Investigation of the Exometabolomic Profiles of Rat Islets of Langerhans Cultured in Microfluidic Biochip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biochip and Fluidic Circuits
2.2. Culture Medium and Reagents
2.3. Pancreatic Islet Culture
2.4. Islet Viability and Functionality
2.5. Exometabolomic Analysis
3. Results
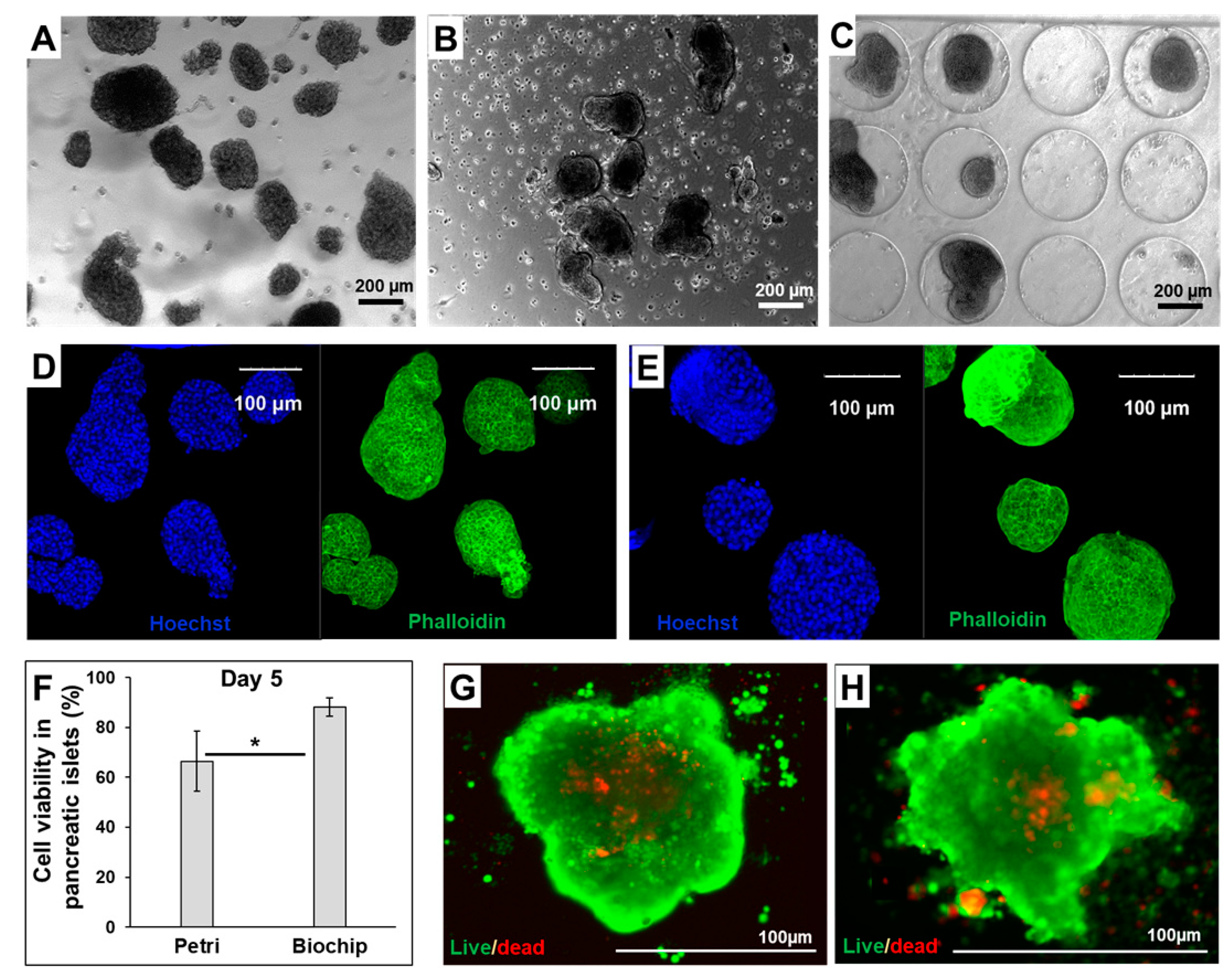
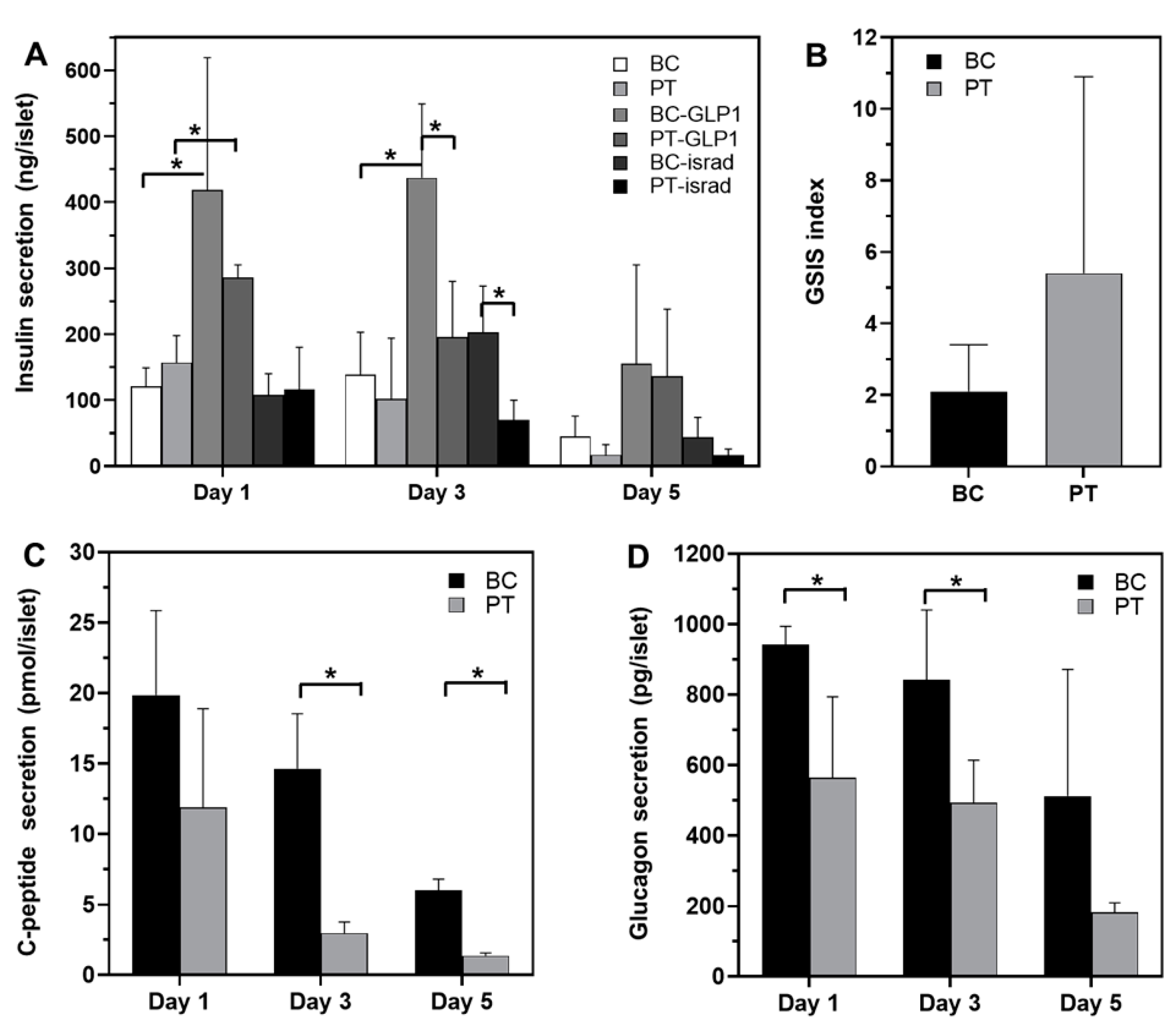
3.1. Morphology and Functionality of Pancreatic Islets in Petri and Biochip
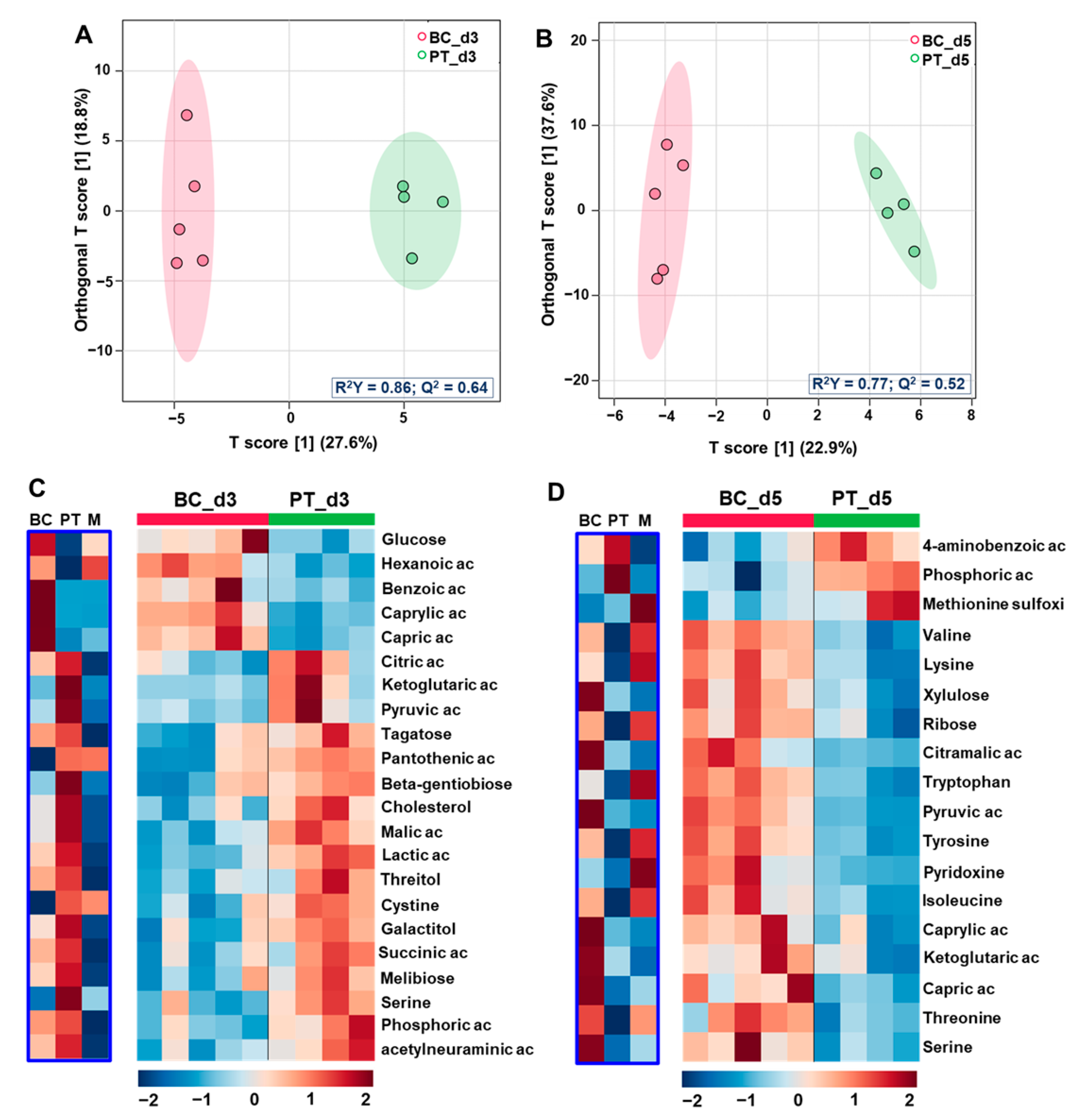
3.2. Comparison of the Exometabolomes of Islets Cultured in Petri and Biochip
3.2.1. Global Multivariate Analysis
3.2.2. Differential Analysis of the Petri vs. Biochip Culture Modes
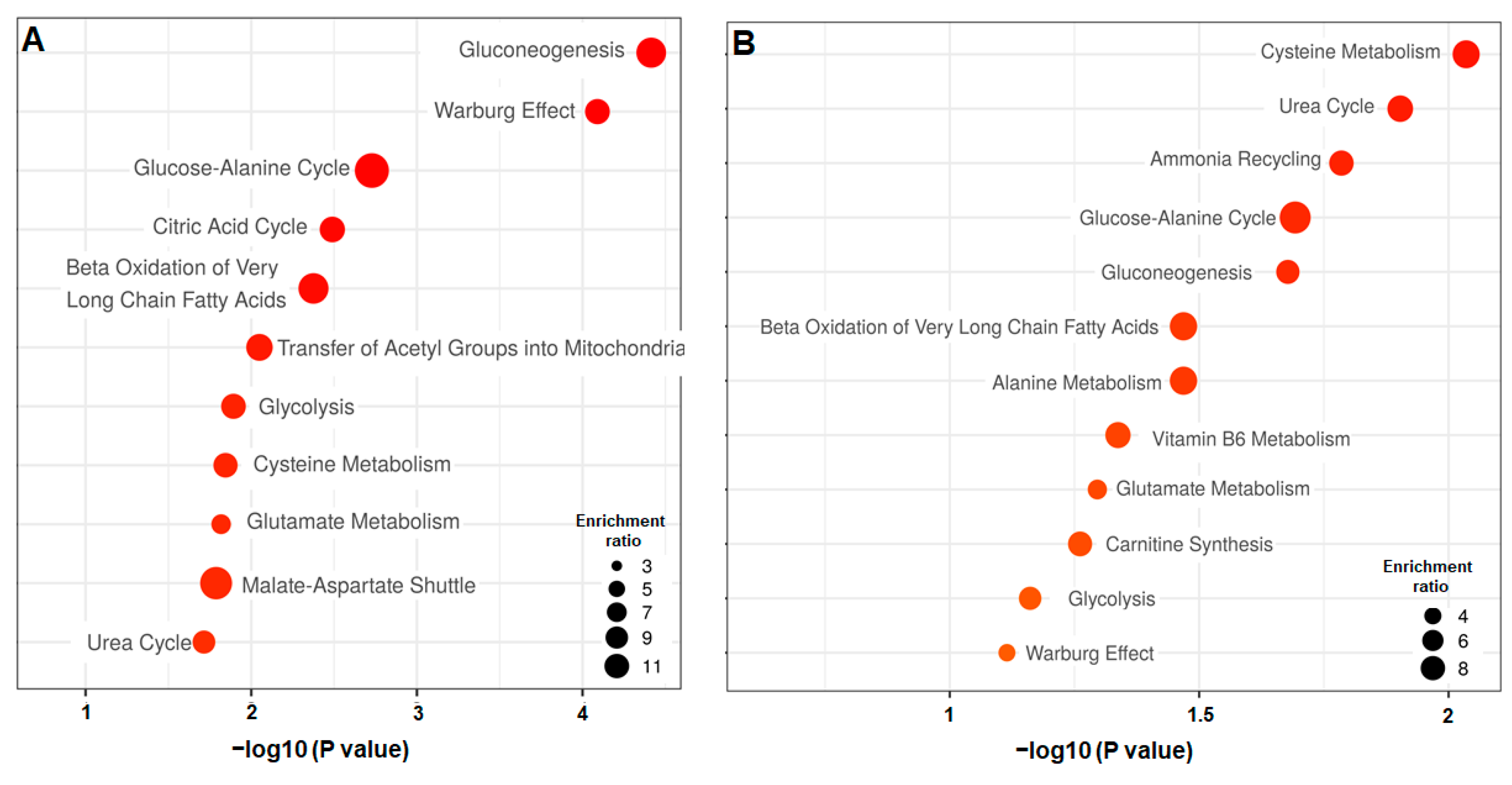
3.2.3. Differential Analysis between the Day 3 vs. Day 5
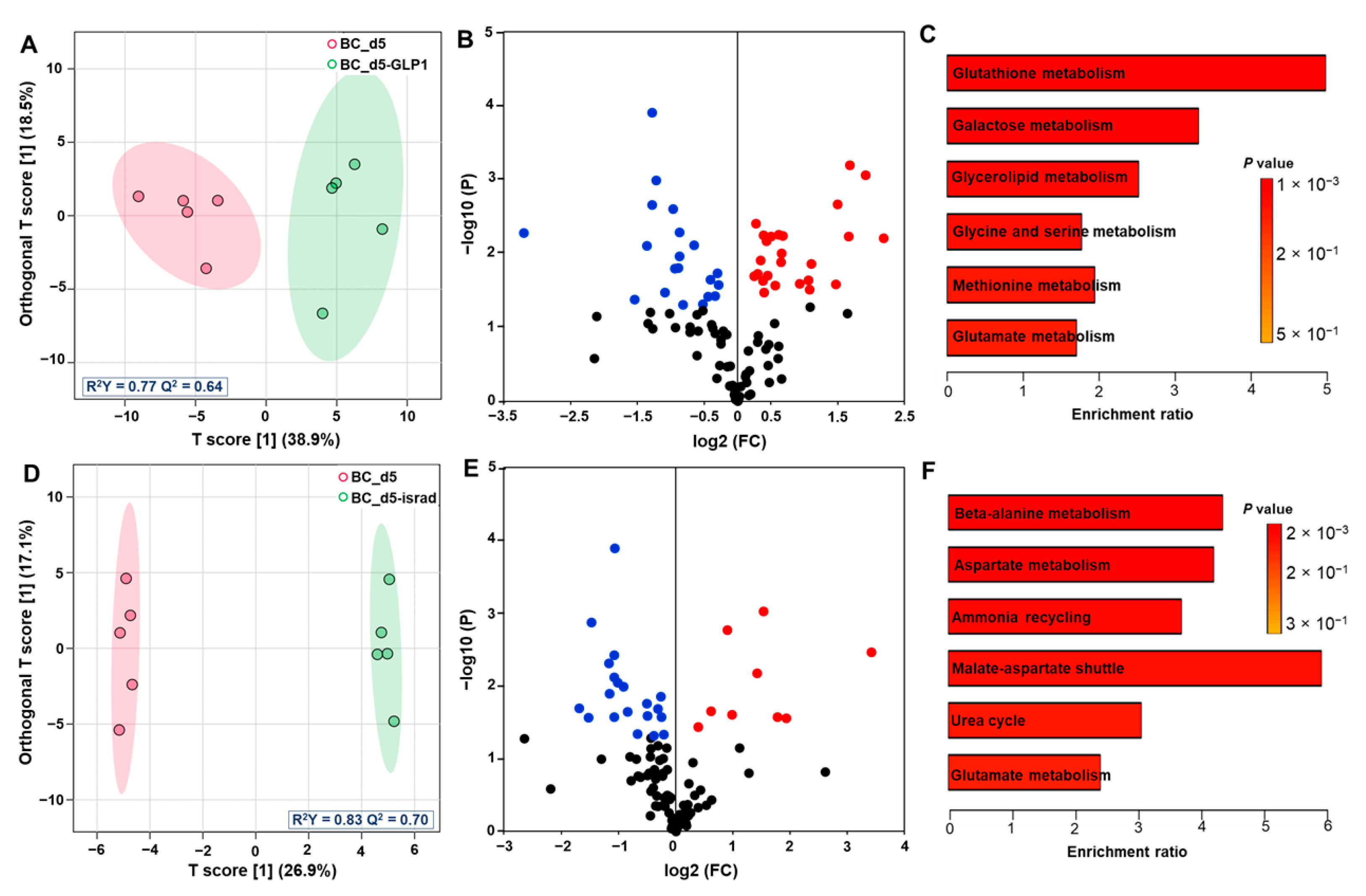
3.3. Effect of GLP1 Treatment on the Pancreas Metabolome
3.4. Effect of Isradipine Treatment on Pancreas Exometabolome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jouvet, N.; Estall, J.L. The pancreas: Bandmaster of glucose homeostasis. Exp. Cell Res. 2017, 360, 19–23. [Google Scholar] [CrossRef]
- Jellali, R.; Essaouiba, A.; Leclerc, E.; Legallais, C. Membrane bioreactors for bio-artificial pancreas. In Current Trends and Future Developments on (Bio-) Membranes; Basile, A., Annesini, M.C., Piemonte, V., Charcosset, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 77–108. [Google Scholar] [CrossRef]
- Sakhneny, L.; Khalifa-Malka, L.; Landsman, L. Pancreas organogenesis: Approaches to elucidate the role of epithelial-mesenchymal interactions. Semin. Cell Dev. Biol. 2019, 92, 89–96. [Google Scholar] [CrossRef]
- Egan, A.M.; Dinneen, S.F. What is diabetes? Medicine 2019, 47, 1–4. [Google Scholar] [CrossRef]
- Rogal, J.; Zbinden, A.; Schenke-Layland, K.; Loskill, P. Stem-cell based organ-on-a-chip models for diabetes research. Adv. Drug Deliv. Rev. 2019, 140, 101–128. [Google Scholar] [CrossRef] [PubMed]
- Punthakee, Z.; Goldenberg, R.; Katz, P. Definition, classification and diagnosis of diabetes, prediabetes and metabolic syndrome. Can. J. Diabetes 2018, 42, S10–S15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szunerits, S.; Melinte, S.; Barras, A.; Pagneux, Q.; Voronova, A.; Abderrahmani, A.; Boukherroub, R. The impact of chemical engineering and technological advances on managing diabetes: Present and future concepts. Chem. Soc. Rev. 2021, 50, 2102–2146. [Google Scholar] [CrossRef]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [Green Version]
