Establishing Imaging Biomarkers of Host Immune System Efficacy during Glioblastoma Therapy Response: Challenges, Obstacles and Future Perspectives
Abstract
1. Introduction
2. Hypothesis Proposal
2.1. Why Do We Need Imaging Biomarkers for Assessing the Efficacy of Immune System Participation in GB Therapy Response?
2.2. Why Are Such Biomarkers of Efficacy Not Yet Available?
2.3. How Can We Produce the Needed Biomarkers?
2.4. Limitations of the Present Hypothesis Proposal
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Tonn, J.C.; Brada, M.; Pentheroudakis, G. High-grade malignant glioma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010, 21, v190–v193. [Google Scholar] [CrossRef] [PubMed]
- Tykocki, T.; Eltayeb, M. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 2018, 54, 7–13. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Zitvogel, L.; Kroemer, G. The secret ally: Immunostimulation by anticancer drugs. Nat. Rev. Drug Discov. 2012, 11, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A.; Middleton, G. The Interplay of Immunotherapy and Chemotherapy: Harnessing Potential Synergies. Cancer Immunol. Res. 2015, 3, 436–443. [Google Scholar] [CrossRef]
- Sologuren, I.; Rodríguez-Gallego, C.; Lara, P.C. Immune effects of high dose radiation treatment: Implications of ionizing radiation on the development of bystander and abscopal effects. Transl. Cancer Res. 2014, 3, 18–31. [Google Scholar] [CrossRef]
- Brown, N.F.; Carter, T.J.; Ottaviani, D.; Mulholland, P. Harnessing the immune system in glioblastoma. Br. J. Cancer 2018, 119, 1171–1181. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Gebremeskel, S.; Johnston, B. Concepts and mechanisms underlying chemotherapy induced immunogenic cell death: Impact on clinical studies and considerations for combined therapies. Oncotarget 2015, 6, 41600–41619. [Google Scholar] [CrossRef]
- Fucikova, J.; Moserova, I.; Urbanova, L.; Bezu, L.; Kepp, O.; Cremer, I.; Salek, C.; Strnad, P.; Kroemer, G.; Galluzzi, L.; et al. Prognostic and predictive value of DAMPs and DAMP-associated processes in cancer. Front. Immunol. 2015, 6, 402. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Buqué Martinez, A.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Sun, C.; Katsube, T.; Wang, B. Biomarkers of Brain Damage Induced by Radiotherapy. Dose-Response 2020, 18, 1559325820938279. [Google Scholar] [CrossRef] [PubMed]
- Ree, A.H.; Meltzer, S.; Flatmark, K.; Dueland, S.; Kalanxhi, E. Biomarkers of treatment toxicity in combined-modality cancer therapies with radiation and systemic drugs: Study design, multiplex methods, molecular networks. Int. J. Mol. Sci. 2014, 15, 22835–22856. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, D.R.; Cascino, T.L.; Schold, S.C.; Cairncross, J.G. Response criteria for phase II studies of supratentorial malignant glioma. J. Clin. Oncol. 1990, 8, 1277–1280. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bent, M.J.; Vogelbaum, M.A.; Wen, P.Y.; Macdonald, D.R.; Chang, S.M. End point assessment in gliomas: Novel treatments limit usefulness of classical Macdonald’s criteria. J. Clin. Oncol. 2009, 27, 2905–2908. [Google Scholar] [CrossRef] [PubMed]
- Hormuth, D.A.; Sorace, A.G.; Virostko, J.; Abramson, R.G.; Bhujwalla, Z.M.; Enriquez-Navas, P.; Gillies, R.; Hazle, J.D.; Mason, R.P.; Quarles, C.C.; et al. Translating preclinical MRI methods to clinical oncology. J. Magn. Reson. Imaging 2019, 50, 1377–1392. [Google Scholar] [CrossRef]
- Essig, M.; Anzalone, N.; Combs, S.E.; Dörfler, A.; Lee, S.K.; Picozzi, P.; Rovira, À.; Weller, M.; Law, M. MR imaging of neoplastic central nervous system lesions: Review and recommendations for current practice. Am. J. Neuroradiol. 2012, 33, 803–817. [Google Scholar] [CrossRef]
- Okada, H.; Weller, M.; Huang, R.; Finocchiaro, G.; Gilbert, M.R.; Wick, W.; Ellingson, B.M.; Hashimoto, N.; Pollack, I.F.; Brandes, A.A.; et al. Immunotherapy response assessment in neuro-oncology: A report of the RANO working group. Lancet Oncol. 2015, 16, e534–e542. [Google Scholar] [CrossRef]
- Juergens, R.A.; Zukotynski, K.A.; Singnurkar, A.; Snider, D.P.; Valliant, J.F.; Gulenchyn, K.Y. Imaging Biomarkers in Immunotherapy. Biomark. Cancer 2016, 8, 1–13. [Google Scholar] [CrossRef]
- Chawla, C.; Shehu, V.; Gupta, P.K.; Nath, K.; Poptani, H. Physiological Imaging Methods for Evaluating Response to Immunotherapies in Glioblastomas. Int. J. Mol. Sci. 2021, 22, 3867. [Google Scholar] [CrossRef]
- Albert, N.L.; Weller, M.; Suchorska, B.; Galldiks, N.; Soffietti, R.; Kim, M.M.; La Fougère, C.; Pope, W.; Law, I.; Arbizu, J.; et al. Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro. Oncol. 2016, 18, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Moreau, A.; Febvey, O.; Mognetti, T.; Frappaz, D.; Kryza, D. Contribution of Different Positron Emission Tomography Tracers in Glioma Management: Focus on Glioblastoma. Front. Oncol. 2019, 9, 1134. [Google Scholar] [CrossRef] [PubMed]
- Tavaré, R.; Escuin-Ordinas, H.; Mok, S.; McCracken, M.N.; Zettlitz, K.A.; Salazar, F.B.; Witte, O.N.; Ribas, A.; Wu, A.M. An effective immuno-PET imaging method to monitor CD8-dependent responses to immunotherapy. Cancer Res. 2016, 76, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Larimer, B.M.; Wehrenberg-Klee, E.; Dubois, F.; Mehta, A.; Kalomeris, T.; Flaherty, K.; Boland, G.; Mahmood, U. Granzyme B PET imaging as a predictive biomarker of immunotherapy response. Cancer Res. 2017, 77, 2318–2327. [Google Scholar] [CrossRef] [PubMed]
- Antonios, J.P.; Soto, H.; Everson, R.G.; Moughon, D.L.; Wang, A.C.; Orpilla, J.; Radu, C.; Ellingson, B.M.; Lee, J.T.; Cloughesy, T.; et al. Detection of immune responses after immunotherapy in glioblastoma using PET and MRI. Proc. Natl. Acad. Sci. USA 2017, 114, 10220–10225. [Google Scholar] [CrossRef]
- Rashidian, M.; LaFleur, M.W.; Verschoor, V.L.; Dongre, A.; Zhang, Y.; Nguyen, T.H.; Kolifrath, S.; Aref, A.R.; Lau, C.J.; Paweletz, C.P.; et al. Immuno-PET identifies the myeloid compartment as a key contributor to the outcome of the antitumor response under PD-1 blockade. Proc. Natl. Acad. Sci. USA 2019, 116, 16971–16980. [Google Scholar] [CrossRef]
- Wang, J.; Wu, J.; Wang, R.; Wang, J.; Chen, D.; Cao, Y.; Fu, W.; Wang, W.; Li, H.; Jiao, Y.; et al. Single-Cell Atlas Reveals Complexity of the Immunosuppressive Microenvironment of Initial and Recurrent Glioblastoma. Front. Immunol. 2020, 11, 835. [Google Scholar] [CrossRef]
- McKelvey, K.J.; Hudson, A.L.; Prasanna Kumar, R.; Wilmott, J.S.; Attrill, G.H.; Long, G.V.; Scolyer, R.A.; Clarke, S.J.; Wheeler, H.R.; Diakos, C.I.; et al. Temporal and spatial modulation of the tumor and systemic immune response in the murine Gl261 glioma model. PLoS ONE 2020, 15, e0226444. [Google Scholar] [CrossRef]
- Gieryng, A.; Pszczolkowska, D.; Walentynowicz, K.A.; Rajan, W.D.; Kaminska, B. Immune microenvironment of gliomas. Lab. Investig. 2017, 97, 498–518. [Google Scholar] [CrossRef]
- Larionova, I.; Cherdyntseva, N.; Liu, T.; Patysheva, M.; Rakina, M.; Kzhyshkowska, J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology 2019, 8, 1596004. [Google Scholar] [CrossRef]
- Nesseler, J.P.; Schaue, D.; McBride, W.H.; Lee, M.H.; Kaprealian, T.; Niclou, S.P.; Nickers, P. Irradiation to Improve the Response to Immunotherapeutic Agents in Glioblastomas. Adv. Radiat. Oncol. 2019, 4, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Ochiai, A. Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors. Cells 2019, 8, 418. [Google Scholar] [CrossRef] [PubMed]
