Abstract
Tumor cells detached from the extracellular matrix (ECM) undergo anoikis resistance and metabolic reprogramming to facilitate cancer cell survival and promote metastasis. During ECM detachment, cancer cells utilize genomic methylation to regulate transcriptional events. One-carbon (1C) metabolism is a well-known contributor of SAM, a global substrate for methylation reactions, especially DNA methylation. DNA methylation-mediated repression of NK cell ligands MICA and MICB during ECM detachment has been overlooked. In the current work, we quantitated the impact of ECM detachment on one-carbon metabolites, expression of 1C regulatory pathway genes, and total methylation levels. Our results showed that ECM detachment promotes the accumulation of one-carbon metabolites and induces regulatory pathway genes and total DNA methylation. Furthermore, we measured the expression of well-known targets of DNA methylation in NK cell ligands in cancer cells, namely, MICA/B, during ECM detachment and observed low expression compared to ECM-attached cancer cells. Finally, we treated the ECM-detached cancer cells with vitamin C (a global methylation inhibitor) and observed a reduction in the promoter methylation of NK cell ligands, resulting in MICA/B re-expression. Treatment with vitamin C was also found to reduce global DNA methylation levels in ECM-detached cancer cells.
1. Introduction
Extracellular matrix (ECM) attachment is required for cell survival [1], whereas ECM detachment induces the programmed cell death called anoikis. Anoikis is considered a cancer hallmark, and during oncogenic progression, cancer cells deregulate anoikis and overcome it; hence, the cells become anoikis-resistant [2,3]. Moreover, ECM detachment intimately assists in cancer metastasis and ensures metabolic reprogramming with metastasis progression [4,5,6]. For example, a research study revealed that ECM detachment could induce the reductive carboxylation metabolism that promotes glycolysis [7]. Furthermore, loss of cell matrix attachment can induce hypoxia [7], which, in turn, is able to adapt tumor metabolism [8,9], though inductive correlations between hypoxia, ECM remodelling, and metastasis exist [10,11]. In addition, a well-founded study also revealed that the ratio of S-adenosyl methionine (SAM) to S-adenosyl homocysteine (SAH) (SAM/SAH ratio) is a tightly regulated epigenetic modification, as is DNA methylation [12]. The distribution of DNA methylation occurs predominantly (60–80%) in CpG dense regions, while 10% occurs in CpG islands (CGIs) [13]. The primary mechanism of DNA methylation underlies the transfer of SAM metabolites’ methyl groups through covalent modification, with one carbon atom attaching to the fifth position at cytosine to form 5-methyl cytosine (5mC) [14,15]. In addition, this covalent modification is mainly driven by a group of DNA methyltransferase enzymes known as DNMT1, DNMT3a, and DNMT3b; DNMT1 maintains the methylation reaction during the replication phase and the other two DNMTs perform de novo methylation [16,17]. Recently, our research team has identified that ECM detachment results in a metabolic rearrangement that could promote levels of metabolites associated with methylation, including S-adenosyl methionine (SAM) and S-adenosyl homocysteine (SAH) [18]. However, the global landscape of pathways related to methylation is still incompletely mapped, with certain pathways missing. Given this situation, our study aims to explore the one-carbon metabolism that supports the methylation of DNA/RNA and proteins.
Natural killer (NK) cells are innate immune cells and, in physiologically damaged conditions, such as tumor cell growth, they act as robust cytotoxic functioning cells. Although healthy cells express MHC-I, an inhibitory ligand for NK cells, cancer cell surfaces lose this molecule [19,20]. Moreover, NK cell-expressed activating receptor (natural killer group 2D) NKG2D captures MHC-I-type ligands on cell surfaces [21], rendering immune surveillance against tumors [22]. One of the most studied and genomically prevalent human ligands is MHC class I polypeptide-related protein A, along with B-MICA [23] and MICB [24]. However, NKG2DL expression has been reported in several pooled scientific data sets as being under tight epigenetic regulation [25,26,27]. A study also revealed that expression may be absent due to DNA methylation-mediated transcriptional silencing [28]. However, epi-transcriptomic silencing could be overcome and expression activated upon treatment with DNMTis, such as DAC and 5azaC [29]. In addition, a recent review revealed that vitamin C could reprogram the epigenome [30,31].
However, the study also showed that MICA expression is reduced when a cell loses its adherence matrix [32]. It has been overlooked whether the upregulated DNA methylation in ECM-detached cancer cells could repress MICA/B expression. ECM detachment can cause hypoxia [7], which mediates the DNA hypermethylation [33]. Therefore, this study has also been designed to investigate DNA methylation status and MICA/B protein expression in both attached and detached conditions with or without 5-aza-dc and vitamin C treatment.
2. Results
2.1. Loss of ECM Attachment Increased Levels of One-Carbon (1C) Metabolites
First, both HeLa and MCF-7 cell lines were grown in poly-HEMA gel-coated 6-well plates for six days to induce the ECM detachment model in cell lines. After that, both HeLa and MCF-7 cell lines, attached and 6 days ECM-detached cell aliquots, were taken for whole experiments (Figure 1a). The extracted metabolites were detected by HPLC coupling with LTQ LC–MS-based high throughput technology used to generate patterns of fragments and identify untargeted metabolites. The metabolite spectrum has been obtained through the data-dependent acquisition (DDA) method, followed by the processing of RAW files. A total of 833 metabolites was detected in both cell lines, and screening was performed to make shorts out of metabolites related to the one-carbon metabolic pathway. The peak intensity of these metabolites was plotted for a box plot and heat map (Figure 1b,c). Our results show that, after 6 days of ECM detachment in the HeLa cell line, higher levels of one-carbon metabolites, such as SAM, methionine, cysteine, homocysteine, etc., except L-serine, were detected. In addition, higher levels of glycerol-3-phosphate, DL-glutamate, pyruvic acid, adenine, spermine, etc., were also detected in the HeLa cell line (Figure 1b).
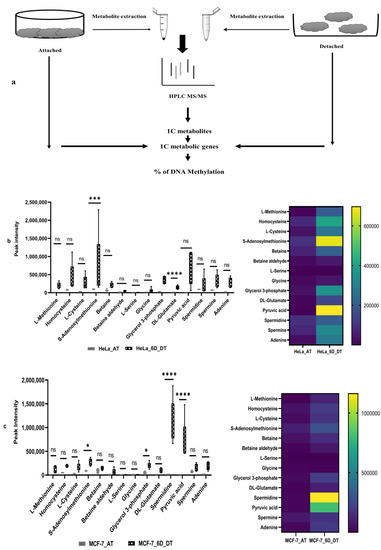
Figure 1.
