The Glycosyltransferase Pathway: An Integrated Analysis of the Cell Metabolome
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Metabolomics
2.2. UGT Enzymatic Assays
2.3. Fatty Acid Synthesis Assay
2.4. Gene Expression Analysis
2.5. Co-Immunoprecipitation
3. Results
3.1. Validation of Catalytic Properties of Ectopically Expressed UGT Enzymes and Alt. Proteins
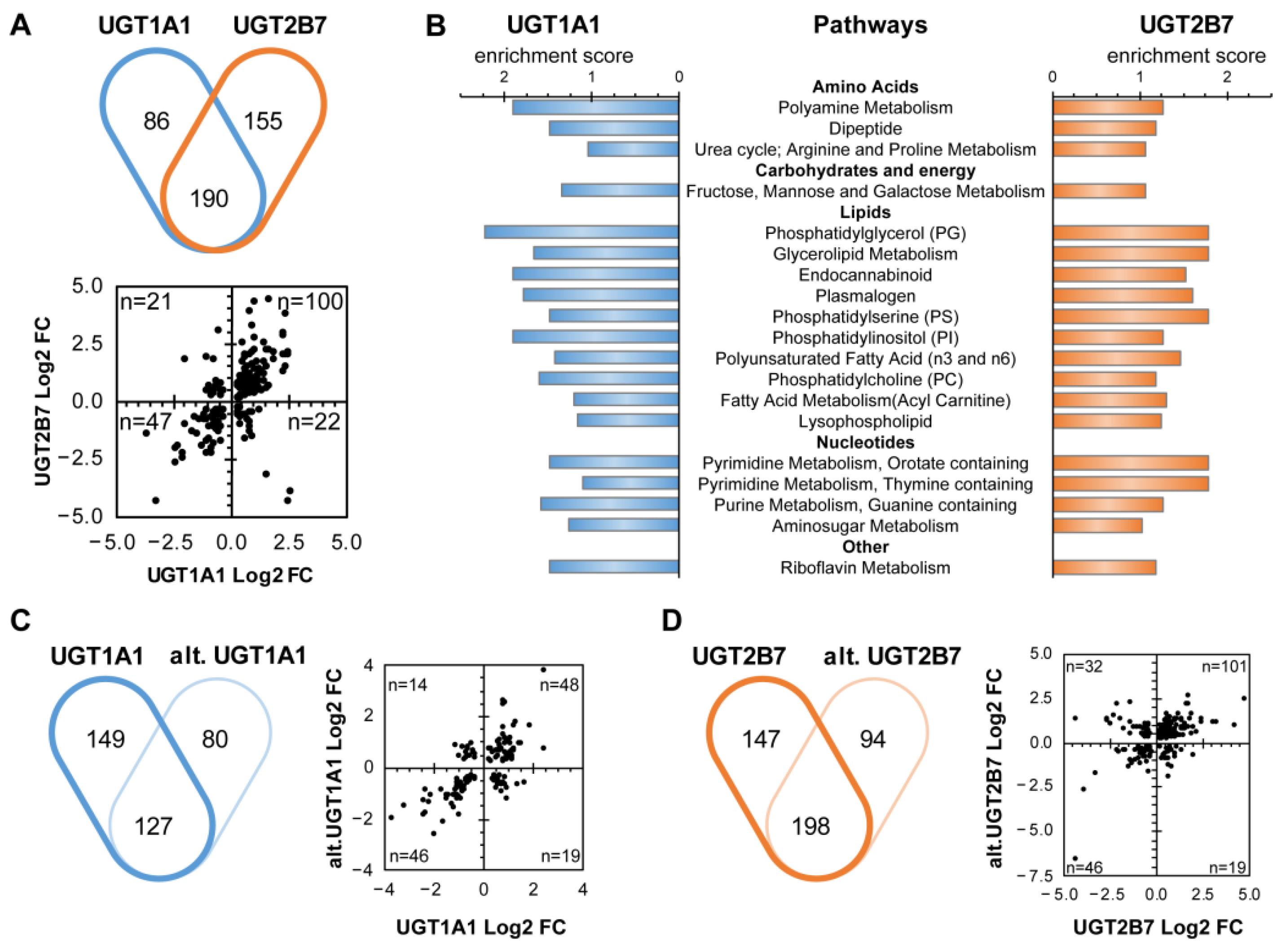
3.2. The Cellular Metabolome Is Broadly Affected by the Expression of Canonical and Alt. UGT Proteins
3.3. Nucleotide Sugar Precursors Are Significantly Altered by UGT Enzymes and Alt. UGT Proteins
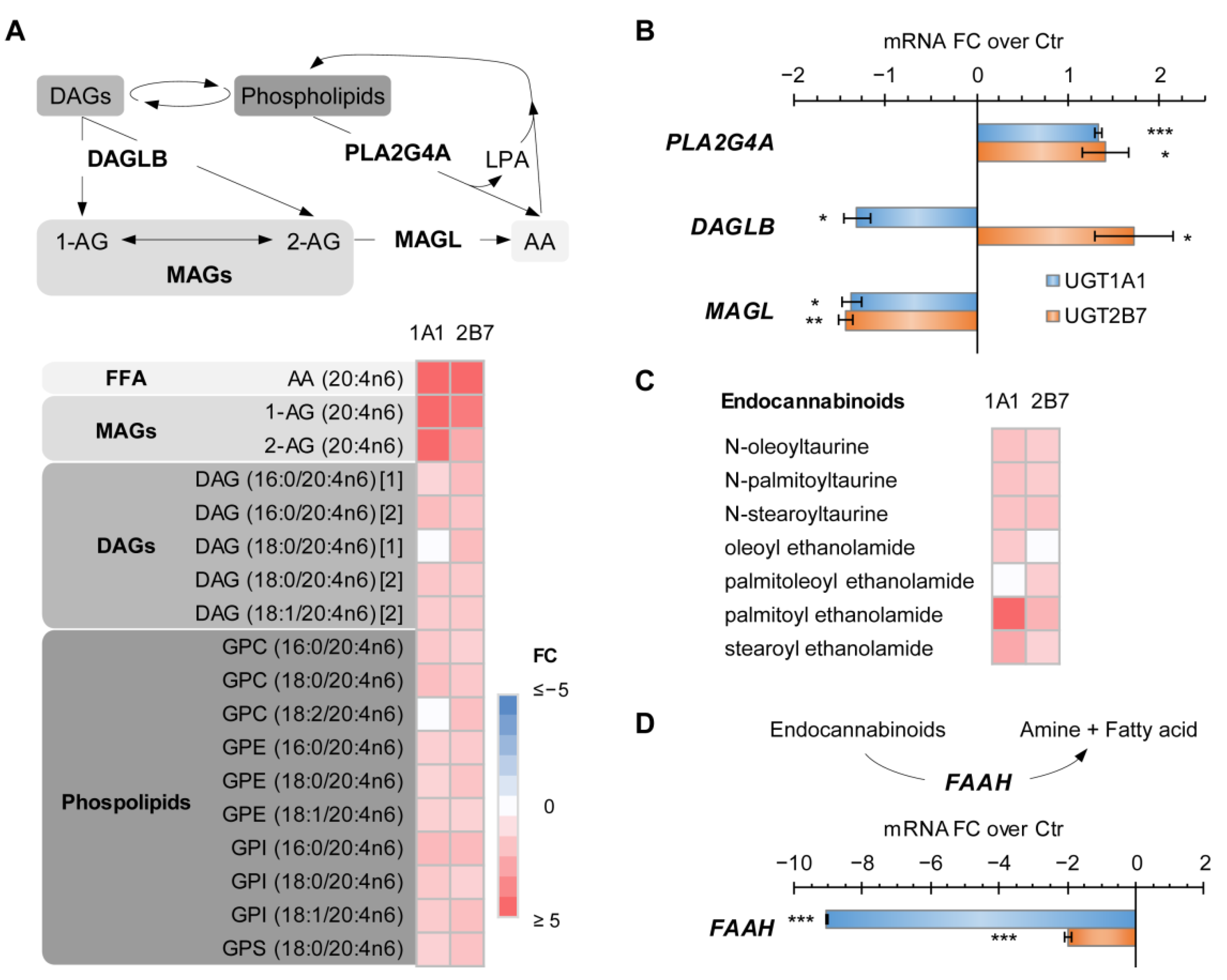
3.4. UGT1A1 and UGT2B7 Enzymes Strongly Affect the Lipidome and Lipid Metabolism-Related Gene Expression
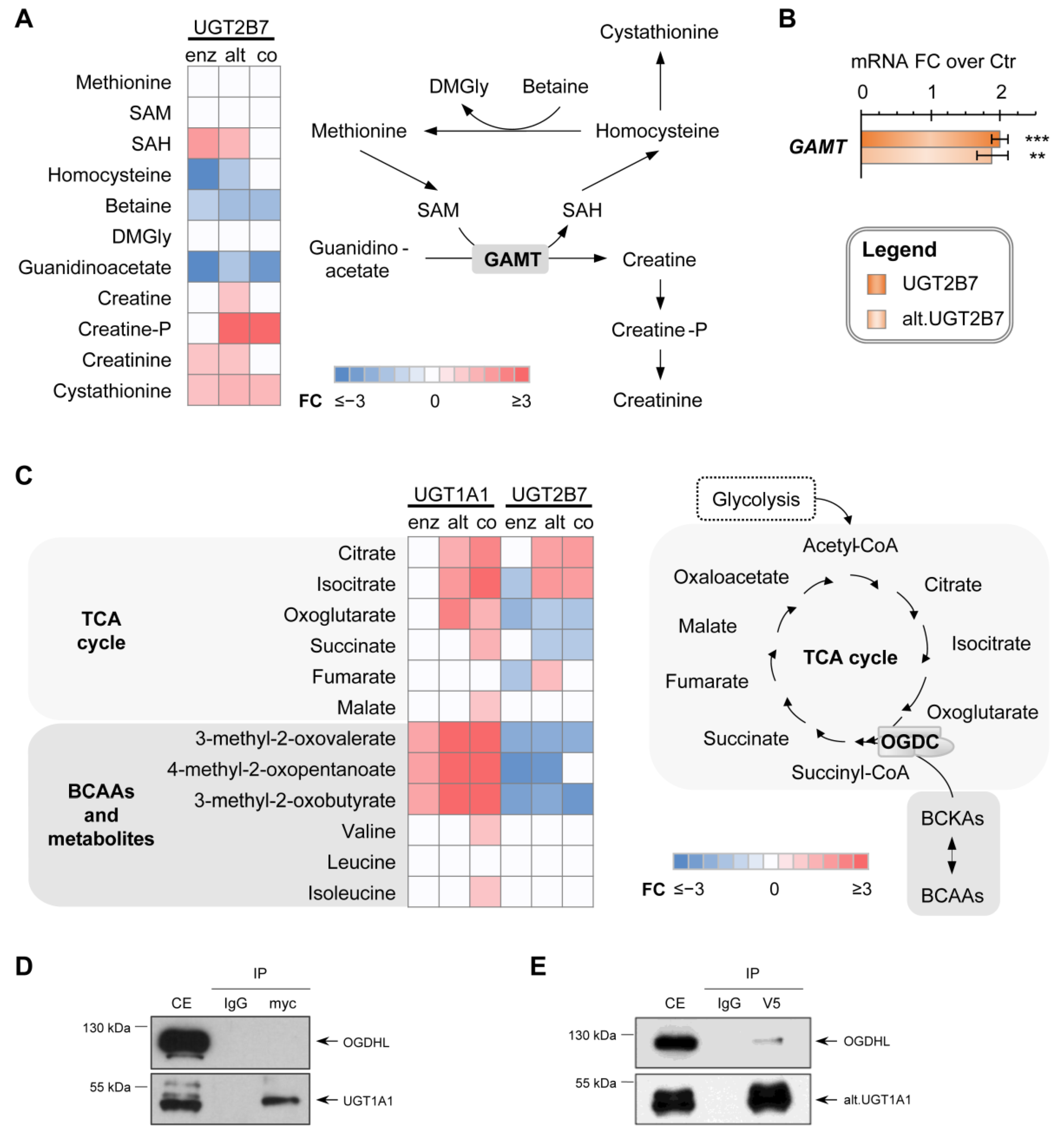
3.5. UGT2B7 Isoforms Affect the Methionine-Creatinine Pathway
3.6. The Metabolic Profile of Alt. UGT Support a Functional Protein–Protein Interaction Leading to Altered Mitochondrial Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guillemette, C.; Levesque, É.; Rouleau, M. Pharmacogenomics of human uridine diphospho-glucuronosyltransferases and clinical implications. Clin. Pharm. Ther. 2014, 96, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef] [PubMed]
- Allain, E.P.; Rouleau, M.; Le, T.; Vanura, K.; Villeneuve, L.; Caron, P.; Turcotte, V.; Levesque, E.; Guillemette, C. Inactivation of Prostaglandin E2 as a Mechanism for UGT2B17-Mediated Adverse Effects in Chronic Lymphocytic Leukemia. Front. Oncol. 2019, 9, 606. [Google Scholar] [CrossRef]
- King, C.D.; Rios, G.R.; Assouline, J.A.; Tephly, T.R. Expression of UDP-glucuronosyltransferases (UGTs) 2B7 and 1A6 in the human brain and identification of 5-hydroxytryptamine as a substrate. Arch. Biochem. Biophys. 