- Masuda, J.; Shigehiro, T.; Matsumoto, T.; Satoh, A.; Mizutani, A.; Umemura, C.; Saito, S.; Kijihira, M.; Takayama, E.; Seno, A.; et al. Cytokine Expression and Macrophage Localization in Xenograft and Allograft Tumor Models Stimulated with Lipopolysaccharide. Int. J. Mol. Sci. 2018, 19, 1261. [Google Scholar] [CrossRef]
- Korsakova, L.; Krasko, J.A.; Stankevicius, E. Metabolic-targeted combination therapy with dichloroacetate and metformin suppresses glioblastoma cell line growth in vitro and in vivo. In Vivo 2021, 35, 341–348. [Google Scholar] [CrossRef]
- Su, Y.-T.; Butler, M.; Zhang, M.; Zhang, W.; Song, H.; Hwang, L.; Tran, A.D.; Bash, R.E.; Schorzman, A.N.; Pang, Y.; et al. MerTK inhibition decreases immune suppressive glioblastoma-associated macrophages and neoangiogenesis in glioblastoma microenvironment. Neuro-Oncology Adv. 2020, 2, vdaa065. [Google Scholar] [CrossRef]
- Tang, B.; Guo, Z.S.; Bartlett, D.L.; Yan, D.Z.; Schane, C.P.; Thomas, D.L.; Liu, J.; McFadden, G.; Shisler, J.L.; Roy, E.J.; et al. Synergistic Combination of Oncolytic Virotherapy and Immunotherapy for Glioma. Clin. Cancer Res. 2020, 26, 2216–2230. [Google Scholar] [CrossRef]
- Oh, T.; Fakurnejad, S.; Sayegh, E.T.; Clark, A.J.; Ivan, M.E.; Sun, M.Z.; Safaee, M.; Bloch, O.; James, C.D.; Parsa, A.T. Immunocompetent murine models for the study of glioblastoma immunotherapy. J. Transl. Med. 2014, 12, 107. [Google Scholar] [CrossRef]
- Reardon, D.A.; Gokhale, P.C.; Klein, S.R.; Ligon, K.L.; Rodig, S.J.; Ramkissoon, S.H.; Jones, K.L.; Conway, A.S.; Liao, X.; Zhou, J.; et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol. Res. 2016, 4, 124–135. [Google Scholar] [CrossRef]
- Ferrer-Font, L.; Arias-Ramos, N.; Lope-Piedrafita, S.; Julià-Sapé, M.; Pumarola, M.; Arús, C.; Candiota, A.P.A.P. Metronomic treatment in immunocompetent preclinical GL261 glioblastoma: Effects of cyclophosphamide and temozolomide. NMR Biomed. 2017, 30, e3748. [Google Scholar] [CrossRef]
- Wu, S.; Calero-Pérez, P.; Arús, C.; Candiota, A.P. Anti-pd-1 immunotherapy in preclinical gl261 glioblastoma: Influence of therapeutic parameters and non-invasive response biomarker assessment with mrsi-based approaches. Int. J. Mol. Sci. 2020, 21, 8775. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Font, L.; Villamañan, L.; Arias-Ramos, N.; Vilardell, J.; Plana, M.; Ruzzene, M.; Pinna, L.A.; Itarte, E.; Arús, C.; Candiota, A.P. Targeting protein kinase CK2: Evaluating CX-4945 potential for GL261 glioblastoma therapy in immunocompetent mice. Pharmaceuticals 2017, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Arias-Ramos, N.; Ferrer-Font, L.; Lope-Piedrafita, S.; Mocioiu, V.; Julià-Sapé, M.; Pumarola, M.; Arús, C.; Candiota, A. Metabolomics of Therapy Response in Preclinical Glioblastoma: A Multi-Slice MRSI-Based Volumetric Analysis for Noninvasive Assessment of Temozolomide Treatment. Metabolites 2017, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Genoud, V.; Marinari, E.; Nikolaev, S.I.; Castle, J.C.; Bukur, V.; Dietrich, P.Y.; Okada, H.; Walker, P.R. Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology 2018, 7, e1501137. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Pich, O.; Muiños, F.; Lolkema, M.P.; Steeghs, N.; Gonzalez-Perez, A.; Lopez-Bigas, N. The mutational footprints of cancer therapies. Nat. Genet. 2019, 51, 1732–1740. [Google Scholar] [CrossRef]
- Puigdelloses, M.; Garcia-Moure, M.; Labiano, S.; Laspidea, V.; Gonzalez-Huarriz, M.; Zalacain, M.; Marrodan, L.; Martinez-Velez, N.; la Nava, D.D.; Ausejo, I.; et al. CD137 and PD-L1 targeting with immunovirotherapy induces a potent and durable antitumor immune response in glioblastoma models. J. Immunother. Cancer 2021, 9, e002644. [Google Scholar] [CrossRef]
- Du, B.; Waxman, D.J. Medium dose intermittent cyclophosphamide induces immunogenic cell death and cancer cell autonomous type I interferon production in glioma models. Cancer Lett. 2020, 470, 170–180. [Google Scholar] [CrossRef]
- Cook, K.M.; Shen, H.; McKelvey, K.J.; Gee, H.E.; Hau, E. Targeting Glucose Metabolism of Cancer Cells with Dichloroacetate to Radiosensitize High-Grade Gliomas. Int. J. Mol. Sci. 2021, 22, 7265. [Google Scholar] [CrossRef]
- Khalsa, J.K.; Cheng, N.; Keegan, J.; Chaudry, A.; Driver, J.; Bi, W.L.; Lederer, J.; Shah, K. Immune phenotyping of diverse syngeneic murine brain tumors identifies immunologically distinct types. Nat. Commun. 2020, 11, 3912. [Google Scholar] [CrossRef]
- Delgado-Goñi, T.; Julià-Sapé, M.; Candiota, A.P.; Pumarola, M.; Arús, C. Molecular imaging coupled to pattern recognition distinguishes response to temozolomide in preclinical glioblastoma. NMR Biomed. 2014, 27, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Goñi, T.; Ortega-Martorell, S.; Ciezka, M.; Olier, I.; Candiota, A.P.; Julià-Sapé, M.; Fernández, F.; Pumarola, M.; Lisboa, P.J.; Arús, C. MRSI-based molecular imaging of therapy response to temozolomide in preclinical glioblastoma using source analysis. NMR Biomed. 2016, 29, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Hakumäki, J.M.; Poptani, H.; Sandmair, A.M.; Ylä-Herttuala, S.; Kauppinen, R.A. 1H MRS detects polyunsaturated fatty acid accumulation during gene therapy of glioma: Implications for the in vivo detection of apoptosis. Nat. Med. 1999, 5, 1323–1327. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed]
- Coward, J.; Harding, A. Size Does Matter: Why Polyploid Tumor Cells are Critical Drug Targets in the War on Cancer. Front. Oncol. 2014, 4, 123. [Google Scholar] [CrossRef] [PubMed]
- Calero-Pérez, P.; Wu, S.; Arús, C.; Candiota, A.P. Immune system-related changes in preclinical GL261 glioblastoma under TMZ treatment: Explaining MRSI-based nosological imaging findings with RT-PCR analyses. Cancers 2021, 13, 2663. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Calero-Pérez, P.; Villamañan, L.; Arias-Ramos, N.; Pumarola, M.; Ortega-Martorell, S.; Julià-Sapé, M.; Arús, C.; Candiota, A.P. Anti-tumour immune response in GL261 glioblastoma generated by Temozolamide Immune-Enhancing Metronomic Schedule monitored with MRSI-based nosological images. NMR Biomed. 2020, 33, e4229. [Google Scholar] [CrossRef]
- Villeneuve, J.; Tremblay, P.; Vallières, L. Tumor necrosis factor reduces brain tumor growth by enhancing macrophage recruitment and microcyst formation. Cancer Res. 2005, 65, 3928–3936. [Google Scholar] [CrossRef]
- Griffin, J.L.; Lehtimäki, K.K.; Valonen, P.K.; Gröhn, O.H.J.; Kettunen, M.I.; Ylä-Herttuala, S.; Pitkänen, A.; Nicholson, J.K.; Kauppinen, R.A. Assignment of 1 H Nuclear Magnetic Resonance Visible Polyunsaturated Fatty Acids in BT4C Gliomas Undergoing Ganciclovir-Thymidine Kinase Gene Therapy-induced Programmed Cell Death 1. Cancer Res. 2003, 63, 3195–3201. [Google Scholar]
- Wibom, C.; Surowiec, I.; Mörén, L.; Bergström, P.; Johansson, M.; Antti, H.; Bergenheim, A.T. Metabolomic patterns in glioblastoma and changes during radiotherapy: A clinical microdialysis study. J. Proteome Res. 2010, 9, 2909–2919. [Google Scholar] [CrossRef]
- Pandey, R.; Caflisch, L.; Lodi, A.; Brenner, A.J.; Tiziani, S. Metabolomic signature of brain cancer. Mol. Carcinog. 2017, 56, 2355–2371. [Google Scholar] [CrossRef] [PubMed]
- Donovan, P.; Cato, K.; Legaie, R.; Jayalath, R.; Olsson, G.; Hall, B.; Olson, S.; Boros, S.; Reynolds, B.A.; Harding, A. Hyperdiploid tumor cells increase phenotypic heterogeneity within Glioblastoma tumors. Mol. Biosyst. 2014, 10, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Fraternale, A.; Brundu, S.; Magnani, M. Polarization and Repolarization of Macrophages. J. Clin. Cell. Immunol. 2015, 6, 100319. [Google Scholar] [CrossRef]