Increasing the level of one-carbon metabolites in the ECM-detached condition. (a) Overall experimental design. (b) The one-carbon metabolites detected in the attached (AT) and 6 days (6D) ECM-detached HeLa cell line. (c) The one-carbon metabolites detected in the attached (AT) and 6 days (6D) ECM-detached MCF-7 cell line. The data are presented as mean ± SEM; ns, not significant; * p < 0.05, *** p < 0.001 and **** p < 0.0001.
In contrast, the peak intensity results for the ECM-detached MCF-7 cell lines showed that they also accumulated higher levels of core one-carbon metabolites, such as SAM, methionine, cysteine, homocysteine, spermidine, pyruvic acid, adenine, etc. Compared with the ECM-detached HeLa cell line, the ECM-detached MCF-7 cell line had accumulated a higher level of core one-carbon metabolites to a lesser extent (Figure 1b,c). However, spermidine was highly detected in ECM-detached MCF-7 cell lines.
2.2. Loss of ECM Attachment Increased Expression of One-Carbon Pathway-Related Metabolic Genes and Global DNA Methylation
We cultured the attached HeLa and MCF-7 cell lines in T75 cm2 flasks. Simultaneously, they were grown in poly-HEMA-coated 24-well plates for 6 days. After extracting RNA, cDNA conversion was performed for conducting qRT-PCR. Our experimental results showed that the methionine cycle-driving genes, such as methionine adenosyltransferase-1A/2A (MAT1A/2A), were significantly upregulated in the HeLa cell line. In addition, two genes (~33%) involved in the folate cycle were induced; the thymidylate synthetase (TYMS) gene was highly induced, while methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) was slightly induced. The cystathionine β-synthase (CBS) gene was marginally induced. The rest of the genes were downregulated (Figure 2a,b). However, the MCF-7 cell line results showed that the genes involved in the methionine cycle, such as betaine homocysteine, S-methyltransferase (BHMT), and S-adenosyl homocysteine hydrolase (AHCY), were upregulated, with marginal induction of MAT1A/2A genes. Only the CBS gene was found to be upregulated among the folate cycles genes, with marginal induction of dihydrofolate reductase (DHFR), TYMS, and MTHFD1 (Figure 2c,d). Moreover, it is well-established that DNA methylation is marked by couples of DNA methyltransferase enzymes that obtain a methyl group from the donor metabolite S-adenosyl methionine (SAM) and covalently add to the 5′ position a cytosine base within the CpG region to form 5′-methylcytosine (5mC) [34]. Therefore, we performed a 5mC level test, in both attached and detached conditions, which revealed that, upon detachment from the ECM, MCF-7 and HeLa cell lines showed higher 5mC levels than attached cell lines (Figure 2e,f). Nevertheless, the levels in HeLa cell lines were about four times higher than those in the MCF-7 cell lines.
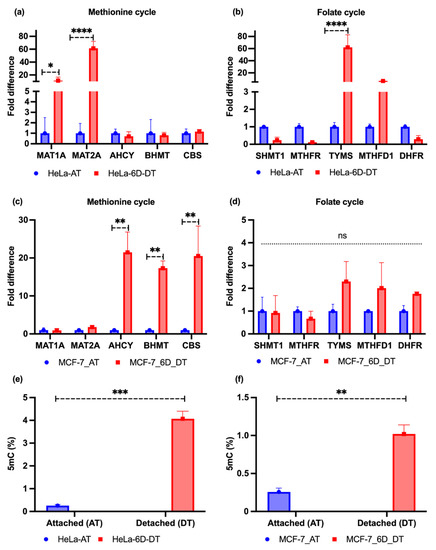
Figure 2.
One-carbon metabolic genes and 5mC levels (%) in ECM-attached and -detached conditions. (a) The mRNA expression levels of one-carbon metabolic genes in the attached (AT) methionine cycles and 6 days detached (6D_DT) conditions in the HeLa cell lines. (b) The mRNA expression levels of one-carbon metabolic genes in folate cycles in attached (AT) and 6 days detached (6D_DT) conditions in the HeLa cell lines. (c) The mRNA expression levels of one-carbon metabolic genes in methionine cycles in attached (AT) and 6 days detached (6D_DT) conditions in the MCF-7 cell lines. (d) The mRNA expression levels of one-carbon metabolic genes in folate cycles in attached (AT) and 6 days detached (6D_DT) conditions in the MCF-7 cell lines. SHMT1, serine hydroxymethyltransferase 1; MTHFR, methylenetetrahydrofolate reductase. (e,f). HeLa and MCF-7 cell lines showed increased 5mC levels in the 6 days (6D_DT) ECM-detached condition compared to the attached (AT) condition. The data are presented as mean ± SEM; * p < 0.05, ** p < 0.01, *** p< 0.001, and **** p < 0.0001.
2.3. Loss of ECM Attachment Represses Expression of NKG2DLs by Inducing Promoter Methylation
As our results showed increased levels of 5mC, global DNA methylation may also increase. Therefore, we checked the mRNA expression levels of DNMT genes. The results for HeLa and MCF-7 cell lines showed that ECM detachment induced mRNA expression of DNMTs in both cell lines (Figure 3a,b). However, MCF-7 cell lines had more DNMT mRNA induction than HeLa cell line (Figure 3b). Moreover, the innate immune system response by NK cells to counter cancer cell tumors necessitates interaction between NKG2D receptors and MICA/B ligands [25,35,36]. Hence, we performed qRT-PCR to assess MICA/B ligand mRNA expression in the 6 days ECM-detached MCF-7 and HeLa tumor cell lines. Our experimental results showed that MICA/B mRNA expression in MCF-7 and HeLa cell lines was repressed in ECM-detached conditions (Figure 3c,d). However, we also explored surface protein (MICA/B) expression through flow cytometry experiments and this showed that ECM detachment downregulated surface protein expression in both cell lines (Figure 3e,f).