1999, 365, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Little, J.M.; Kurkela, M.; Sonka, J.; Jantti, S.; Ketola, R.; Bratton, S.; Finel, M.; Radominska-Pandya, A. Glucuronidation of oxidized fatty acids and prostaglandins B1 and E2 by human hepatic and recombinant UDP-glucuronosyltransferases. J. Lipid Res. 2004, 45, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Battistone, M.A.; Mendelsohn, A.C.; Spallanzani, R.G.; Allegretti, A.S.; Liberman, R.N.; Sesma, J.; Kalim, S.; Wall, S.M.; Bonventre, J.V.; Lazarowski, E.R.; et al. Proinflammatory P2Y14 receptor inhibition protects against ischemic acute kidney injury in mice. J. Clin. Investig. 2020, 130, 3734–3749. [Google Scholar] [CrossRef] [PubMed]
- Mederacke, I.; Filliol, A.; Affo, S.; Nair, A.; Hernandez, C.; Sun, Q.; Hamberger, F.; Brundu, F.; Chen, Y.; Ravichandra, A.; et al. The purinergic P2Y14 receptor links hepatocyte death to hepatic stellate cell activation and fibrogenesis in the liver. Sci. Transl. Med. 2022, 14, eabe5795. [Google Scholar] [CrossRef]
- Bock, K.W. Roles of human UDP-glucuronosyltransferases in clearance and homeostasis of endogenous substrates, and functional implications. Biochem. Pharmacol. 2015, 96, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Meech, R.; McKinnon, R.A.; Mackenzie, P.I. Transcriptional regulation of human UDP-glucuronosyltransferase genes. Drug Metab. Rev. 2014, 46, 421–458. [Google Scholar] [CrossRef] [PubMed]
- Audet-Delage, Y.; Rouleau, M.; Rouleau, M.; Roberge, J.; Miard, S.; Picard, F.; Têtu, B.; Guillemette, C. Cross-Talk between Alternatively Spliced UGT1A Isoforms and Colon Cancer Cell Metabolism. Mol. Pharmacol. 2017, 91, 167–177. [Google Scholar] [CrossRef]
- Rouleau, M.; Roberge, J.; Bellemare, J.; Guillemette, C. Dual roles for splice variants of the glucuronidation pathway as regulators of cellular metabolism. Mol. Pharmacol. 2014, 85, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, M.; Tourancheau, A.; Girard-Bock, C.; Villeneuve, L.; Vaucher, J.; Duperre, A.M.; Audet-Delage, Y.; Gilbert, I.; Popa, I.; Droit, A.; et al. Divergent Expression and Metabolic Functions of Human Glucuronosyltransferases through Alternative Splicing. Cell Rep. 2016, 17, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.F.; Bernard, O.; Villeneuve, L.; Têtu, B.; Guillemette, C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clin. Cancer Res. 2006, 12, 1850–1858. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, E.; Labriet, A.; Hovington, H.; Allain, E.P.; Melo-Garcia, L.; Rouleau, M.; Brisson, H.; Turcotte, V.; Caron, P.; Villeneuve, L.; et al. Alternative promoters control UGT2B17-dependent androgen catabolism in prostate cancer and its influence on progression. Br. J. Cancer 2020, 122, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Riches, Z.; Collier, A.C. Posttranscriptional regulation of uridine diphosphate glucuronosyltransferases. Expert Opin. Drug Metab. Toxicol. 2015, 11, 949–965. [Google Scholar] [CrossRef] [PubMed]
- Émond, J.P.; Labriet, A.; Desjardins, S.; Rouleau, M.; Villeneuve, L.; Hovington, H.; Brisson, H.; Lacombe, L.; Simonyan, D.; Caron, P.; et al. Factors Affecting Interindividual Variability of Hepatic UGT2B17 Protein Expression Examined Using a Novel Specific Monoclonal Antibody. Drug Metab. Dispos. 2019, 47, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Girard, H.; Lévesque, E.; Bellemare, J.; Journault, K.; Caillier, B.; Guillemette, C. Genetic diversity at the UGT1 locus is amplified by a novel 3′ alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenet. Genom. 2007, 17, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, E.; Girard, H.; Journault, K.; Lepine, J.; Guillemette, C. Regulation of the UGT1A1 bilirubin-conjugating pathway: Role of a new splicing event at the UGT1A locus. Hepatology 2007, 45, 128–138. [Google Scholar] [CrossRef]
- Menard, V.; Collin, P.; Margaillan, G.; Guillemette, C. Modulation of the UGT2B7 enzyme activity by C-terminally truncated proteins derived from alternative splicing. Drug Metab. Dispos. 2013, 41, 2197–2205. [Google Scholar] [CrossRef] [PubMed]