- Geisenberger, C.; Mock, A.; Warta, R.; Rapp, C.; Schwager, C.; Korshunov, A.; Nied, A.K.; Capper, D.; Brors, B.; Jungk, C.; et al. Molecular profiling of long-term survivors identifies a subgroup of glioblastoma characterized by chromosome 19/20 co-gain. Acta Neuropathol. 2015, 130, 419–434. [Google Scholar] [CrossRef]
- Zhang, M.; Hutter, G.; Kahn, S.A.; Azad, T.D.; Gholamin, S.; Xu, C.Y.; Liu, J.; Achrol, A.S.; Richard, C.; Sommerkamp, P.; et al. Anti-CD47 Treatment Stimulates Phagocytosis of Glioblastoma by M1 and M2 Polarized Macrophages and Promotes M1 Polarized Macrophages In Vivo. PLoS ONE 2016, 11, e0153550. [Google Scholar] [CrossRef]
- Shi, Y.; Ping, Y.F.; Zhou, W.; He, Z.C.; Chen, C.; Bian, B.S.J.; Zhang, L.; Chen, L.; Lan, X.; Zhang, X.C.; et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 2017, 8, 15080. [Google Scholar] [CrossRef]
- Wang, Q.; He, Z.; Huang, M.; Liu, T.; Wang, Y.; Xu, H.; Duan, H.; Ma, P.; Zhang, L.; Zamvil, S.S.; et al. Vascular niche IL-6 induces alternative macrophage activation in glioblastoma through HIF-2α. Nat. Commun. 2018, 9, 559. [Google Scholar] [CrossRef]
- Gabrusiewicz, K.; Hossain, M.B.; Cortes-Santiago, N.; Fan, X.; Kaminska, B.; Marini, F.C.; Fueyo, J.; Gomez-Manzano, C. Macrophage Ablation Reduces M2-Like Populations and Jeopardizes Tumor Growth in a MAFIA-Based Glioma Model. Neoplasia 2015, 17, 374–384. [Google Scholar] [CrossRef]
- Garofalo, S.; Porzia, A.; Mainiero, F.; Di Angelantonio, S.; Cortese, B.; Basilico, B.; Pagani, F.; Cignitti, G.; Chece, G.; Maggio, R.; et al. Environmental stimuli shape microglial plasticity in glioma. eLife 2017, 6, e33415. [Google Scholar] [CrossRef]
- Salazar, N.; Carlson, J.C.; Huang, K.; Zheng, Y.; Oderup, C.; Gross, J.; Jang, A.D.; Burke, T.M.; Lewén, S.; Scholz, A.; et al. A Chimeric Antibody against ACKR3/CXCR7 in Combination with TMZ Activates Immune Responses and Extends Survival in Mouse GBM Models. Mol. Ther. 2018, 26, 1354–1365. [Google Scholar] [CrossRef]
- Morshed, N.; Ralvenius, W.T.; Nott, A.; Watson, L.A.; Rodriguez, F.H.; Akay, L.A.; Joughin, B.A.; Pao, P.C.; Penney, J.; LaRocque, L.; et al. Phosphoproteomics identifies microglial Siglec-F inflammatory response during neurodegeneration. Mol. Syst. Biol. 2020, 16, e9819. [Google Scholar] [CrossRef] [PubMed]
- Villamañan, L.; Martínez-escardó, L.; Arús, C.; Yuste, V.J.; Candiota, A.P. Successful partnerships: Exploring the potential of immunogenic signals triggered by TMZ, CX-4945, and combined treatment in Gl261 glioblastoma cells. Int. J. Mol. Sci. 2021, 22, 3453. [Google Scholar] [CrossRef] [PubMed]
- De Edelenyi, F.S.; Rubin, C.; Estève, F.; Grand, S.; Décorps, M.; Lefournier, V.; Le Bas, J.F.; Rémy, C. A new approach for analyzing proton magnetic resonance spectroscopic images of brain tumors: Nosologic images. Nat. Med. 2000, 6, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Martorell, S.; Lisboa, P.J.G.; Vellido, A.; Simões, R.V.; Pumarola, M.; Julià-Sapé, M.; Arús, C. Convex Non-Negative Matrix Factorization for Brain Tumor Delimitation from MRSI Data. PLoS ONE 2012, 7, e47824. [Google Scholar] [CrossRef] [PubMed]
- Karman, J.; Ling, C.; Sandor, M.; Fabry, Z. Initiation of Immune Responses in Brain Is Promoted by Local Dendritic Cells. J. Immunol. 2004, 173, 2353–2361. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Haus, E.; Lakatua, D.J.; Swoyer, J.; Sackett-Lundeen, L. Chronobiology in hematology and immunology. Am. J. Anat. 1983, 168, 467–517. [Google Scholar] [CrossRef]
- Walker, D.G.; Shakya, R.; Morrison, B.; Neller, M.A.; Matthews, K.K.; Nicholls, J.; Smith, C.; Khanna, R. Impact of pre-therapy glioblastoma multiforme microenvironment on clinical response to autologous CMV-specific T-cell therapy. Clin. Transl. Immunol. 2019, 8, e01088. [Google Scholar] [CrossRef]
- Liang, H.; Deng, L.; Chmura, S.; Burnette, B.; Liadis, N.; Darga, T.; Beckett, M.A.; Lingen, M.W.; Witt, M.; Weichselbaum, R.R.; et al. Radiation-Induced Equilibrium Is a Balance between Tumor Cell Proliferation and T Cell–Mediated Killing. J. Immunol. 2013, 190, 5874–5881. [Google Scholar] [CrossRef]
- Vrabec, M.; Van Cauter, S.; Himmelreich, U.; Van Gool, S.W.; Sunaert, S.; De Vleeschouwer, S.; Šuput, D.; Demaerel, P. MR perfusion and diffusion imaging in the follow-up of recurrent glioblastoma treated with dendritic cell immunotherapy: A pilot study. Neuroradiology 2011, 53, 721–731. [Google Scholar] [CrossRef]
- Ceschin, R.; Kurland, B.F.; Abberbock, S.R.; Ellingson, B.M.; Okada, H.; Jakacki, R.I.; Pollack, I.F.; Panigrahy, A. Parametric response mapping of apparent diffusion coefficient as an imaging biomarker to distinguish pseudoprogression from true tumor progression in peptide-based vaccine therapy for pediatric diffuse intrinsic pontine glioma. Am. J. Neuroradiol. 2015, 36, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Li, X.; Stroiney, A.; Qu, J.; Helgager, J.; Reardon, D.A.; Young, G.S. Advanced MRI assessment to predict benefit of anti-programmed cell death 1 protein immunotherapy response in patients with recurrent glioblastoma. Neuroradiology 2017, 59, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Matulewicz, ł.; Sokół, M.; Michnik, A.; Wydmański, J. Long-term normal-appearing brain tissue monitoring after irradiation using proton magnetic resonance spectroscopy in vivo: Statistical analysis of a large group of patients. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Ina Ly, K.; Vakulenko-Lagun, B.; Emblem, K.E.; Ou, Y.; Da, X.; Betensky, R.A.; Kalpathy-Cramer, J.; Duda, D.G.; Jain, R.K.; Chi, A.S.; et al. Probing tumor microenvironment in patients with newly diagnosed glioblastoma during chemoradiation and adjuvant temozolomide with functional MRI. Sci. Rep. 2018, 8, 17062. [Google Scholar] [CrossRef]
- Zhou, Y.; Larsen, P.H.; Hao, C.; Wee Yong, V. CXCR4 Is a Major Chemokine Receptor on Glioma Cells and Mediates Their Survival. J. Biol. Chem. 2002, 277, 49481–49487. [Google Scholar] [CrossRef]
- Booth, T.C.; Thompson, G.; Bulbeck, H.; Boele, F.; Buckley, C.; Cardoso, J.; Canas Dos Santos, L.; Jenkinson, D.; Ashkan, K.; Kreindler, J.; et al. A Position Statement on the Utility of Interval Imaging in Standard of Care Brain Tumour Management: Defining the Evidence Gap and Opportunities for Future Research. Front. Oncol. 2021, 11, 620070. [Google Scholar] [CrossRef]
- Rühle, P.F.; Goerig, N.; Wunderlich, R.; Fietkau, R.; Gaipl, U.S.; Strnad, A.; Frey, B. Modulations in the peripheral immune system of glioblastoma patient is connected to therapy and tumor progression-A case report from the IMMO-GLIO-01 trial. Front. Neurol. 2017, 8, 296. [Google Scholar] [CrossRef]
- Alban, T.J.; Alvarado, A.G.; Sorensen, M.D.; Bayik, D.; Volovetz, J.; Serbinowski, E.; Mulkearns-Hubert, E.E.; Sinyuk, M.; Hale, J.S.; Onzi, G.R.; et al. Global immune fingerprinting in glioblastoma patient peripheral blood reveals immune-suppression signatures associated with prognosis. JCI Insight 2018, 3, e122264. [Google Scholar] [CrossRef]
- Coventry, B.J.; Ashdown, M.L.; Quinn, M.A.; Markovic, S.N.; Yatomi-Clarke, S.L.; Robinson, A.P. CRP identifies homeostatic immune oscillations in cancer patients: A potential treatment targeting tool? J. Transl. Med. 2009, 7, 102. [Google Scholar] [CrossRef]
- Nehring, S.M.; Goyal, A.; Bansal, P.; Patel, B.C. C Reactive Protein. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021; Volume 65, pp. 237–244. Available online: https://www.ncbi.nlm.nih.gov/books/NBK441843/ (accessed on 21 October 2021).