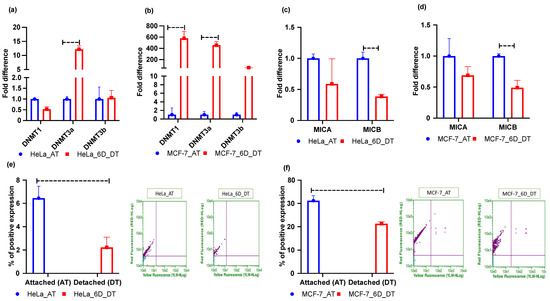
Figure 3.
ECM detachment represses NKG2DLs–MICA/B expression. (a) Fold changes in mRNA expression of DNMTs in attached (AT) and 6 days ECM-detached (6D_DT) HeLa cell lines. (b) Fold changes in mRNA expression of DNMTs in attached (AT) and 6 days ECM-detached (6D_DT) MCF-7 cell lines. (c) Fold changes in mRNA expression of MICA/B in attached (AT) and 6 days ECM-detached (6D_DT) conditions in HeLa cell lines. (d) Fold changes in mRNA expression of MICA/B in attached (AT) and 6 days ECM-detached (6D_DT) conditions in MCF-7 cell lines. (e,f). MICA/B surface expression is repressed in the 6 days ECM-detached (6D_DT) condition compared to the attached (AT) condition in HeLa and MCF-7 cell lines. The data are presented as mean ± SEM; ns, not significant.
2.4. Vitamin C—A Global Methylation Inhibitor Reduces DNA Methylation and Induces MICA/B Expression during ECM Detachment
Vitamin C was found to be a potent epigenetic re-programmer in cancer and stem cells [37,38]; hence, we first assessed the 5mC levels of vitamin C-treated 6 days ECM-detached cells. Our results show that vitamin C treatment reduced the global methylation level in the HeLa cell line (Figure 4a). Furthermore, we also checked DNMTs mRNA expression by qRT-PCR for vitamin C-treated ECM-detached MCF-7 and HeLa cell lines. The results showed that treatment reduced DNMT3a and DNMT1 expression in HeLa cell lines but that the treatment did not affect DNMT3b mRNA expression. The results were not statistically significant (Figure 4b). However, DNMTi treatment reduced mRNA expression of all the DNMTs (Figure S2). We also performed methylation-specific PCR experiments to explore and correlate methylation status. Our results showed that vitamin C did not reduce methylation-indicative signal intensity in HeLa compared to untreated 6 days ECM-detached cell lines (Figure 4c). After that, we also checked the status of the expression of NKG2DLs in vitamin C-treated ECM-detached cell lines and found that MICA/B mRNA and surface protein expression were induced in vitamin C-treated HeLa cell line (Figure 4d,e).
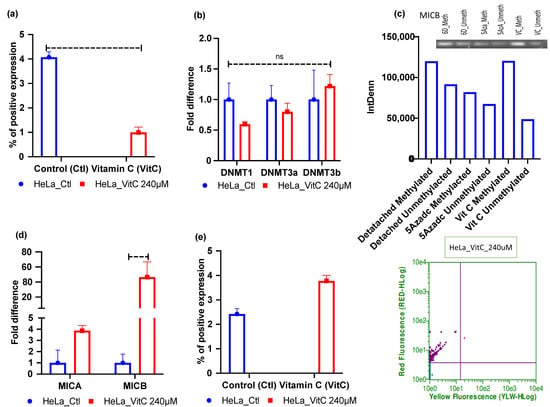
Figure 4.
Vitamin C treatment mediated reductions in global DNA methylation, DNA methyltransferase (DNMT1/3a), and induction of MICA/B expression in ECM-detached HeLa cell lines. (a) Vitamin C treatment reduced 5mC levels. (b) Vitamin C treatment repressed DNMT1/3a mRNA expression. (c) HeLa cell line showing that 5-aza-dc and vitamin C did not reduce the methylation signal in gel electrophoresis compared to the untreated 6 Days ECM-detached condition. (d) Vitamin C treatment increased the fold changes in mRNA expression of MICA/B in the 6 days ECM-detached condition. (e) Vitamin C treatment increased surface MICA/B expression. 6D_Meth, 6 days detached, methylated; 6D_Unmrth, 6 days detached, unmethylated; 5Azadc_Meth, 5-aza-dc methyl-treated; 5Azadc_Unmeth, 5-aza-dc unmethylated; VC_meth, vitamin C methyl-treated; VC_Unmeth, vitamin C unmethylated. The data are presented as mean ± SEM; ns, not significant.
In contrast, in MCF-7 cell lines, vitamin C significantly repressed the global methylation level (Figure 5a). Not only that, vitamin C treatment also significantly hit and reduced mRNA expression of all DNMTs (Figure 5b). Moreover, the reduced methylation level in the MICB gene was confirmed by MSPCR experiments (Figure 5c). Additionally, vitamin C treatment-induced MICA/B mRNA and protein expression (Figure 5d,e). However, we could not detect methylation levels in MICA gene amplification on MSPCR. Since 5Aza-2-deoxycytidine (5-aza-dc) is known as a potent DNMT inhibitor [39], we chose 5-aza-dc as a positive control for our experimental treatments. (The data are presented in Supplementary Figures S1–S3.)

Figure 5.
Vitamin C treatment-mediated reduction of global DNA methylation, DNA methyltransferase, and induction of MICA/B expression in ECM-detached MCF-7 cell lines. (a) Vitamin C treatment reduced 5mC levels. (b) Vitamin C treatment repressed DNMTs mRNA expression. (c) MCF-7 cell line showing 5-aza-dc and vitamin C reducing methylation signal in gel electrophoresis compared to untreated 6 Days ECM-detached condition. (d) Vitamin C treatment increased the fold change of MICA/B mRNA expression in the 6 days ECM-detached condition. (e) Vitamin C treatment increased surface MICA/B expression. 6D_Meth, 6 days detached methylated; 6D_Unmrth, 6 days detached unmethylated; 5Azadc_Meth, 5-aza-dc methyl-treated; 5Azadc_Unmeth, 5-aza-dc unmethylated; VC_meth, vitamin C methyl-treated; VC_Unmeth, vitamin C unmethylated. The data are presented as mean ± SEM; ns, not significant.
