang, Y.; Chen, Y. Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J. 2014, 8, 1069–1078. [Google Scholar] [CrossRef] [Green Version]
- Richards, M.A.; Lie, T.J.; Zhang, J.; Ragsdale, S.W.; Leigh, J.A.; Price, N.D. Exploring Hydrogenotrophic Methanogenesis: A Genome Scale Metabolic Reconstruction of Methanococcus maripaludis. J. Bacteriol. 2016, 198, 3379–3390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benedict, M.N.; Gonnerman, M.C.; Metcalf, W.W.; Price, N.D. Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A. J. Bacteriol. 2012, 194, 855–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazem-Bokaee, H.; Gopalakrishnan, S.; Ferry, J.G.; Wood, T.K.; Maranas, C.D. Assessing methanotrophy and carbon fixation for biofuel production by Methanosarcina acetivorans. Microb. Cell Fact. 2016, 15, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z.; Ferry, J.G. Electron Bifurcation and Confurcation in Methanogenesis and Reverse Methanogenesis. Front. Microbiol. 2018, 9, 1322. [Google Scholar] [CrossRef] [Green Version]
- Timmers, P.H.; Welte, C.U.; Koehorst, J.J.; Plugge, C.M.; Jetten, M.S.; Stams, A.J. Reverse Methanogenesis and Respiration in Methanotrophic Archaea. Archaea 2017, 2017, 1654237. [Google Scholar] [CrossRef]
- Thor, S.; Peterson, J.R.; Luthey-Schulten, Z. Genome-Scale Metabolic Modeling of Archaea Lends Insight into Diversity of Metabolic Function. Archaea 2017, 2017, 9763848. [Google Scholar] [CrossRef] [Green Version]
- Carey, M.A.; Dräger, A.; Beber, M.E.; Papin, J.A.; Yurkovich, J.T. Community standards to facilitate development and addresschallenges in metabolic modeling. Mol. Syst. Biol. 2020, 16, 9235. [Google Scholar] [CrossRef]
- Lieven, C.; Beber, M.E.; Olivier, B.G.; Bergmann, F.T.; Ataman, M.; Babaei, P.; Bartell, J.A.; Blank, L.M.; Chauhan, S.; Correia, K.; et al. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020, 38, 272–276. [Google Scholar] [CrossRef] [Green Version]
- Feist, A.M.; Scholten, J.C.; Palsson, B.Ø.; Brockman, F.J.; Ideker, T. Modeling methanogenesis with a genome-scale metabolicreconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2006, 2, 2006-0004. [Google Scholar] [CrossRef] [Green Version]
- Gonnerman, M.C.; Benedict, M.N.; Feist, A.M.; Metcalf, W.W.; Price, N.D. Genomically and biochemically accurate metabolicreconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotechnol. J. 2013, 8, 1070–1079. [Google Scholar] [CrossRef]
- Satish Kumar, V.; Ferry, J.G.; Maranas, C.D. Metabolic reconstruction of the archaeon methanogen Methanosarcina Acetivorans. BMC Syst. Biol. 2011, 5, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, T.; Koch, J.; Ermler, U.; Shima, S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 2017, 357, 699–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nauhaus, K.; Albrecht, M.; Elvert, M.; Boetius, A.; Widdel, F. In vitro cell growth of marine archaeal-bacterial consortia duringanaerobic oxidation of methane with sulfate. Environ. Microbiol. 2007, 9, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Welte, C.; Deppenmeier, U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 2014, 1837, 1130–1147. [Google Scholar] [CrossRef] [Green Version]
- Henry, C.S.; Broadbelt, L.J.; Hatzimanikatis, V. Thermodynamics-based metabolic flux analysis. Biophys. J. 2007, 92, 1792–1805. [Google Scholar] [CrossRef] [Green Version]
- Thiele, I.; Palsson, B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; Dejongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [Green Version]
- Henry, C.S.; Dejongh, M.; Best, A.A.; Frybarger, P.M.; Linsay, B.; Stevens, R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010, 28, 977–982. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef] [Green Version]