- Harding, J.L.; Pavkov, M.E.; Magliano, D.J.; Shaw, J.E.; Gregg, E.W. Global trends in diabetes complications: A review of current evidence. Diabetologia 2019, 62, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Saeedi, P.; Salpea, P.; Karuranga, S.; Petersohn, I.; Malanda, B.; Gregg, E.W.; Unwin, N.; Wild, S.H.; Williams, R. Mortality attributable to diabetes in 20–79 years old adults, 2019 estimates: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2020, 162, 108086. [Google Scholar] [CrossRef]
- Zbinden, A.; Marzi, J.; Schlünder, K.; Probst, C.; Urbanczyk, M.; Black, S.; Brauchle, E.M.; Layland, S.L.; Kraushaar, U.; Duffy, G.; et al. Non-invasive marker-independent high content analysis of a microphysiological human pancreas-on-a-chip model. Matrix Biol. 2020, 85–86, 205–220. [Google Scholar] [CrossRef]
- King, A.; Bowe, J. Animal models for diabetes: Understanding the pathogenesis and finding new treatments. Biochem. Pharmacol. 2016, 99, 1–10. [Google Scholar] [CrossRef]
- Gao, B.; Wang, L.; Han, S.; Pingguan-Murphy, B.; Zhang, X.; Xu, F. Engineering of microscale three-dimensional pancreatic islet models in vitro and their biomedical applications. Crit. Rev. Biotechnol. 2016, 36, 619–629. [Google Scholar] [CrossRef]
- Sokolowska, P.; Janikiewicz, J.; Jastrzebska, E.; Brzozka, Z.; Dobrzyn, A. Combinations of regenerative medicine and Lab-on-a-chip systems: New hope to restoring the proper function of pancreatic islets in diabetes. Biosens. Bioelectron. 2020, 167, 112451. [Google Scholar] [CrossRef] [PubMed]
- Salg, G.A.; Giese, N.A.; Schenk, M.; Hüttner, F.J.; Felix, K.; Probst, P.; Diener, M.K.; Hackert, T.; Kenngott, H.G. The emerging field of pancreatic tissue engineering: A systematic review and evidence map of scaffold materials and scaffolding techniques for insulin-secreting cells. J. Tissue Eng. 2019, 10, 2041731419884708. [Google Scholar] [CrossRef]
- Lewis, P.L.; Wells, J.M. Engineering-inspired approaches to study β-cell function and diabetes. Stem Cells 2021, 39, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Messelmani, T.; Morisseau, L.; Sakai, Y.; Legallais, C.; Le Goff, A.; Leclerc, E.; Jellali, R. Liver organ-on-chip models for toxicity studies and risk assessment. Lab Chip 2022, 22, 2423–2450. [Google Scholar] [CrossRef] [PubMed]
- Merlier, F.; Jellali, R.; Leclerc, E. Online hepatic rat metabolism by coupling liver biochip and mass spectrometry. Analyst 2017, 142, 3747–3757. [Google Scholar] [CrossRef]