- Tubin, S.; Ashdown, M.; Jeremic, B. Time-synchronized immune-guided SBRT partial bulky tumor irradiation targeting hypoxic segment while sparing the peritumoral immune microenvironment. Radiat. Oncol. 2019, 14, 220. [Google Scholar] [CrossRef]
- Kitano, Y.; Aoki, K.; Ohka, F.; Yamazaki, S.; Motomura, K.; Tanahashi, K.; Hirano, M.; Naganawa, T.; Iida, M.; Shiraki, Y.; et al. Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds. ACS Appl. Mater. Interfaces 2021, 13, 17316–17329. [Google Scholar] [CrossRef] [PubMed]
- Tandle, A.T.; Shankavaram, U.; Brown, M.V.; Ho, J.; Graves, C.; Lita, E.; Pfohl, J.; Mohney, R.; Tofilon, P.; Camphausen, K. Urinary Metabolomic Profiling of Patients with Glioblastoma Multiforme) Urinary Metabolomic Profiling of Patients with Glioblastoma Multiforme. J. Proteom. Bioinform. 2013, S6, 003. [Google Scholar] [CrossRef]
- Lazovic, J.; Jensen, M.C.; Ferkassian, E.; Aguilar, B.; Raubitschek, A.; Jacobs, R.E. Imaging immune response in vivo: Cytolytic action of genetically altered T cells directed to glioblastoma multiforme. Clin. Cancer Res. 2008, 14, 3832–3839. [Google Scholar] [CrossRef] [PubMed]
- Brekke Rygh, C.; Wang, J.; Thuen, M.; Gras Navarro, A.; Marie Huuse, E.; Thorsen, F.; Poli, A.; Zimmer, J.; Haraldseth, O.; Atle Lie, S.; et al. Dynamic contrast enhanced MRI detects early response to adoptive NK cellular immunotherapy targeting the NG2 proteoglycan in a rat model of glioblastoma. PLoS ONE 2014, 9, e108414. [Google Scholar] [CrossRef]
- Miyai, M.; Tomita, H.; Soeda, A.; Yano, H.; Iwama, T.; Hara, A. Current trends in mouse models of glioblastoma. J. Neurooncol. 2017, 135, 423–432. [Google Scholar] [CrossRef]
- Irtenkauf, S.M.; Sobiechowski, S.; Hasselbach, L.A.; Nelson, K.K.; Transou, A.D.; Carlton, E.T.; Mikkelsen, T.; De Carvalho, A.C. Optimization of glioblastoma mouse orthotopic xenograft models for translational research. Comp. Med. 2017, 67, 300–314. [Google Scholar]
- Patrizii, M.; Bartucci, M.; Pine, S.R.; Sabaawy, H.E. Utility of glioblastoma patient-derived orthotopic xenografts in drug discovery and personalized therapy. Front. Oncol. 2018, 8, 23. [Google Scholar] [CrossRef]
- Su, Y.B.; Sohn, S.; Krown, S.E.; Livingston, P.O.; Wolchok, J.D.; Quinn, C.; Williams, L.; Foster, T.; Sepkowitz, K.A.; Chapman, P.B. Selective CD4+ lymphopenia in melanoma patients treated with temozolomide: A toxicity with therapeutic implications. J. Clin. Oncol. 2004, 22, 610–616. [Google Scholar] [CrossRef]
- Niewald, M.; Berdel, C.; Fleckenstein, J.; Licht, N.; Ketter, R.; Rübe, C. Toxicity after radiochemotherapy for glioblastoma using temozolomide-a retrospective evaluation. Radiat. Oncol. 2011, 6, 141. [Google Scholar] [CrossRef]
- Kim, S.S.; Rait, A.; Kim, E.; DeMarco, J.; Pirollo, K.F.; Chang, E.H. Encapsulation of temozolomide in a tumor-targeting nanocomplex enhances anti-cancer efficacy and reduces toxicity in a mouse model of glioblastoma. Cancer Lett. 2015, 369, 250–258. [Google Scholar] [CrossRef]
- Lynes, J.P.; Nwankwo, A.K.; Sur, H.P.; Sanchez, V.E.; Sarpong, K.A.; Ariyo, O.I.; Dominah, G.A.; Nduom, E.K. Biomarkers for immunotherapy for treatment of glioblastoma. J. Immunother. Cancer 2020, 8, 348. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Maxwell, R.; Xia, Y.; Cardarelli, P.; Oyasu, M.; Belcaid, Z.; Kim, E.; Hung, A.; Luksik, A.S.; Garzon-Muvdi, T.; et al. Combination anti-CXCR4 and anti-PD-1 immunotherapy provides survival benefit in glioblastoma through immune cell modulation of tumor microenvironment. J. Neurooncol. 2019, 143, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Saenz-Antoñanzas, A.; Auzmendi-Iriarte, J.; Carrasco-Garcia, E.; Moreno-Cugnon, L.; Ruiz, I.; Villanua, J.; Egaña, L.; Otaegui, D.; Samprón, N.; Matheu, A. Liquid biopsy in glioblastoma: Opportunities, applications and challenges. Cancers 2019, 11, 950. [Google Scholar] [CrossRef] [PubMed]
- Narang, S.; Lehrer, M.; Yang, D.; Lee, J.; Rao, A. Radiomics in glioblastoma: Current status, challenges and potential opportunities. Transl. Cancer Res. 2016, 5, 383–397. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Elshafeey, N.; Kotrotsou, A.; Hassan, A.; Elshafei, N.; Hassan, I.; Ahmed, S.; Abrol, S.; Agarwal, A.; El Salek, K.; Bergamaschi, S.; et al. Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat. Commun. 2019, 10, 3170. [Google Scholar] [CrossRef]