- Tourancheau, A.; Margaillan, G.; Rouleau, M.; Gilbert, I.; Villeneuve, L.; Lévesque, E.; Droit, A.; Guillemette, C. Unravelling the transcriptomic landscape of the major phase II UDP-glucuronosyltransferase drug metabolizing pathway using targeted RNA sequencing. Pharm. J. 2016, 16, 60–70. [Google Scholar] [CrossRef][Green Version]
- Hu, D.G.; Marri, S.; Mackenzie, P.I.; Hulin, J.A.; McKinnon, R.A.; Meech, R. The Expression Profiles and Deregulation of UDP-Glycosyltransferase (UGT) Genes in Human Cancers and Their Association with Clinical Outcomes. Cancers 2021, 13, 4491. [Google Scholar] [CrossRef] [PubMed]
- Tourancheau, A.; Rouleau, M.; Guauque-Olarte, S.; Villeneuve, L.; Gilbert, I.; Droit, A.; Guillemette, C. Quantitative profiling of the UGT transcriptome in human drug-metabolizing tissues. Pharm. J. 2018, 18, 251–261. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Dong, W.; Qu, X.; Huang, C.; Yan, T.; Du, J. Combination of hesperetin and platinum enhances anticancer effect on lung adenocarcinoma. Biomed. Pharmacother. 2019, 113, 108779. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Du, M.; Ge, Y.; Gao, F.; Xin, J.; Lv, Q.; Qin, C.; Zhu, Y.; Gu, C.; Wang, M.; et al. Identification of low-frequency variants of UGT1A3 associated with bladder cancer risk by next-generation sequencing. Oncogene 2021, 40, 2382–2394. [Google Scholar] [CrossRef]
- Zahreddine, H.A.; Culjkovic-Kraljacic, B.; Assouline, S.; Gendron, P.; Romeo, A.A.; Morris, S.J.; Cormack, G.; Jaquith, J.B.; Cerchietti, L.; Cocolakis, E.; et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014, 511, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Weh, K.M.; Howard, C.L.; Riethoven, J.J.; Clarke, J.L.; Lagisetty, K.H.; Lin, J.; Reddy, R.M.; Chang, A.C.; Beer, D.G.; et al. Characterizing isoform switching events in esophageal adenocarcinoma. Mol. Ther. Nucleic Acids 2022, 29, 749–768. [Google Scholar] [CrossRef]