- Morisseau, L.; Messelmani, T.; Essaouiba, A.; Sakai, Y.; Le Goff, A.; Legallais, C.; Leclerc, E.; Jellali, R. Microfluidic and organ-on-a-chip-based technologies for diabetes therapy and research. In Nanotechnology for Diabetes Management; Abderrahmani, A., Szunerits, S., Boukerroub, R., El Ouaamari, A., Eds.; Royal Society of Chemistry: Cambridge, UK, 2022; pp. 188–232. [Google Scholar] [CrossRef]
- Sankar, K.S.; Green, B.J.; Crocker, A.R.; Verity, J.E.; Altamentova, S.M.; Rocheleau, J.V. Culturing pancreatic islets in microfluidic flow enhances morphology of the associated endothelial cells. PLoS ONE 2011, 6, e24904. [Google Scholar] [CrossRef]
- Li, X.; Brooks, J.C.; Hu, J.; Ford, K.I.; Easley, C.J. 3D-templated, fully automated microfluidic input/output multiplexer for endocrine tissue culture and secretion sampling. Lab Chip 2017, 17, 341–349. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Hong, S.; Song, J.; Cho, B.; Han, E.J.; Kondapavulur, S.; Kim, D.; Lee, L.P. Microphysiological analysis platform of pancreatic islet β-cell spheroids. Adv. Healthc. Mater. 2018, 7, 1701111. [Google Scholar] [CrossRef]
- Jun, Y.; Lee, J.; Choi, S.; Yang, J.H.; Sander, M.; Chung, S.; Lee, S.H. In vivo-mimicking microfluidic perfusion culture of pancreatic islet spheroids. Sci. Adv. 2019, 5, eaax4520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, T.; Wang, Y.; Chen, W.; Li, Z.; Su, W.; Guo, Y.; Deng, P.; Qin, J. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip 2019, 19, 948–958. [Google Scholar] [CrossRef]
- Schulze, T.; Mattern, K.; Erfle, P.; Brüning, D.; Scherneck, S.; Dietzel, A.; Rustenbeck, I. A parallel perifusion slide from glass for the functional and morphological analysis of pancreatic islets. Front. Bioeng. Biotechnol. 2021, 9, 615639. [Google Scholar] [CrossRef]
- Essaouiba, A.; Jellali, R.; Shinohara, M.; Scheidecker, B.; Legallais, C.; Sakai, Y.; Leclerc, E. Analysis of the behavior of 2D monolayers and 3D spheroid human pancreatic beta cells derived from induced pluripotent stem cells in a microfluidic environment. J. Biotechnol. 2021, 330, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Essaouiba, A.; Okitsu, T.; Jellali, R.; Shinohara, M.; Danoy, M.; Tauran, Y.; Legallais, C.; Sakai, Y.; Leclerc, E. Microwell-based pancreas-on-chip model enhances genes expression and functionality of rat islets of Langerhans. Mol. Cell Endocrinol. 2020, 514, 110892. [Google Scholar] [CrossRef] [PubMed]