- Núñez, L.M.; Romero, E.; Julià-Sapé, M.; Ledesma-Carbayo, M.J.; Santos, A.; Arús, C.; Candiota, A.P.; Vellido, A. Unraveling response to temozolomide in preclinical GL261 glioblastoma with MRI/MRSI using radiomics and signal source extraction. Sci. Rep. 2020, 10, 19699. [Google Scholar] [CrossRef]
- Chang, Y.-C.C.; Ackerstaff, E.; Tschudi, Y.; Jimenez, B.; Foltz, W.; Fisher, C.; Lilge, L.; Cho, H.; Carlin, S.; Gillies, R.J.; et al. Delineation of Tumor Habitats based on Dynamic Contrast Enhanced MRI. Sci. Rep. 2017, 7, 9746. [Google Scholar] [CrossRef]
- Salzillo, T.C.; Mawoneke, V.; Weygand, J.; Shetty, A.; Gumin, J.; Zacharias, N.M.; Gammon, S.T.; Piwnica-Worms, D.; Fuller, G.N.; Logothetis, C.J.; et al. Measuring the Metabolic Evolution of Glioblastoma throughout Tumor Development, Regression, and Recurrence with Hyperpolarized Magnetic Resonance. Cells 2021, 10, 2621. [Google Scholar] [CrossRef]
- Dutta, P.; Salzillo, T.C.; Pudakalakatti, S.; Gammon, S.T.; Kaipparettu, B.A.; McAllister, F.; Wagner, S.; Frigo, D.E.; Logothetis, C.J.; Zacharias, N.M.; et al. Assessing Therapeutic Efficacy in Real-time by Hyperpolarized Magnetic Resonance Metabolic Imaging. Cells 2019, 8, 340. [Google Scholar] [CrossRef]
- Kurhanewicz, J.; Vigneron, D.B.; Ardenkjaer-Larsen, J.H.; Bankson, J.A.; Brindle, K.; Cunningham, C.H.; Gallagher, F.A.; Keshari, K.R.; Kjaer, A.; Laustsen, C.; et al. Hyperpolarized 13C MRI: Path to Clinical Translation in Oncology. Neoplasia 2019, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Leftin, A.; Ben-Chetrit, N.; Klemm, F.; Joyce, J.A.; Koutcher, J.A. Iron imaging reveals tumor and metastasis macrophage hemosiderin deposits in breast cancer. PLoS ONE 2017, 12, e0184765. [Google Scholar] [CrossRef] [PubMed]
- Janssen, B.; Vugts, D.; Windhorst, A.; Mach, R. PET Imaging of Microglial Activation—Beyond Targeting TSPO. Molecules 2018, 23, 607. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.G.; Nieman, L.T.; Stemmer-Rachamimov, A.O.; Zheng, X.S.; Stafford, K.; Nagashima, H.; Miller, J.J.; Kiyokawa, J.; Ting, D.T.; Wakimoto, H.; et al. IDH-mutant gliomas harbor fewer regulatory T cells in humans and mice. Oncoimmunology 2020, 9, 1806662. [Google Scholar] [CrossRef] [PubMed]
- Pellegatta, S.; Valletta, L.; Corbetta, C.; Patanè, M.; Zucca, I.; Riccardi Sirtori, F.; Bruzzone, M.G.; Fogliatto, G.; Isacchi, A.; Pollo, B.; et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol. Commun. 2015, 3, 4. [Google Scholar] [CrossRef]
- Hu, C.; Wang, K.; Damon, C.; Fu, Y.; Ma, T.; Kratz, L.; Lal, B.; Ying, M.; Xia, S.; Cahill, D.P.; et al. ATRX loss promotes immunosuppressive mechanisms in IDH1 mutant glioma. Neuro. Oncol. 2021, noab292. [Google Scholar] [CrossRef]
- Cai, J.; Chen, J.; Zhang, W.; Yang, P.; Zhang, C.; Li, M.; Yao, K.; Wang, H.; Li, Q.; Jiang, C.; et al. Loss of ATRX, associated with DNA methylation pattern of chromosome end, impacted biological behaviors of astrocytic tumors. Oncotarget 2015, 6, 18105. [Google Scholar] [CrossRef][Green Version]
- Thust, S.C.; Heiland, S.; Falini, A.; Jäger, H.R.; Waldman, A.D.; Sundgren, P.C.; Godi, C.; Katsaros, V.K.; Ramos, A.; Bargallo, N.; et al. Glioma imaging in Europe: A survey of 220 centres and recommendations for best clinical practice. Eur. Radiol. 2018, 28, 3306–3317. [Google Scholar] [CrossRef]
- Stefan, D.; De Cesare, F.; Andrasescu, A.; Popa, E.; Lazariev, A.; Vescovo, E.; Strbak, O.; Williams, S.; Starcuk, Z.; Cabanas, M.; et al. Quantitation of magnetic resonance spectroscopy signals: The jMRUI software package. Meas Sci. Technol. 2009, 20, 104035. [Google Scholar] [CrossRef]
- Naressi, A.; Couturier, C.; Devos, J.M.; Janssen, M.; Mangeat, C.; de Beer, R.; Graveron-Demilly, D. Java-based graphical user interface for the MRUI quantitation package. Magn. Reson. Mater. Phys. Biol. Med. 2001, 122, 141–152. [Google Scholar] [CrossRef]
- Wilson, M.; Andronesi, O.; Barker, P.B.; Bartha, R.; Bizzi, A.; Bolan, P.J.; Brindle, K.M.; Choi, I.Y.; Cudalbu, C.; Dydak, U.; et al. Methodological consensus on clinical proton MRS of the brain: Review and recommendations. Magn. Reson. Med. 2019, 82, 527–550. [Google Scholar] [CrossRef]
- Hellström, J.; Zapata, R.R.; Libard, S.; Wikström, J.; Ortiz-Nieto, F.; Alafuzoff, I.; Raininko, R. The value of magnetic resonance spectroscopy as a supplement to MRI of the brain in a clinical setting. PLoS ONE 2018, 13, e0207336. [Google Scholar] [CrossRef]