- Caspi, R.; Billington, R.; Ferrer, L. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D459–D471. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, I.; Chang, A.; Ebeling, C.; Gremse, M.; Heldt, C.; Huhn, G.; Schomburg, D. Brenda the enzyme database: Updates and major new developments. Nucleic Acids Res. 2004, 32, 431–434. [Google Scholar] [CrossRef] [Green Version]
- Consortium, U. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2007, 35, D154–D159. [Google Scholar]
- Schellenberger, J.; Park, J.O.; Conrad, T.M.; Palsson, B.Ø. BiGG: A Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinform. 2010, 11, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberhardt, M.A.; Palsson, B.Ø.; Papin, J.A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 2009, 5, 320. [Google Scholar] [CrossRef]
- Schellenberger, J.; Que, R.; Fleming, R.M. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat. Protoc. 2011, 6, 1290–1307. [Google Scholar] [CrossRef] [Green Version]
- King, Z.A.; Dräger, A.; Ebrahim, A.; Sonnenschein, N.; Lewis, N.E.; Palsson, B.O. Escher: A Web Application for Building, Sharing, and Embedding Data-Rich Visualizations of Biological Pathways. PLoS Comput. Biol. 2015, 11, e1004321. [Google Scholar] [CrossRef] [Green Version]


| Organism | Model | Mets | Rxns | Central Metabolic Pathway | Main Energy-Conserving Enzymes | Soluble Heterodisufide (HdrABC) | Citations |
|---|---|---|---|---|---|---|---|
| M. nitroreducens M. barkeri | iMN22HE iAF698 | 684 558 | 813 619 | Reverse methanogenesis Hydrogenotrophic methano-genesis; | Fqo Fpo Ech Vho | Electron confurcation NR | [41] |
| Methylotrophic methanogenesis | |||||||
| iMG746 | 718 | 815 | Hydrogenotrophic methanogenesis; | Fpo Ech Vho | Electron bifurcation | [42] | |
| Methylotrophic methanogenesis | |||||||
| iVS941 | 708 | 705 | Acetoclastic methanogenesis; Methylotrophic methanogenesis | Fpo Rnf | NR | [43] | |
| M. acetivorans | iMB745 | 715 | 818 | Acetoclastic methanogenesis; Methylotrophic methanogenesis | Fpo Rnf | Electron bifurcation | [34] |
| iMAC868 | 707 | 839 | Acetoclastic methanogenesis; Methylotrophic methanogenesis; | Fpo Rnf | Electron bifurcation | [35] | |
| M. maripaludis | iMR539 | 605 | 570 | Reverse methanogenesis Hydrogenotrophic methano-genesis | Eha/Ehb | Electron bifurcation | [33] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, B.; Cai, C.; McCubbin, T.; Muriel, J.C.; Sonnenschein, N.; Hu, S.; Yuan, Z.; Marcellin, E. A Genome-Scale Metabolic Model of Methanoperedens nitroreducens: Assessing Bioenergetics and Thermodynamic Feasibility. Metabolites 2022, 12, 314. https://doi.org/10.3390/metabo12040314
He B, Cai C, McCubbin T, Muriel JC, Sonnenschein N, Hu S, Yuan Z, Marcellin E. A Genome-Scale Metabolic Model of Methanoperedens nitroreducens: Assessing Bioenergetics and Thermodynamic Feasibility. Metabolites. 2022; 12(4):314. https://doi.org/10.3390/metabo12040314
Chicago/Turabian StyleHe, Bingqing, Chen Cai, Tim McCubbin, Jorge Carrasco Muriel, Nikolaus Sonnenschein, Shihu Hu, Zhiguo Yuan, and Esteban Marcellin. 2022. "A Genome-Scale Metabolic Model of Methanoperedens nitroreducens: Assessing Bioenergetics and Thermodynamic Feasibility" Metabolites 12, no. 4: 314. https://doi.org/10.3390/metabo12040314
APA StyleHe, B., Cai, C., McCubbin, T., Muriel, J. C., Sonnenschein, N., Hu, S., Yuan, Z., & Marcellin, E. (2022). A Genome-Scale Metabolic Model of Methanoperedens nitroreducens: Assessing Bioenergetics and Thermodynamic Feasibility. Metabolites, 12(4), 314. https://doi.org/10.3390/metabo12040314