- Jellali, R.; Bricks, T.; Jacques, S.; Fleury, M.J.; Paullier, P.; Merlier, F.; Leclerc, E. Long-term human primary hepatocyte cultures in a microfluidic liver biochip show maintenance of mRNA levels and higher drug metabolism compared with Petri cultures. Biopharm. Drug. Dispos. 2016, 37, 264–275. [Google Scholar] [CrossRef]
- Essaouiba, A.; Okitsu, T.; Kinoshita, R.; Jellali, R.; Shinohara, M.; Danoy, M.; Legallais, C.; Sakai, Y.; Leclerc, E. Development of a pancreas-liver organ-on-chip coculture model for organ-to-organ interaction studies. Biochem. Eng. J. 2020, 164, 107783. [Google Scholar] [CrossRef]
- Gu, W.; Tong, Z.; Zhong, T. Clinical application of metabolomics in pancreatic diseases: A mini-review. J. Clin. Chem. Lab. Med. 2020, 2, 1000122. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; von Bergen, M.; Buesen, R.; Hackermüller, J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef] [Green Version]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.; Hurton, S.; Ayloo, S.; Molinari, M. Advances in pancreatic cancer: The role of metabolomics. J. Pancreas 2015, 16, 244–248. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for biomarker discovery: Moving to the clinic. Biomed. Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef] [Green Version]
- Bain, J.; Stevens, R.; Wenner, B.; Ilkayeva, O.; Muoio, D.; Newgard, C. Metabolomics applied to diabetes research, moving from information to knowledge. Diabetes 2009, 58, 2429–2443. [Google Scholar] [CrossRef] [Green Version]
- Gooding, J.R.; Jensen, M.V.; Newgard, C.B. Metabolomics applied to the pancreatic islet. Arch. Biochem. Biophys. 2016, 589, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Mehta, K.; Wu, H.J.; Menon, S.; Fallah, Y.; Zhong, X.; Rizk, N.; Unger, K.; Mapstone, M.; Fiandaca, M.; Federoff, H.; et al. Metabolomic biomarkers of pancreatic cancer: A meta-analysis study. Oncotarget 2017, 15, 68899–68915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choucha Snouber, L.; Bunescu, A.; Legallais, C.; Brochot, C.; Dumas, M.E.; Elena-Herrmann, B.; Leclerc, E. Metabolomics-on-a-chip of hepatotoxicity induced by anticancer drug flutamide and its active metabolite hydroxyflutamide using HepG2/C3a microfluidic biochips. Toxicol. Sci. 2013, 132, 8–20. [Google Scholar] [CrossRef]
- Jellali, R.; Gilard, F.; Pandolfi, V.; Legendre, A.; Fleury, M.J.; Paullier, P.; Legallais, C.; Leclerc, E. Metabolomics-on-a-chip approach to study hepatotoxicity of DDT, permethrin and their mixtures. J. Appl. Toxicol. 2018, 38, 1121–1134. [Google Scholar] [CrossRef]