- Julià-Sapé, M.; Coronel, I.; Majós, C.; Candiota, A.P.; Serrallonga, M.; Cos, M.; Aguilera, C.; Acebes, J.J.; Griffiths, J.R.; Arús, C. Prospective diagnostic performance evaluation of single-voxel 1H MRS for typing and grading of brain tumours. NMR Biomed. 2012, 25, 661–673. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Rapalino, O.; Gerstner, E.; Chi, A.; Batchelor, T.T.; Cahill, D.P.; Sorensen, A.G.; Rosen, B.R. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J. Clin. Investig. 2013, 123, 3659–3663. [Google Scholar] [CrossRef]
- Maudsley, A.A.; Domenig, C.; Govind, V.; Darkazanli, A.; Studholme, C.; Arheart, K.; Bloomer, C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI). Magn. Reson. Med. 2009, 61, 548–559. [Google Scholar] [CrossRef]
- Franco, P.; Huebschle, I.; Simon-gabriel, C.P.; Dacca, K.; Schnell, O.; Beck, J.; Mast, H.; Urbach, H.; Wuertemberger, U.; Prinz, M.; et al. Mapping of metabolic heterogeneity of glioma using mr-spectroscopy. Cancers 2021, 13, 2417. [Google Scholar] [CrossRef]
- Öz, G.; Deelchand, D.K.; Wijnen, J.P.; Mlynárik, V.; Xin, L.; Mekle, R.; Noeske, R.; Scheenen, T.W.J.; Tkáč, I.; Andronesi, O.; et al. Advanced single voxel 1H magnetic resonance spectroscopy techniques in humans: Experts’ consensus recommendations. NMR Biomed. 2021, 34, e4236. [Google Scholar] [CrossRef]
- Bhogal, A.A.; Broeders, T.A.A.; Morsinkhof, L.; Edens, M.; Nassirpour, S.; Chang, P.; Klomp, D.W.J.; Vinkers, C.H.; Wijnen, J.P. Lipid-suppressed and tissue-fraction corrected metabolic distributions in human central brain structures using 2D 1H magnetic resonance spectroscopic imaging at 7 T. Brain Behav. 2020, 10, e01852. [Google Scholar] [CrossRef]
- Wijnen, J.P.; Idema, A.J.S.; Stawicki, M.; Lagemaat, M.W.; Wesseling, P.; Wright, A.J.; Scheenen, T.W.J.; Heerschap, A. Quantitative short echo time 1H MRSI of the peripheral edematous region of human brain tumors in the differentiation between glioblastoma, metastasis, and meningioma. J. Magn. Reson. Imaging 2012, 36, 1072–1082. [Google Scholar] [CrossRef]
- Hangel, G.; Jain, S.; Springer, E.; Hečková, E.; Strasser, B.; Považan, M.; Gruber, S.; Widhalm, G.; Kiesel, B.; Furtner, J.; et al. High-resolution metabolic mapping of gliomas via patch-based super-resolution magnetic resonance spectroscopic imaging at 7T. Neuroimage 2019, 191, 587–595. [Google Scholar] [CrossRef]
- Hangel, G.; Cadrien, C.; Lazen, P.; Furtner, J.; Lipka, A.; Hečková, E.; Hingerl, L.; Motyka, S.; Gruber, S.; Strasser, B.; et al. High-resolution metabolic imaging of high-grade gliomas using 7T-CRT-FID-MRSI. NeuroImage Clin. 2020, 28, 102433. [Google Scholar] [CrossRef]
- Bogner, W.; Otazo, R.; Henning, A. Accelerated MR spectroscopic imaging—a review of current and emerging techniques. NMR Biomed. 2021, 34, e4314. [Google Scholar] [CrossRef]
- Horská, A.; Barker, P.B. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin. N. Am. 2010, 20, 293–310. [Google Scholar] [CrossRef]
- Laino, M.E.; Young, R.; Beal, K.; Haque, S.; Mazaheri, Y.; Corrias, G.; Bitencourt, A.G.; Karimi, S.; Thakur, S.B. Magnetic resonance spectroscopic imaging in gliomas: Clinical diagnosis and radiotherapy planning. BJR|Open 2020, 2, 20190026. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Esmaeili, M.; Borra, R.J.H.; Emblem, K.; Gerstner, E.R.; Pinho, M.C.; Plotkin, S.R.; Chi, A.S.; Eichler, A.F.; Dietrich, J.; et al. Early changes in glioblastoma metabolism measured by MR spectroscopic imaging during combination of anti-angiogenic cediranib and chemoradiation therapy are associated with survival. NPJ Precis. Oncol. 2017, 1, 20. [Google Scholar] [CrossRef]
- Nelson, S.J.; Li, Y.; Lupo, J.M.; Olson, M.; Crane, J.C.; Molinaro, A.; Roy, R.; Clarke, J.; Butowski, N.; Prados, M.; et al. Serial analysis of 3D H-1 MRSI for patients with newly diagnosed GBM treated with combination therapy that includes bevacizumab. J. Neurooncol. 2016, 130, 171–179. [Google Scholar] [CrossRef]
- Nelson, S.J.; Kadambi, A.K.; Park, I.; Li, Y.; Crane, J.; Olson, M.; Molinaro, A.; Roy, R.; Butowski, N.; Cha, S.; et al. Association of early changes in 1H MRSI parameters with survival for patients with newly diagnosed glioblastoma receiving a multimodality treatment regimen. Neuro. Oncol. 2017, 19, 430–439. [Google Scholar] [CrossRef]
- Muruganandham, M.; Clerkin, P.P.; Smith, B.J.; Anderson, C.M.; Morris, A.; Capizzano, A.A.; Magnotta, V.; McGuire, S.M.; Smith, M.C.; Bayouth, J.E.; et al. 3-dimensional magnetic resonance spectroscopic imaging at 3 Tesla for early response assessment of glioblastoma patients during external beam radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, 181–189. [Google Scholar] [CrossRef]