- Rosario, S.R.; Long, M.D.; Affronti, H.C.; Rowsam, A.M.; Eng, K.H.; Smiraglia, D.J. Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat. Commun. 2018, 9, 5330. [Google Scholar] [CrossRef] [PubMed]
- Allain, E.P.; Rouleau, M.; Levesque, E.; Guillemette, C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br. J. Cancer 2020, 122, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Allain, E.P.; Rouleau, M.; Vanura, K.; Tremblay, S.; Vaillancourt, J.; Bat, V.; Caron, P.; Villeneuve, L.; Labriet, A.; Turcotte, V.; et al. UGT2B17 modifies drug response in chronic lymphocytic leukaemia. Br. J. Cancer 2020, 123, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Belledant, A.; Hovington, H.; Garcia, L.; Caron, P.; Brisson, H.; Villeneuve, L.; Simonyan, D.; Têtu, B.; Fradet, Y.; Lacombe, L.; et al. The UGT2B28 Sex-steroid Inactivation Pathway Is a Regulator of Steroidogenesis and Modifies the Risk of Prostate Cancer Progression. Eur. Urol. 2016, 69, 601–609. [Google Scholar] [CrossRef]
- Bhoi, S.; Baliakas, P.; Cortese, D.; Mattsson, M.; Engvall, M.; Smedby, K.E.; Juliusson, G.; Sutton, L.A.; Mansouri, L. UGT2B17 expression: A novel prognostic marker within IGHV-mutated chronic lymphocytic leukemia? Haematologica 2016, 101, e63–e65. [Google Scholar] [CrossRef] [PubMed]
- Gruber, M.; Bellemare, J.; Hoermann, G.; Gleiss, A.; Porpaczy, E.; Bilban, M.; Le, T.; Zehetmayer, S.; Mannhalter, C.; Gaiger, A.; et al. Overexpression of uridine diphospho glucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood 2013, 121, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef]
- Lepine, J.; Bernard, O.; Plante, M.; Têtu, B.; Pelletier, G.; Labrie, F.; Belanger, A.; Guillemette, C. Specificity and regioselectivity of the conjugation of estradiol, estrone, and their catecholestrogen and methoxyestrogen metabolites by human uridine diphospho-glucuronosyltransferases expressed in endometrium. J. Clin. Endocrinol. Metab. 2004, 89, 5222–5232. [Google Scholar] [CrossRef]