- Harden, S.L.; Zhou, J.; Gharanei, S.; Diniz-da-Costa, M.; Lucas, E.S.; Cui, L.; Murakami, K.; Fang, J.; Chen, Q.; Brosens, J.J.; et al. Exometabolomic analysis of decidualizing human endometrial stromal and perivascular cells. Front. Cell Dev. Biol. 2021, 9, 626619. [Google Scholar] [CrossRef]
- Balcerczyk, A.; Damblon, C.; Elena-Herrmann, B.; Panthu, B.; Rautureau, G.J.P. Metabolomic approaches to study chemical exposure-related metabolism alterations in mammalian cell cultures. Int. J. Mol. Sci. 2020, 21, 6843. [Google Scholar] [CrossRef] [PubMed]
- Klauser, R.; Prager, R.; Gaube, S.; Gisinger, C.; Schnack, C.; Küenburg, E.; Schernthaner, G. Metabolic effects of isradipine versus hydrochlorothiazide in diabetes mellitus. Hypertension 1991, 17, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Rorsman, P.; Ashcroft, F.M. Pancreatic β-cell electrical activity and insulin secretion: Of mice and men. Physiol. Rev. 2018, 98, 117–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, M.; Ramracheya, R.; Bengtsson, M.; Zhang, Q.; Karanauskaite, J.; Partridge, C.; Johnson, P.R.; Rorsman, P. Voltage-gated ion channels in human pancreatic beta-cells: Electrophysiological characterization and role in insulin secretion. Diabetes 2008, 57, 1618–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, J.D.; Dula, S.B.; Corbin, K.L.; Wu, R.; Nunemaker, C.S. A practical guide to rodent islet isolation and assessment. Biol. Proced. Online 2009, 11, 3–31. [Google Scholar] [CrossRef] [Green Version]
- Yonekawa, Y.; Okitsu, T.; Wake, K.; Iwanaga, Y.; Noguchi, H.; Nagata, H.; Liu, X.; Kobayashi, N.; Matsumoto, N. A new mouse model for intraportal islet transplantation with limited hepatic lobe as a graft site. Transplantation 2006, 82, 712–715. [Google Scholar] [CrossRef]
- Kiba, T.; Tanemura, M.; Yagyu, K. High-quality RNA extraction from rat pancreatic islet. Cell Biol. Int. Rep. 2013, 20, 1–4. [Google Scholar] [CrossRef]
- Jellali, R.; Poulain, S.; Lereau-Bernier, M.; Gilard, F.; Tauran, Y.; Kato, S.; Danoy, M.; Segard, B.D.; Kido, T.; Miyajima, A.; et al. Integration of metabolomic and transcriptomic profiling to compare two protocols of differentiation of human induced pluripotent stem cells into hepatocytes. Process Biochem. 2020, 88, 138–147. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Tran, R.; Moraes, C.; Hoesli, C.A. Developmentally-inspired biomimetic culture models to produce functional islet-like cells from pluripotent precursors. Front. Bioeng. Biotechnol. 2020, 8, 583970. [Google Scholar] [CrossRef]