- Hattingen, E.; Bähr, O.; Rieger, J.; Blasel, S.; Steinbach, J.; Pilatus, U. Phospholipid Metabolites in Recurrent Glioblastoma: In Vivo Markers Detect Different Tumor Phenotypes before and under Antiangiogenic Therapy. PLoS ONE 2013, 8, e56439. [Google Scholar] [CrossRef]
- Wright, A.J.; Fellows, G.; Byrnes, T.J.; Opstad, K.S.; McIntyre, D.J.O.; Griffiths, J.R.; Bell, B.A.; Clark, C.A.; Barrick, T.R.; Howe, F.A. Pattern recognition of MRSI data shows regions of glioma growth that agree with DTI markers of brain tumor infiltration. Magn. Reson. Med. 2009, 62, 1646–1651. [Google Scholar] [CrossRef]
- Julià-Sapé, M.; Pons, A.; Arús, C.; Gil-Gil, M.; Majós, C. MRSI and a supervised classifier predict treatment response in glioblastoma: Preliminary results. Magn. Reson. Mater. Phy. 2017, 30, S480. [Google Scholar] [CrossRef]
- Li, Y.; Sima, D.M.; Van Cauter, S.; Himmelreich, U.; Sava, A.R.C.; Pi, Y.; Liu, Y.; Van Huffel, S. Unsupervised nosologic imaging for glioma diagnosis. IEEE Trans. Biomed. Eng. 2013, 60, 1760–1763. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Li, T.; Jordan, M. Convex and Semi-Nonnegative Matrix Factorizations. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Martorell, S.; Lisboa, P.J.; Vellido, A.; Julià-Sapé, M.; Arús, C. Non-negative matrix factorisation methods for the spectral decomposition of MRS data from human brain tumours. BMC Bioinform. 2012, 13, 38. [Google Scholar] [CrossRef]
- Von Roemeling, C.A.; Wang, Y.; Qie, Y.; Yuan, H.; Zhao, H.; Liu, X.; Yang, Z.; Yang, M.; Deng, W.; Bruno, K.A.; et al. Therapeutic modulation of phagocytosis in glioblastoma can activate both innate and adaptive antitumour immunity. Nat. Commun. 2020, 11, 1508. [Google Scholar] [CrossRef]
- Wouters, R.; Bevers, S.; Riva, M.; De Smet, F.; Coosemans, A. Immunocompetent Mouse Models in the Search for Effective Immunotherapy in Glioblastoma. Cancers 2020, 13, 19. [Google Scholar] [CrossRef]
- Leten, C.; Struys, T.; Dresselaers, T.; Himmelreich, U. In vivo and ex vivo assessment of the blood brain barrier integrity in different glioblastoma animal models. J. Neuro-Oncol. 2014, 119, 297–306. [Google Scholar] [CrossRef]
- Zagzag, D.; Miller, U.C.; Chiriboga, L.; Yee, H.; Newcomb, E.W. Green fluorescent protein immunohistochemistry as a novel experimental tool for the detection of glioma cell invasion in vivo. Brain Pathol. 2006, 13, 34–37. [Google Scholar] [CrossRef]
- Jacobs, V.L.; Valdes, P.A.; Hickey, W.F.; De Leo, J.A. Current Review of in Vivo GBM Rodent Models: Emphasis on the CNS-1 Tumour Model. ASN Neuro. 2011, 3, AN20110014. [Google Scholar] [CrossRef]
- Stevenson, C.B.; Ehtesham, M.; McMillan, K.M.; Valadez, J.G.; Edgeworth, M.L.; Price, R.R.; Abel, T.W.; Mapara, K.Y.; Thompson, R.C. CXCR4 Expression is Elevated in Glioblastoma Multiforme and Correlates with an Increase in Intensity and Extent of Peritumoral T2-weighted Magnetic Resonance Imaging Signal Abnormalities. Neurosurgery 2008, 63, 560–570. [Google Scholar] [CrossRef]
- Musumeci, G.; Castorina, A.; Magro, G.; Cardile, V.; Castorina, S.; Ribatti, D. Enhanced expression of CD31/platelet endothelial cell adhesion molecule 1 (PECAM1) correlates with hypoxia inducible factor-1 alpha (HIF-1α) in human glioblastoma multiforme. Exp. Cell Res. 2015, 339, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bolduc, A.R.; Hoda, N.; Gamble, D.N.; Dolisca, S.-B.; Bolduc, A.K.; Hoang, K.; Ashley, C.; McCall, D.; Rojiani, A.M.; et al. The indoleamine 2,3-dioxygenase pathway controls complement-dependent enhancement of chemo-radiation therapy against murine glioblastoma. J. Immunother. Cancer 2014, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Shen, Y.; Tang, Q.; Chen, L.; Gao, H.; Zhu, J. BCNU/PLGA microspheres: A promising strategy for the treatment of gliomas in mice. Chin. J. Cancer Res. 2014, 26, 81–88. [Google Scholar] [CrossRef]
- Newcomb, E.W.; Zagzag, D. The Murine GL261 Glioma Experimental Model to Assess Novel Brain Tumor Treatments. In CNS Cancer; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2009; pp. 227–241. [Google Scholar]
- Wu, A.; Oh, S.; Wiesner, S.M.; Ericson, K.; Chen, L.; Hall, W.A.; Champoux, P.E.; Low, W.C.; Ohlfest, J.R. Persistence of CD133+Cells in Human and Mouse Glioma Cell Lines: Detailed Characterization of GL261 Glioma Cells with Cancer Stem Cell-Like Properties. Stem Cells Dev. 2008, 17, 173–184. [Google Scholar] [CrossRef]