3. Discussion
Anoikis-resistant ECM-detached cancer cells undergo various metabolic adaptations and reprogramming to survive. For example, upon loss of ECM attachment, AMPK is induced to promote the uptake of glutamine, which lowers excessive oxidative stress. This metabolic reprogramming helps ECM-detached cancer cells to survive [40,41]. Moreover, tumor cells undergo epigenetic regulation, and the one-carbon metabolic pathway is linked to epigenetic modification, for instance, in DNA methylation reactions. This being so, the main aim of this study was to explore the relation between the one-carbon metabolic profiling and methylation-mediated transcriptomic regulation of the major innate immune ligands-MICA/B, in ECM-detached conditions. For this purpose, we performed untargeted metabolomics and sorted out the metabolites for the methionine and folate cycles of one-carbon metabolism. A recent study showed that loss of ECM attachment induced SAM and homocysteine levels in HCT116 and 22RV1 cancer cells. Although the SAM is a methyl group donor that activates methylation reactions in DNA, the study did not reveal the one-carbon metabolic pathway-mediated global methylation level in DNA [18]. Our study results demonstrated increased levels of SAM, methionine, and homocysteine in HeLa and MCF-7 cell lines. Most of the metabolites associated with 1C metabolism, such as glutamate, arginine, spermidine, and pyruvic acid, were also detected as higher metabolites.
Moreover, several previous studies have measured the expression levels of one-carbon metabolic genes among healthy cohorts, inflammatory macrophages, and liver cancer cells [42,43,44]. Another study elaborately revealed that one-carbon metabolites—methionine, SAM, m-THF, serine, etc.—and their associated enzymes are required by cancer cells for cell proliferation, DNA methylation reactions, and nucleotide synthesis [45]. Another study showed that controlling pyruvate by blocking its lactate-converting enzymes can slow cell proliferation in gastric cardia cancer clinical samples [46]. One-carbon metabolic pathway-mediated DNA methylation has already been targeted in cancer therapy, supported by several studies. For example, cell proliferation in HeLa cell lines decreased upon the downregulation of the folate-dependent DNA synthesis enzyme TYMS after 0.5 µM fluorouracil treatment with miRNA [47]. Similarly, the deregulation of folate cycle enzymes, such as the high expression of the MTHFD1 gene, affects survival rates in acute leukemia patients [48]. Even with the knockdown of the SHMT1 gene, there was evidence of apoptosis induction in lung cancer [49].
Moreover, therapeutic aspects have previously been revealed by several studies. For example, a study showed that 5-fluorouracil (5-FU—an inhibitor of TYMS) and methotrexate target the DHFR in ALL as a chemotherapeutic drug. Combined treatment with cyclophosphamide, 5-FU, and methotrexate showed effectiveness in node-negative breast cancer patients [50]. Another study revealed that 1C metabolism mediated DNA synthesis and DNA methylation can be used for cancer biomarkers, as well as one-carbon metabolic inhibitors can be useful in cancer therapy [51]. We examined these one-carbon metabolic genes in ECM-detached conditions. Our results indicated that expression levels of the genes responsible for SAM, m-THF, and dTUMP synthesis in ECM-detached HeLa cell lines were significantly high. However, methionine and homocysteine synthesis were slightly downregulated. The overall folate cycle was downregulated, except for trihydro-folate (THF) synthesis.
Similarly, the ECM-detached MCF-7 cell line also had high methionine, SAM, DHFR, and cystathionine synthesizing gene expression levels. The cell line also showed the slight induction of dTUMP, m-THF, and DHFR synthesizing gene expression. Moreover, both MCF-7 and HeLa cell lines showed that loss of ECM attachment induced 5mC levels, which indicated that DNMT activity could be induced [52]. A previous study also found increased 5mC levels in suspension carcinoma cells [53]. Hence, we aimed to check this effect of higher DNA methylation on the major innate immune cell ligands, especially the NK cell ligands MICA/B in the ECM-detached condition. We observed that ECM detachment induced DNMT mRNA expression and repressed MICA/B mRNA expression. Even the expression of surface proteins was downregulated after ECM detachment.
However, currently, natural compounds-based oncological therapy is not so widely available for cancer patients. These therapies have been clinically neglected for a long time, though they have been found to have very negligible side effects. Considering this, we decided to check the epigenetic effect on cancer cells of one of the most promising natural compounds, vitamin C. We treated ECM-detached MCF-7 and HeLa cell lines with 5-aza-dc as a positive control and vitamin C for 6 Days. In the ECM-detached MCF-7 cell line, vitamin C treatment-induced NKG2D ligands and significantly downregulated DNMTs expression and the global DNA methylation (5mC) level. However, vitamin C treatment-induction of NKG2D ligands did not significantly affect DNMT3b mRNA levels in ECM-detached HeLa cell line.
Nevertheless, vitamin C treatment downregulated DNMT1/3a mRNA expression in the HeLa cell line. A previous study may explain this controversy, for it was demonstrated there that ascorbate (vitamin C) could inhibit only DNMT enzyme activity in melanoma cells [54]. Additional analyses showed that DNMTi treatment slightly reduced DNMT expression [55].
Nevertheless, 5mC level assessment revealed that both cell lines had reduced global methylation levels after vitamin C treatment. Additionally, the MSPCR result for the MICB gene showed that vitamin C reduced the methylation signal only in the MCF-7 but not in the HeLa cell line. However, in both cell lines, MICA gene methylation was not detected in MSPCR.
Hence, our overall results demonstrate that the two cell lines showed almost similar outcomes. MICA/B gene and surface protein expression were induced in DNMTi- and vitamin C-treated MCF-7 cell lines, with significant downregulation of DNMT and global methylation (5mC) levels. This means that MICA/MICB genes may have been suppressed or downregulated by epigenetic mechanisms in ECM-detached MCF-7 cell lines. On the other hand, the DNMTi- and vitamin C-treated ECM-detached HeLa cell line had reduced global DNA methylation. Though treatment reduced DNMT1/3a expression while not affecting DNMT3b, DNMTi and vitamin C induced MICA/B expression, suggesting that the MICA/B genes in the ECM-detached HeLa cell line are probably regulated by other epigenetic mechanisms. A previous study also suggested this probability, having shown that MICA/B promoters are silenced by histone hypo-acetylation [56]. Even DNMTi treatment can induce 5hmC [57], which is also methylated; hence, hydroxyl methylation in MSPCR bands may have been present in the vitamin C-treated ECM-detached HeLa cell line, such that the reduction of methylation was not shown after treatment.