- Chotard-Ghodsnia, R.; Drochon, A.; Faucheux, N.; Nagel, M.D.; Grebe, R. Effect of shear stress and of transmural pressure on cAMP-dependent responses of cells adhering to a biomaterial. Eur. Phys. J. AP 2002, 17, 155–162. [Google Scholar] [CrossRef]
- Silva, P.N.; Green, B.J.; Altamentova, S.M.; Rocheleau, J.V. A microfluidic device designed to induce media flow throughout pancreatic islets while limiting shear-induced damage. Lab Chip 2013, 13, 4374–4384. [Google Scholar] [CrossRef]
- Costes, S.; Bertrand, G.; Ravier, M.A. Mechanisms of Beta-cell apoptosis in type 2 diabetes-prone situations and potential protection by GLP-1-based therapies. Int. J. Mol. Sci. 2021, 22, 5303. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, N.; Vaziri, N.D.; Dafoe, D.C.; Ichii, H. The role of oxidative stress in pancreatic β cell dysfunction in diabetes. Int. J. Mol. Sci. 2021, 22, 1509. [Google Scholar] [CrossRef] [PubMed]
- Evans-Molina, C.; Garmey, J.C.; Ketchum, R.; Brayman, K.L.; Deng, S.; Mirmira, R.G. Glucose regulation of insulin gene transcription and pre-mRNA processing in human islets. Diabetes 2007, 56, 827–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreozzi, F.; D’Alessandris, C.; Federici, M.; Laratta, E.; Del Guerra, S.; Del Prato, S.; Marchetti, P.; Lauro, R.; Perticone, F.; Sesti, G. Activation of the hexosamine pathway leads to phosphorylation of insulin receptor substrate-1 on Ser307 and Ser612 and impairs the phosphatidylinositol 3 kinase/Akt/mammalian target of rapamycin insulin biosynthetic pathway in RIN pancreatic beta-cells. Endocrinology 2004, 145, 2845–2857. [Google Scholar] [CrossRef]
- Swierczynski, J.; Hebanowska, A.; Sledzinski, T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J. Gastroenterol. 2014, 20, 2279–2303. [Google Scholar] [CrossRef]
- Ribas, V.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondria, cholesterol and cancer cell metabolism. Clin. Transl. Med. 2016, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Sunami, Y.; Rebelo, A.; Kleeff, J. Lipid metabolism and lipid droplets in pancreatic cancer and stellate cells. Cancers 2017, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Yost, T.J.; Eckel, R. Hypocaloric feeding in obese women: Metabolic effects of medium-chain triglyceride substitution. Am. J. Clin. Nutr. 1989, 49, 326–330. [Google Scholar] [CrossRef] [Green Version]
- Sung, M.H.; Liao, F.H.; Chien, Y.W. Medium-chain triglycerides lower blood lipids and body weight in streptozotocin-induced type 2 diabetes rats. Nutrients 2018, 10, 963. [Google Scholar] [CrossRef] [Green Version]