- Belanger, A.S.; Caron, P.; Harvey, M.; Zimmerman, P.A.; Mehlotra, R.K.; Guillemette, C. Glucuronidation of the antiretroviral drug efavirenz by UGT2B7 and an in vitro investigation of drug-drug interaction with zidovudine. Drug Metab. Dispos. 2009, 37, 1793–1796. [Google Scholar] [CrossRef] [PubMed]
- Gagne, J.F.; Montminy, V.; Belanger, P.; Journault, K.; Gaucher, G.; Guillemette, C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol. Pharmacol. 2002, 62, 608–617. [Google Scholar] [CrossRef]
- Vazquez-Martin, A.; Colomer, R.; Brunet, J.; Lupu, R.; Menendez, J.A. Overexpression of fatty acid synthase gene activates HER1/HER2 tyrosine kinase receptors in human breast epithelial cells. Cell Prolif. 2008, 41, 59–85. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, M.; Audet-Delage, Y.; Desjardins, S.; Rouleau, M.; Girard-Bock, C.; Guillemette, C. Endogenous Protein Interactome of Human UDP-Glucuronosyltransferases Exposed by Untargeted Proteomics. Front. Pharmacol. 2017, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Bray, B.J.; Rosengren, R.J. Retinol potentiates acetaminophen-induced hepatotoxicity in the mouse: Mechanistic studies. Toxicol. Appl. Pharmacol. 2001, 173, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Gregus, Z.; Madhu, C.; Goon, D.; Klaassen, C.D. Effect of galactosamine-induced hepatic UDP-glucuronic acid depletion on acetaminophen elimination in rats. Dispositional differences between hepatically and extrahepatically formed glucuronides of acetaminophen and other chemicals. Drug Metab. Dispos. 1988, 16, 527–533. [Google Scholar] [PubMed]
- Hjelle, J.J. Hepatic UDP-glucuronic acid regulation during acetaminophen biotransformation in rats. J. Pharmacol. Exp. Ther. 1986, 237, 750–756. [Google Scholar] [PubMed]
- Kultti, A.; Pasonen-Seppänen, S.; Jauhiainen, M.; Rilla, K.J.; Kärnä, R.; Pyöriä, E.; Tammi, R.H.; Tammi, M.I. 4-Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP-glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp. Cell Res. 2009, 315, 1914–1923. [Google Scholar] [CrossRef]