4. Methods
4.1. Cell Culture and Viability Assay
Poly-HEMA gel was purchased from Sigma, dissolved in 95% ethanol at 65 °C, and incubated overnight. Cells were then seeded in a poly-HEMA- (lot no. SLBV7383, Sigma, India) coated plate. MCF-7 and HeLa cell lines were collected and grown and subcultured in DMEM media (UFS Biotech, Riyadh, Saudi Arabia) supplemented with 10% FBS and 1% penicillin and incubated in a CO2 incubator. Upon 85–90% confluence, cells were trypsinized and seeded in poly-HEMA-coated 96-well plates and incubated overnight to ensure that cells were healthy without any contamination. Then, cells were treated with 5-aza-dc (lot no. S1782, Selleckchem, Houston, TX, USA) at 500 nM, 1.5 µM, and 3 µM for 48 h. For vitamin C (lot no. S13114, Selleckchem) treatment, cells were treated at 5 µM, 10 µM, 15 µM, 20 µM, 25 µM, and 30 µM. After 48 h, WST-1 was added, and a cell viability assay was performed with a BioTek ELISA microplate reader at 450 nm. Based on the cell viability assay, we selected doses of 5-aza-dc of 60 µM and 240 µM for vitamin C treatment for 6 days.
4.2. Metabolites Extraction
Metabolites were extracted from attached and detached cells with methanol: acetonitrile: water at a ratio of 2:2:1 (v/v). Then, 1 mL of ice-cold solvent was added to the cell supernatant, followed by quick vortexing for 30s and incubation for 1 h at −20 °C. The supernatant was then spun for 15 min at 13,000 rpm at 4 °C, after which it was removed and the sample was dried, reconstituted in 100 μL of acetonitrile: water (1:1, v/v), vortexed for 10 min, and spun for 15 min at 13,000 rpm at 4 °C to remove insoluble debris. Finally, supernatants were taken for LC–MS/MS [58].
4.3. Analysis of HPLC Coupled LC–MS/MS
For LC–MS/MS, 10 µL of the sample was taken for injection into the HPLC column of an LTQ XL™ linear ion trap instrument (Thermo Fisher Scientific, Waltham, MA, USA). The full scan range was chosen from 100 to 1000 m/z, with spray voltage at −3.0 kV and capillary voltage at 4.0 V. The temperature was fixed at 270 °C. Helium was used as a buffer gas, nitrogen as a sheath gas. The flow rate was fixed at 40 arbitrary units. Next, the obtained data were processed with XCMS online server data processing software [18].
4.4. Quantitative Real-Time Reverse Transcription PCR
Total RNA was extracted using a PureLink™ RNA Mini Kit (lot no. 1944999, Thermo Fisher Scientific). Then, 100 ng/uL RNA was transcribed into cDNA using a high-capacity cDNA synthesis kit (lot no. 00656567, Applied Biosystems, Waltham, MA, USA). AHCY, MICA, MICB, and DNMTs primers sequences were obtained from the UCSC genome browser. One-carbon metabolic gene primers were obtained from other studies [47,48,49,59,60,61,62]. All the primer sequences are enlisted in Table 1. qRT-PCR was carried out by PowerUp SYBR Green Master Mix (lot no. 1805029, Applied Biosystems). The conditions for qRT-PCR were: 50 °C for 2 min, 95 °C for 2 min, 95 °C for 15 s, and 60 °C for 1 min. After that, qRT-PCR was performed in an ABI 7300 Prism. The numbers of transcripts were normalized with RPLP0 and calculated by applying the 2−(ΔΔCt) method [63].

Table 1.
List of qRT-PCR primers.
4.5. Quantitative Assay of DNA Methylcytosine (5mC) Level
DNA was extracted using an easy DNA kit (lot no. 155152507, MQ Genomic DNA isolation Kit, Molequle On, Auckland, New Zealand). The 5mC kit was purchased from Abcam (lot no. ab117128, Abcam, Trumpington, UK). The extracted DNA was seeded for binding the DNA to the assay well. Then, the wells were washed and the capture antibody was added. After that, the wells were rewashed, and detection antibody, enhancer solution, and developing solution were added. Finally, absorbance was observed at 450 nm. The whole procedure and data calculation were performed as per the manufacturer’s protocol.
4.6. Flow Cytometry
Both HeLa and MCF-7 cell lines were seeded in poly-HEMA-coated 24-well plates and treated with 5-aza-dc at 60 µM and 240 µM for vitamin C treatment for 6 days. Then, the collected cells were washed with 1X PBS, followed by thorough mixing with 100 uL 2% FBS. Anti-Hu MICA/B Ab (lot no. 2115636, Invitrogen, eBioscienceTM, Waltham, MA, USA) was diluted as 1:100 in 2% FBS for staining. Stained samples were put in a shaker for 1 h, followed by 1X PBS wash. After that, the surface expression of MICA/B was analyzed by Guava easyCyte HT.
4.7. Bisulfite Conversion and Methylation-Specific PCR (MSP)
Using an EpiJET DNA bisulfite conversion kit (lot no. 00596381, Thermo Fischer Scientific, Waltham, MA, USA), bisulfite conversion of extracted DNA was performed according to the supplier’s protocol. The bisulfite-modified DNA template was used for further PCR reactions using the DNA Master Mix Kit (Molequle On). For PCR reactions, whole gene sequences were selected for both MICA/B genes and submitted to Metprimer online software (https://www.urogene.org/methprimer/, accessed on 15 January 2020) for methylated and unmethylated primer design (listed below, Table 2). During the primer design, CpG islands were selected. Bisulfite-converted DNA was taken for qPCR-mediated amplification using MSP primers; the qRT-PCR conditions were 50 °C for 2 min, 95 °C for 2 min, 95 °C for 15 s, and 60 °C for 1 min, and amplified products were visualized in the UVP iBOX 500 imaging system after gel electrophoresis (SCIE PLAS, Cambridge, UK). The signal intensity (IntDen) of the MSPCR amplicon was measured using Image J software.

Table 2.
List of MSPCR primers.
4.8. Statistical Analysis
The statistical analysis was carried out by multiple t-tests or two ANOVA mixed models using GraphPad Prism 9 software. The Holme–Sidak and Tukey test methods were chosen, and the alpha was set as ≤0.5 for considering statistical significance. The data were presented as mean ± SEM.