- Murata, Y.; Harada, N.; Yamane, S.; Iwasaki, K.; Ikeguchi, E.; Kanemaru, Y.; Harada, T.; Sankoda, A.; Shimazu-Kuwahara, S.; Joo, E.; et al. Medium-chain triglyceride diet stimulates less GIP secretion and suppresses body weight and fat mass gain compared with long-chain triglyceride diet. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E53–E64. [Google Scholar] [CrossRef]
- Sugiyama, C.; Akter, K.N.; Morishita, S.; Miura, A.; Takase, S. Effects of medium-chain triglycerides on gluconeogenesis and ureagenesis in weaned rats fed a high fat diet. J. Nutr. Intermed. Metab. 2015, 2, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Cornell, S. A review of GLP-1 receptor agonists in type 2 diabetes: A focus on the mechanism of action of once-weekly agents. J. Clin. Pharm. Ther. 2020, 45, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Molnos, S.; Wahl, S.; Haid, M.; Eekhoff, E.; Pool, R.; Floegel, A.; Deelen, J.; Much, D.; Prehn, C.; Breier, M.; et al. Metabolite ratios as potential biomarkers for type 2 diabetes: A DIRECT study. Diabetologia 2018, 61, 117–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, M.V.; Gooding, J.R.; Ferdaoussi, M.; Dai, X.Q.; Peterson, B.S.; MacDonald, P.E.; Newgard, C.B. Metabolomics applied to islet nutrient sensing mechanisms. Diabetes Obes. Metab. 2017, 19, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Connell, N.J.; Houtkooper, R.H.; Schrauwen, P. NAD+ metabolism as a target for metabolic health: Have we found the silver bullet? Diabetologia 2019, 62, 888–899. [Google Scholar] [CrossRef] [Green Version]
- Ellis, J.; Athersuch, T.; Cavill, R.; Radford, R.; Slattery, C.; Jennings, P.; McMorrow, T.; Ryan, M.; Ebbels, T.; Keun, H. Metabolic response to low-level toxicant exposure in a novel renal tubule epithelial cell system. Mol. BioSyst. 2011, 7, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Okitsu, T.; Xibao, L.; Teramae, H.; Okonogi, A.; Toyoda, K.; Uemoto, S.; Fukushima, M. Improved hypothermic short-term storage of isolated mouse islets by adding serum to preservation solutions. Islets 2013, 5, 45–52. [Google Scholar] [CrossRef]
- Kneteman, N.M.; Warnock, G.L.; Evans, M.G.; Dawidson, I.; Rajotte, R.V. Islet isolation from human pancreas stored in UW solution for 6 to 26 hours. Transplant. Proc. 1990, 22, 763–764. [Google Scholar]


| Metabolites | |
|---|---|