- Haddad, A.F.; Young, J.S.; Amara, D.; Berger, M.S.; Raleigh, D.R.; Aghi, M.K.; Butowski, N.A. Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neuro-Oncol. Adv. 2021, 3, 215. [Google Scholar] [CrossRef]
- Zagzag, D.; Salnikow, K.; Chiriboga, L.; Yee, H.; Lan, L.; Ali, M.A.; Garcia, R.; Demaria, S.; Newcomb, E.W. Downregulation of major histocompatibility complex antigens in invading glioma cells: Stealth invasion of the brain. Lab. Investig. 2005, 85, 328–341. [Google Scholar] [CrossRef]
- Anderson, R.C.; Anderson, D.E.; Elder, J.B.; Brown, M.D.; Mandigo, C.E.; Parsa, A.T.; Goodman, R.R.; McKhann, G.M.; Sisti, M.B.; Bruce, J.N. Lack of B7 expression, not human leukocyte antigen expression, facilitates immune evasion by human malignant gliomas. Neurosurgery 2007, 60, 1129–1136. [Google Scholar] [CrossRef]
- Kayabolen, A.; Yilmaz, E.; Bagci-Onder, T. IDH Mutations in Glioma: Double-Edged Sword in Clinical Applications? Biomedicines 2021, 9, 799. [Google Scholar] [CrossRef]
- Liimatainen, T.J.; Erkkilä, A.T.; Valonen, P.; Vidgren, H.; Lakso, M.; Wong, G.; Gröhn, O.H.; Ylä-Herttuala, S.; Hakumäki, J.M. 1H MR spectroscopic imaging of phospholipase-mediated membrane lipid release in apoptotic rat glioma in vivo. Magn. Reson. Med. 2008, 59, 1232–1238. [Google Scholar] [CrossRef]
- Mirbahai, L.; Wilson, M.; Shaw, C.S.; McConville, C.; Malcomson, R.D.G.; Kauppinen, R.A.; Peet, A.C. Lipid biomarkers of glioma cell growth arrest and cell death detected by1H magic angle spinning MRS. NMR Biomed. 2012, 25, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wilson, M.; McConville, C.; Arvanitis, T.N.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. Increased unsaturation of lipids in cytoplasmic lipid droplets in DAOY cancer cells in response to cisplatin treatment. Metabolomics 2012, 9, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, F.; Deleyrolle, L.P.; Karachi, A.; Mitchell, D.A.; Rahman, M. Metabolomics Monitoring of Treatment Response to Brain Tumor Immunotherapy. Front. Oncol. 2021, 11, 250. [Google Scholar] [CrossRef]
- Lim, H.; Martínez-Santiesteban, F.; Jensen, M.D.; Chen, A.; Wong, E.; Scholl, T.J. Monitoring Early Changes in Tumor Metabolism in Response to Therapy Using Hyperpolarized 13C MRSI in a Preclinical Model of Glioma. Tomography 2020, 6, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Hvinden, I.C.; Berg, H.E.; Sachse, D.; Skaga, E.; Skottvoll, F.S.; Lundanes, E.; Sandberg, C.J.; Vik-Mo, E.O.; Rise, F.; Wilson, S.R. Nuclear Magnetic Resonance Spectroscopy to Identify Metabolite Biomarkers of Nonresponsiveness to Targeted Therapy in Glioblastoma Tumor Stem Cells. J. Proteome Res. 2019, 18, 2012–2020. [Google Scholar] [CrossRef]
- Qu, J.R.; Jiang, T.; Dai, J.P.; Li, H.L.; Luo, J.P.; Li, S.W.; Ai, L.; Jiang, T.Z. Proton magnetic resonance spectroscopy predicts radiotherapy response and time-to-progression in high-grade gliomas after surgery. Chin. Med. 2012, 125, 4334–4337. [Google Scholar] [CrossRef]
- Wei, L.; Hong, S.; Yoon, Y.; Hwang, S.N.; Park, J.C.; Zhang, Z.; Olson, J.J.; Hu, X.; Shim, H. Early prediction of response to Vorinostat in an orthotopic rat glioma model. NMR Biomed. 2012, 25, 1104–1111. [Google Scholar] [CrossRef]
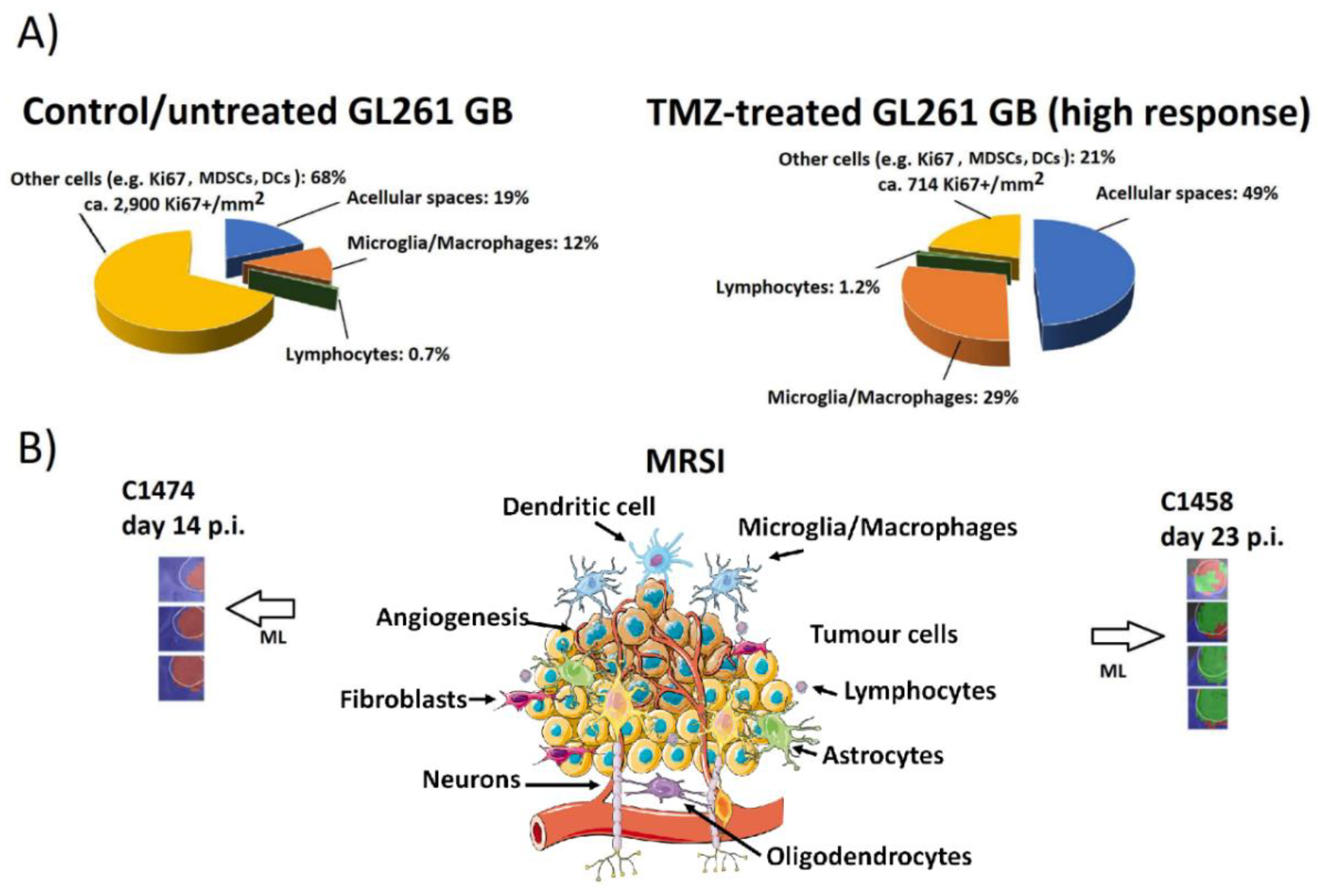
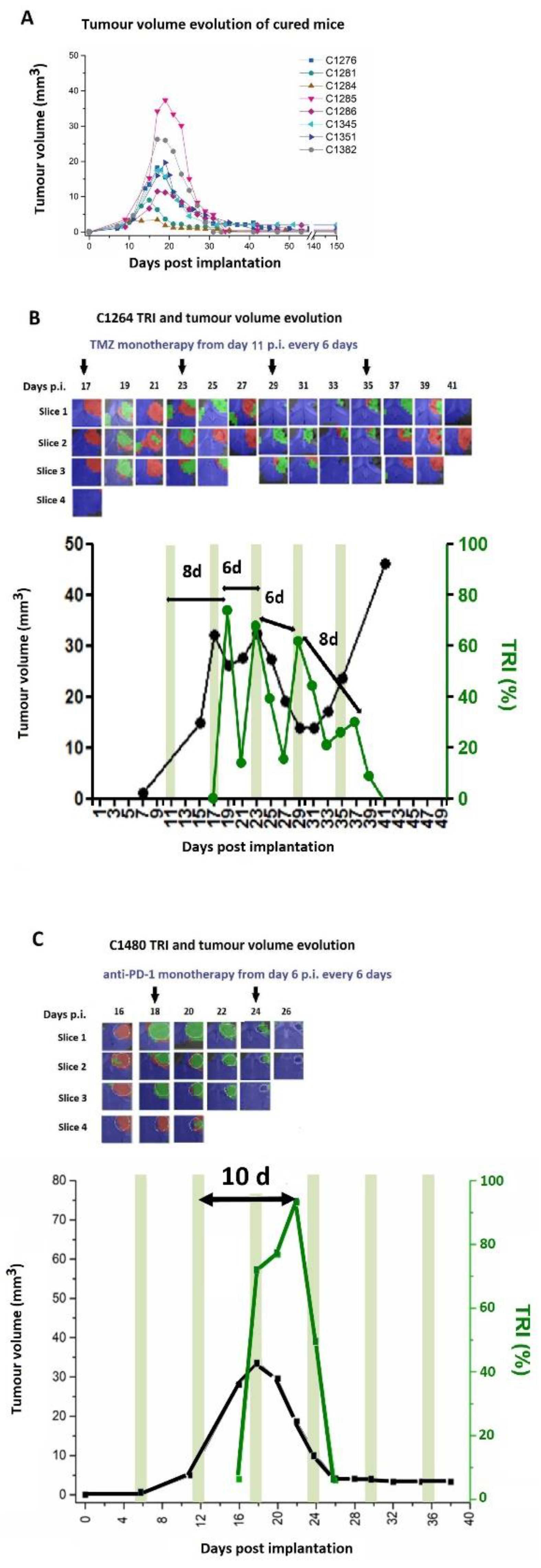
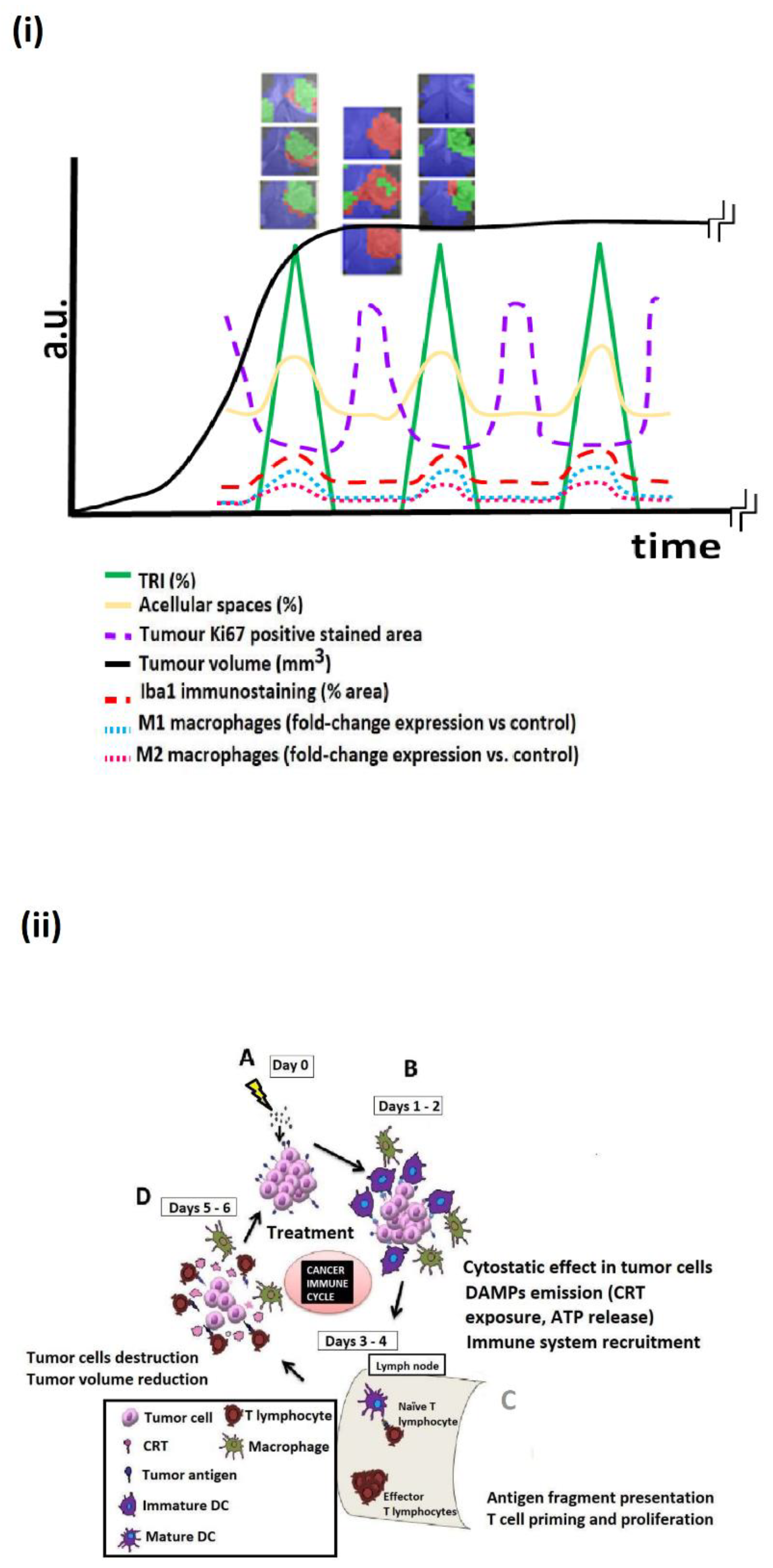
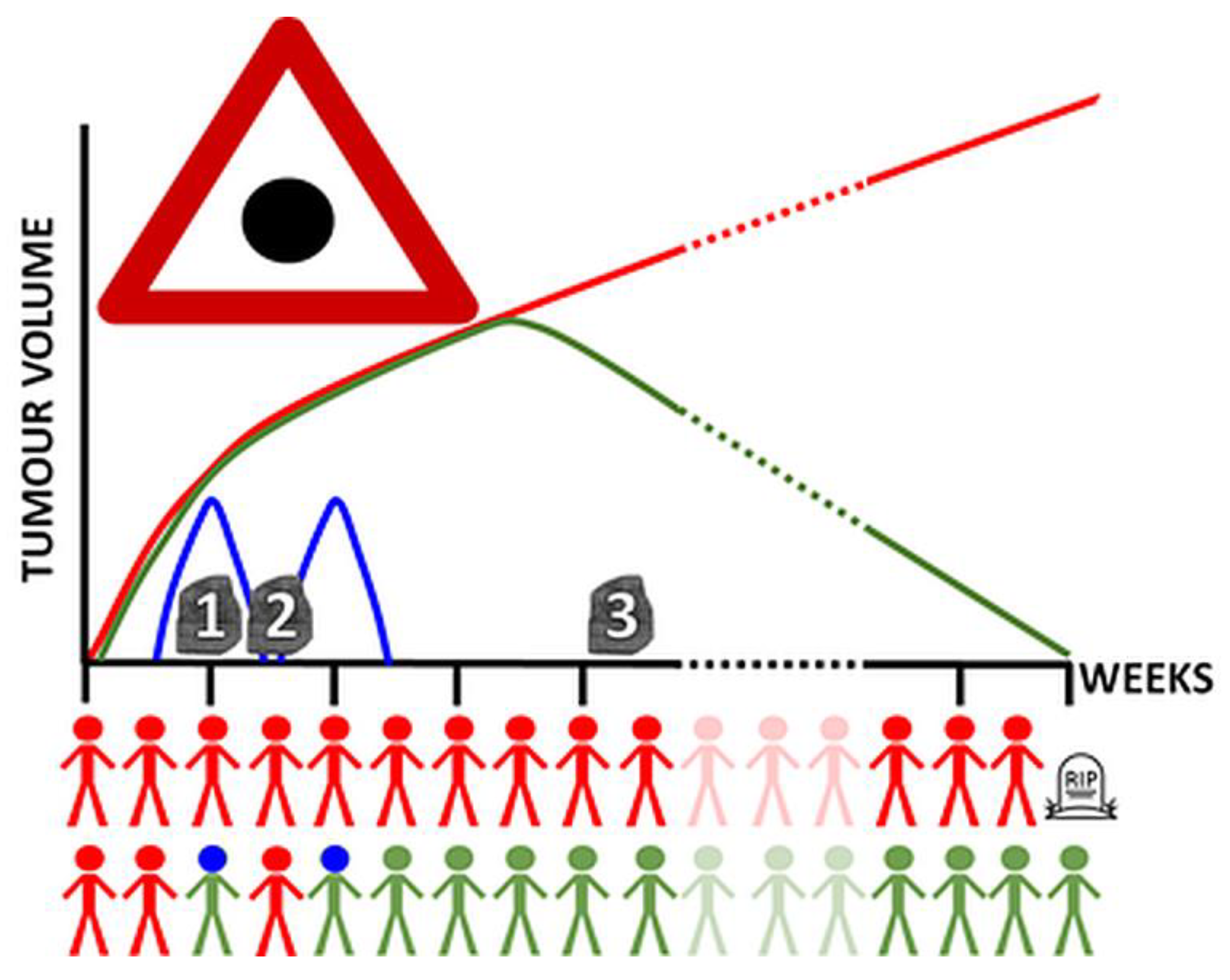
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Candiota, A.P.; Arús, C. Establishing Imaging Biomarkers of Host Immune System Efficacy during Glioblastoma Therapy Response: Challenges, Obstacles and Future Perspectives. Metabolites 2022, 12, 243. https://doi.org/10.3390/metabo12030243
Candiota AP, Arús C. Establishing Imaging Biomarkers of Host Immune System Efficacy during Glioblastoma Therapy Response: Challenges, Obstacles and Future Perspectives. Metabolites. 2022; 12(3):243. https://doi.org/10.3390/metabo12030243
Chicago/Turabian StyleCandiota, Ana Paula, and Carles Arús. 2022. "Establishing Imaging Biomarkers of Host Immune System Efficacy during Glioblastoma Therapy Response: Challenges, Obstacles and Future Perspectives" Metabolites 12, no. 3: 243. https://doi.org/10.3390/metabo12030243
APA StyleCandiota, A. P., & Arús, C. (2022). Establishing Imaging Biomarkers of Host Immune System Efficacy during Glioblastoma Therapy Response: Challenges, Obstacles and Future Perspectives. Metabolites, 12(3), 243. https://doi.org/10.3390/metabo12030243