5. Conclusions
ECM-detached cancer cells possess high levels of one-carbon metabolites that mediate the induction of global methylation in these conditions. Higher global DNA methylation levels may suppress one of the major NK cell immune ligands-MICB. Hence, targeting the one-carbon metabolic pathway and/or global methylation levels constitutes a beneficial strategy for chemotherapy outcomes in patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/metabo12030267/s1, Figure S1: Cell viability assay for 5-aza-dc-treated monolayer and spheroid cell lines; Figure S2: 5-aza-dc-mediated expression of NKG2DLs and status of DNA methylation in HeLa cell lines; Figure S3: 5-aza-dc-mediated expression of NKG2DLs and status of DNA methylation in MCF-7 cell lines.
Author Contributions
Conceptualization, M.I.K., M.A.Z. and H.C.; methodology, S.M.N. and M.R.S.M.; software, S.M.N. and I.A.R.; validation, S.M.N. and M.R.S.M.; formal analysis, M.I.K. and I.A.R.; investigation, M.I.K.; resources, M.I.K., M.A.Z. and H.C.; data curation, S.M.N. and I.A.R.; writing—original draft preparation, S.M.N.; writing—review and editing, A.A., B.A. and I.A.R.; visualization, A.A. and B.A.; supervision, M.I.K.; project administration, M.A.Z.; funding acquisition, M.A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University (KAU), Jeddah, under grant no. KEP-15-130-38. The authors, therefore, acknowledge with thanks DSR technical and financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We would like to acknowledge the Deanship of Scientific Research (DSR), the Department of Biochemistry, and Hani’s Lab, KFMRC, King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia for cordial scientific support and assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Buchheit, C.L.; Weigel, K.J.; Schafer, Z.T. Cancer Cell Survival during Detachment from the ECM: Multiple Barriers to Tumour Progression. Nat. Rev. Cancer 2014, 14, 632–641. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis Molecular Pathways and Its Role in Cancer Progression. Biochim. Et. Biophys. Acta-Mol. Cell Res. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, J.A.; Davison-Versagli, C.A.; Leliaert, A.K.; Pape, D.J.; McCallister, C.; Zuo, J.; Durbin, S.M.; Buchheit, C.L.; Zhang, S.; Schafer, Z.T. Oncogenic Ras Differentially Regulates Metabolism and Anoikis in Extracellular Matrix-Detached Cells. Cell Death Differ. 2016, 23, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranganathan, S.; Kumar, S.; Mohanty, S.S.; Jolly, M.K.; Rangarajan, A. Cellular Plasticity in Matrix-Attached and -Detached Cells: Implications in Metastasis. J. Indian Inst. Sci. 2020, 100, 525–536. [Google Scholar] [CrossRef]
- Sousa, B.; Pereira, J.; Paredes, J. The Crosstalk between Cell Adhesion and Cancer Metabolism. Int. J. Mol. Sci. 2019, 20, 1933. [Google Scholar] [CrossRef] [Green Version]
- Elia, I.; Doglioni, G.; Fendt, S.M. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 2018, 28, 673–684. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; Cheung, E.C.; Blagih, J.; Domart, M.C.; Vousden, K.H. Cell Clustering Promotes a Metabolic Switch That Supports Metastatic Colonization. Cell Metab. 2019, 30, 720–734. [Google Scholar] [CrossRef] [Green Version]
- Eales, K.L.; Hollinshead, K.E.R.; Tennant, D.A. Hypoxia and Metabolic Adaptation of Cancer Cells. Oncogenesis 2016, 5, e190. [Google Scholar] [CrossRef] [Green Version]
- Bacigalupa, Z.A.; Rathmell, W.K. Beyond Glycolysis: Hypoxia Signaling as a Master Regulator of Alternative Metabolic Pathways and the Implications in Clear Cell Renal Cell Carcinoma. Cancer Lett. 2020, 489, 19–28. [Google Scholar] [CrossRef]
- Rankin, E.B.; Nam, J.M.; Giaccia, A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer 2016, 2, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the Extracellular Matrix: Drivers of Tumour Metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Zhang, J.; Li, F.; Du, W.; Zhou, X.; Wan, M.; Fan, Y.; Xu, X.; Zhou, X.; Zheng, L.; et al. One-Carbon Metabolism Links Nutrition Intake to Embryonic Development via Epigenetic Mechanisms. Stem Cells Int. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA Methylation: Roles in Mammalian Development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Cahill, C.M.; Huang, X.; Roffman, J.L.; Lamon-Fava, S.; Fava, M.; Mischoulon, D.; Rogers, J.T. S-Adenosyl Methionine and Transmethylation Pathways in Neuropsychiatric Diseases Throughout Life. Neurotherapeutics 2018, 15, 156–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reik, W.; Dean, W.; Walter, J. Epigenetic Reprogramming in Mammalian Development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.M.; Ali, M.M. Methyl Donor Micronutrients That Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [Green Version]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Shait Mohammed, M.R.; Alghamdi, R.A.; Alzahrani, A.M.; Zamzami, M.A.; Choudhry, H.; Khan, M.I. Compound C, a Broad Kinase Inhibitor Alters Metabolic Fingerprinting of Extra Cellular Matrix Detached Cancer Cells. Front. Oncol. 2021, 11, 1. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Eagle, R.; Jafferji, I.; Barrow, A. Beyond Stressed Self: Evidence for NKG2D Ligand Expression on Healthy Cells. Curr. Immunol. Rev. 2009, 5, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, A.M.; Diefenbach, A.; McMahon, C.W.; Xiong, N.; Carlyle, J.R.; Raulet, D.H. The Role of the NKG2D Immunoreceptor in Immune Cell Activation and Natural Killing. Immunity 2002, 17, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Nausch, N.; Cerwenka, A. NKG2D Ligands in Tumor Immunity. Oncogene 2008, 27, 5944–5958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghadially, H.; Brown, L.; Lloyd, C.; Lewis, L.; Lewis, A.; Dillon, J.; Sainson, R.; Jovanovic, J.; Tigue, N.J.; Bannister, D.; et al. MHC Class i Chain-Related Protein A and B (MICA and MICB) Are Predominantly Expressed Intracellularly in Tumour and Normal Tissue. Br. J. Cancer 2017, 116, 1208–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingoni, A.; Molfetta, R.; Fionda, C.; Soriani, A.; Paolini, R.; Cippitelli, M.; Cerboni, C.; Santoni, A. NKG2D and Its Ligands: “One for All, All for One”. Front. Immunol. 2018, 9, 476. [Google Scholar] [CrossRef]
- Bugide, S.; Green, M.R.; Wajapeyee, N. Inhibition of Enhancer of Zeste Homolog 2 (EZH2) Induces Natural Killer Cell-Mediated Eradication of Hepatocellular Carcinoma Cells. Proc. Natl. Acad. Sci. USA 2018, 115, E3509–E3518. [Google Scholar] [CrossRef] [Green Version]
- Kato, N.; Tanaka, J.; Sugita, J.; Toubai, T.; Miura, Y.; Ibata, M.; Syono, Y.; Ota, S.; Kondo, T.; Asaka, M.; et al. Regulation of the Expression of MHC Class I-Related Chain A, B (MICA, MICB) via Chromatin Remodeling and Its Impact on the Susceptibility of Leukemic Cells to the Cytotoxicity of NKG2D-Expressing Cells. Leukemia 2007, 21, 2103–2108. [Google Scholar] [CrossRef]
- Sers, C.; Kuner, R.; Falk, C.S.; Lund, P.; Sueltmann, H.; Braun, M.; Buness, A.; Ruschhaupt, M.; Conrad, J.; Mang-Fatehi, S.; et al. Down-Regulation of HLA Class I and NKG2D Ligands through a Concerted Action of MAPK and DNA Methyltransferases in Colorectal Cancer Cells. Int. J. Cancer 2009, 125, 1626–1639. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Baraganõ Raneros, A.; Martín-Palanco, V.; Fernandez, A.F.; Rodriguez, R.M.; Fraga, M.F.; Lopez-Larrea, C.; Suarez-Alvarez, B. Methylation of NKG2D Ligands Contributes to Immune System Evasion in Acute Myeloid Leukemia. Genes Immun. 2015, 16, 71–82. [Google Scholar] [CrossRef]
- Lee Chong, T.; Ahearn, E.L.; Cimmino, L. Reprogramming the Epigenome with Vitamin C. Front. Cell Dev. Biol. 2019, 7, 128. [Google Scholar] [CrossRef]
- Young, J.I.; Züchner, S.; Wang, G. Regulation of the Epigenome by Vitamin C. Annu. Rev. Nutr. 2015, 35, 545–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moncayo, G.; Lin, D.; McCarthy, M.T.; Watson, A.A.; O’Callaghan, C.A. MICA Expression Is Regulated by Cell Adhesion and Contact in a FAK/Src-Dependent Manner. Front. Immunol. 2017, 7, 687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquière, B.; Van Dyck, L.; Boeckx, B.; Schoonjans, L.; et al. Tumour Hypoxia Causes DNA Hypermethylation by Reducing TET Activity. Nature 2016, 537, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Fouad, M.A.; Salem, S.E.; Hussein, M.M.; Zekri, A.R.N.; Hafez, H.F.; El Desouky, E.D.; Shouman, S.A. Impact of Global DNA Methylation in Treatment Outcome of Colorectal Cancer Patients. Front. Pharmacol. 2018, 9, 1173. [Google Scholar] [CrossRef] [PubMed]
- Guerra, N.; Tan, Y.X.; Joncker, N.T.; Choy, A.; Gallardo, F.; Xiong, N.; Knoblaugh, S.; Cado, D.; Greenberg, N.R.; Raulet, D.H. NKG2D-Deficient Mice Are Defective in Tumor Surveillance in Models of Spontaneous Malignancy. Immunity 2008, 28, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Raulet, D.H.; Gasser, S.; Gowen, B.G.; Deng, W.; Jung, H. Regulation of Ligands for the NKG2D Activating Receptor. Annu. Rev. Immunol. 2013, 31, 413–441. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, L.; Neel, B.G.; Aifantis, I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol. 2018, 28, 698–708. [Google Scholar] [CrossRef]
- Huff, T.C.; Sant, D.W.; Camarena, V.; Van Booven, D.; Andrade, N.S.; Mustafi, S.; Monje, P.V.; Wang, G. Vitamin C Regulates Schwann Cell Myelination by Promoting DNA Demethylation of Pro-Myelinating Genes. J. Neurochem. 2020, 157, 1759–1773. [Google Scholar] [CrossRef]
- Palii, S.S.; Van Emburgh, B.O.; Sankpal, U.T.; Brown, K.D.; Robertson, K.D. DNA Methylation Inhibitor 5-Aza-2′-Deoxycytidine Induces Reversible Genome-Wide DNA Damage That Is Distinctly Influenced by DNA Methyltransferases 1 and 3B. Mol. Cell. Biol. 2008, 28, 752–771. [Google Scholar] [CrossRef] [Green Version]
- Mason, J.A.; Hagel, K.R.; Hawk, M.A.; Schafer, Z.T. Metabolism during ECM Detachment: Achilles Heel of Cancer Cells? Trends Cancer 2017, 3, 475–481. [Google Scholar] [CrossRef]
- Endo, H.; Owada, S.; Inagaki, Y.; Shida, Y.; Tatemichi, M. Metabolic Reprogramming Sustains Cancer Cell Survival Following Extracellular Matrix Detachment. Redox Biol. 2020, 36, 101643. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.L.; Xu, X.; Wallenstein, S.; Chen, J. Gene Expression Profiles of the One-Carbon Metabolism Pathway. J. Genet. Genom. 2009, 36, 277–282. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Wang, Z.; Zhang, K.; Chi, Z.; Xu, T.; Jiang, D.; Chen, S.; Li, W.; Yang, X.; Zhang, X.; et al. One-Carbon Metabolism Supports S-Adenosylmethionine and Histone Methylation to Drive Inflammatory Macrophages. Mol. Cell 2019, 75, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Mao, Z.; Hwang, J.J.; Lu, S.C. Differential Expression of Methionine Adenosyltransferase Genes Influences the Rate of Growth of Human Hepatocellular Carcinoma Cells. Cancer Res. 1998, 58, 1444–1450. [Google Scholar]