| BC_d5-GLP1 vs. BC_d5 | Trehalose, galactonic ac, Beta-gentiobiose, n-acetylneuraminic acid, galactitol, melibiose, cysteinylglycine, trans-4-hydroxyproline, methionine sulfoxide, trans-13-octadecenoic acid, glycerol 1-phosphate, glucosaminic acid, oleic acid, kynurenine, hippuric acid, xanthine, mannose (gluconic ac lactone), arabinose, glyceric acid, 4-aminobenzoic acid Fructose, myo-inositol, serine, tryptophan, ribose, isoleucine, lysine, tyrosine, norvaline, xylulose, histidine, L-pyroglutamic acid, glycine, glutamine, pyridoxine, creatinine, proline, oxalic acid, alanine, leucine, methionine, valine, glycerol, glutaric acid, cysteine |
| PT_d5-GLP1 vs. PT_d5 | Threitol, hypoxanthine, tagatose, citramalic acid, serine, fructose, pyridoxine, Beta-hydroxyisovalerate, tyrosine, tryptophan, pyruvic ac |
| BC_d5-israd vs. BC_d5 | Uridine, arachidic acid, trans-4-hydroxyproline, aspartic acid, oleic acid, stearic acid, phenylalanine, mannose (gluconic ac lactone), Beta-alanine, trans-13-octadecenoic acid, palmitic acid, eicosapentaenoic acid, pantothenic acid, kynurenine, xanthine, galacturonic acid, sorbitol, citric acid, glyceric acid, uracil Glutamine, glutamic acid, 2-phenylacetamide, asparagine, oxalic acid, methionine, valine, proline, cysteine, nicotinic acid |
| PT_d5-israd vs. PT_d5 | Benzoic ac Glutaric ac |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Essaouiba, A.; Jellali, R.; Gilard, F.; Gakière, B.; Okitsu, T.; Legallais, C.; Sakai, Y.; Leclerc, E. Investigation of the Exometabolomic Profiles of Rat Islets of Langerhans Cultured in Microfluidic Biochip. Metabolites 2022, 12, 1270. https://doi.org/10.3390/metabo12121270
Essaouiba A, Jellali R, Gilard F, Gakière B, Okitsu T, Legallais C, Sakai Y, Leclerc E. Investigation of the Exometabolomic Profiles of Rat Islets of Langerhans Cultured in Microfluidic Biochip. Metabolites. 2022; 12(12):1270. https://doi.org/10.3390/metabo12121270
Chicago/Turabian StyleEssaouiba, Amal, Rachid Jellali, Françoise Gilard, Bertrand Gakière, Teru Okitsu, Cécile Legallais, Yasuyuki Sakai, and Eric Leclerc. 2022. "Investigation of the Exometabolomic Profiles of Rat Islets of Langerhans Cultured in Microfluidic Biochip" Metabolites 12, no. 12: 1270. https://doi.org/10.3390/metabo12121270
APA StyleEssaouiba, A., Jellali, R., Gilard, F., Gakière, B., Okitsu, T., Legallais, C., Sakai, Y., & Leclerc, E. (2022). Investigation of the Exometabolomic Profiles of Rat Islets of Langerhans Cultured in Microfluidic Biochip. Metabolites, 12(12), 1270. https://doi.org/10.3390/metabo12121270