- Newman, A.C.; Maddocks, O.D.K. One-Carbon Metabolism in Cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Zhao, J.S.; Li, J.J.; Peng, D.N.; Wang, X.Y.; Chen, T.L.; Qiu, Y.P.; Chen, P.P.; Li, W.J.; Xu, L.Y.; et al. A Combined Proteomics and Metabolomics Profiling of Gastric Cardia Cancer Reveals Characteristic Dysregulations in Glucose Metabolism. Mol. Cell. Proteom. 2010, 9, 2617–2628. [Google Scholar] [CrossRef] [Green Version]
- Gotanda, K.; Hirota, T.; Matsumoto, N.; Ieiri, I. MicroRNA-433 Negatively Regulates the Expression of Thymidylate Synthase (TYMS) Responsible for 5-Fluorouracil Sensitivity in HeLa Cells. BMC Cancer 2013, 13, 369. [Google Scholar] [CrossRef] [Green Version]
- Organista-Nava, J.; Gómez-Gómez, Y.; Del Moral-Hernandez, O.; Illades-Aguiar, B.; Gómez-Santamaria, J.; Rivera-Ramírez, A.B.; Saavedra-Herrera, M.V.; Jimenez-López, M.A.; Leyva-Vázquez, M.A. Deregulation of Folate Pathway Gene Expression Correlates with Poor Prognosis in Acute Leukemia. Oncol. Lett. 2019, 18, 3115–3127. [Google Scholar] [CrossRef]
- Paone, A.; Marani, M.; Fiascarelli, A.; Rinaldo, S.; Giardina, G.; Contestabile, R.; Paiardini, A.; Cutruzzolà, F. SHMT1 Knockdown Induces Apoptosis in Lung Cancer Cells by Causing Uracil Misincorporation. Cell Death Dis. 2014, 5, e1525. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, J. One-Carbon Metabolism and Breast Cancer: An Epidemiological Perspective. J. Genet. Genom. Yi Chuan Xue Bao 2009, 36, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Asai, A.; Konno, M.; Koseki, J.; Taniguchi, M.; Vecchione, A.; Ishii, H. One-Carbon Metabolism for Cancer Diagnostic and Therapeutic Approaches. Cancer Lett. 2020, 470, 141–148. [Google Scholar] [CrossRef] [PubMed]
- van der Wijst, M.G.P.; Venkiteswaran, M.; Chen, H.; Xu, G.L.; Plösch, T.; Rots, M.G. Local Chromatin Microenvironment Determines DNMT Activity: From DNA Methyltransferase to DNA Demethylase or DNA Dehydroxymethylase. Epigenetics 2015, 10, 671–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desjobert, C.; El Maï, M.; Gérard-Hirne, T.; Guianvarćh, D.; Carrier, A.; Pottier, C.; Arimondo, P.B.; Riond, J. Combined Analysis of DNA Methylation and Cell Cycle in Cancer Cells. Epigenetics 2015, 10, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Sinnberg, T.W.; Berger, A.; Noor, S.; Levesque, M.; Böcker, A.; Niessner, H.; Lauer, U.M.; Bitzer, M.; Garbe, C.; et al. Epigenetic Impacts of Ascorbate on Human Metastatic Melanoma Cells. Front. Oncol. 2014, 4, 227. [Google Scholar] [CrossRef] [Green Version]
- Rocha, M.A.; Veronezi, G.M.B.; Felisbino, M.B.; Gatti, M.S.V.; Tamashiro, W.M.S.C.; Mello, M.L.S. Sodium Valproate and 5-Aza-2′-Deoxycytidine Differentially Modulate DNA Demethylation in G1 Phase-Arrested and Proliferative HeLa Cells. Sci. Rep. 2019, 9, 18236. [Google Scholar] [CrossRef]
- Ritter, C.; Fan, K.; Paulson, K.G.; Nghiem, P.; Schrama, D.; Becker, J.C. Reversal of Epigenetic Silencing of MHC Class i Chain-Related Protein A and B Improves Immune Recognition of Merkel Cell Carcinoma. Sci. Rep. 2016, 6, 21678. [Google Scholar] [CrossRef] [Green Version]
- Sajadian, S.O.; Ehnert, S.; Vakilian, H.; Koutsouraki, E.; Damm, G.; Seehofer, D.; Thasler, W.; Dooley, S.; Baharvand, H.; Sipos, B.; et al. Induction of Active Demethylation and 5hmC Formation by 5-Azacytidine Is TET2 Dependent and Suggests New Treatment Strategies against Hepatocellular Carcinoma. Clin. Epigenetics 2015, 7, 98. [Google Scholar] [CrossRef] [Green Version]
- AlGhamdi, A.A.; Mohammed, M.R.S.; Zamzami, M.A.; Al-Malki, A.L.; Qari, M.H.; Khan, M.I.; Choudhry, H. Untargeted Metabolomics Identifies Key Metabolic Pathways Altered by Thymoquinone in Leukemic Cancer Cells. Nutrients 2020, 12, 1792. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Yu, W.; Li, C.; Gui, Y.; Cai, Z. Expression of Methionine Adenosyltransferase 2A in Renal Cell Carcinomas and Potential Mechanism for Kidney Carcinogenesis. BMC Cancer 2014, 14, 196. [Google Scholar] [CrossRef] [Green Version]
- Avila, M.A.; Berasain, C.; Torres, L.; Martín-Duce, A.; Corrales, F.J.; Yang, H.; Prieto, J.; Lu, S.C.; Caballería, J.; Rodés, J.; et al. Reduced MRNA Abundance of the Main Enzymes Involved in Methionine Metabolism in Human Liver Cirrhosis and Hepatocellular Carcinoma. J. Hepatol. 2000, 33, 907–914. [Google Scholar] [CrossRef] [Green Version]
- Bing, Y.; Zhu, S.; Yu, G.; Li, T.; Liu, W.; Li, C.; Wang, Y.; Qi, H.; Guo, T.; Yuan, Y.; et al. Glucocorticoid-Induced S-Adenosylmethionine Enhances the Interferon Signaling Pathway by Restoring STAT1 Protein Methylation in Hepatitis B Virus-Infected Cells. J. Biol. Chem. 2014, 289, 32639–32655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, K.; Ooyama, A.; Ruszkiewicz, A.; Jin, M.; Watanabe, G.; Moore, J.; Oka, T.; Iacopetta, B.; Minamoto, T. Low Expression of γ-Glutamyl Hydrolase MRNA in Primary Colorectal Cancer with the CpG Island Methylator Phenotype. Br. J. Cancer 2008, 98, 1555–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).