- Grancharov, K.; Naydenova, Z.; Lozeva, S.; Golovinsky, E. Natural and synthetic inhibitors of UDP-glucuronosyltransferase. Pharmacol. Ther. 2001, 89, 171–186. [Google Scholar] [CrossRef]
- Ishii, Y.; Nurrochmad, A.; Yamada, H. Modulation of UDP-glucuronosyltransferase activity by endogenous compounds. Drug Metab. Pharmacokinet. 2010, 25, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Buchheit, D.; Dragan, C.A.; Schmitt, E.I.; Bureik, M. Production of ibuprofen acyl glucosides by human UGT2B7. Drug Metab. Dispos. 2011, 39, 2174–2181. [Google Scholar] [CrossRef]
- Chau, N.; Elliot, D.J.; Lewis, B.C.; Burns, K.; Johnston, M.R.; Mackenzie, P.I.; Miners, J.O. Morphine glucuronidation and glucosidation represent complementary metabolic pathways that are both catalyzed by UDP-glucuronosyltransferase 2B7: Kinetic, inhibition, and molecular modeling studies. J. Pharmacol. Exp. Ther. 2014, 349, 126–137. [Google Scholar] [CrossRef]
- Mackenzie, P.; Little, J.M.; Radominska-Pandya, A. Glucosidation of hyodeoxycholic acid by UDP-glucuronosyltransferase 2B7. Biochem. Pharmacol. 2003, 65, 417–421. [Google Scholar] [CrossRef]
- Karcz, T.P.; Whitehead, G.S.; Nakano, K.; Nakano, H.; Grimm, S.A.; Williams, J.G.; Deterding, L.J.; Jacobson, K.A.; Cook, D.N. UDP-glucose and P2Y14 receptor amplify allergen-induced airway eosinophilia. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Zimmer, B.M.; Howell, M.E.; Wei, Q.; Ma, L.; Romsdahl, T.; Loughman, E.G.; Markham, J.E.; Seravalli, J.; Barycki, J.J.; Simpson, M.A. Loss of exogenous androgen dependence by prostate tumor cells is associated with elevated glucuronidation potential. Horm. Cancer 2016, 7, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Caputo, M.; Zirpoli, H.; Torino, G.; Tecce, M.F. Selective regulation of UGT1A1 and SREBP-1c mRNA expression by docosahexaenoic, eicosapentaenoic, and arachidonic acids. J. Cell. Physiol. 2011, 226, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Dates, C.R.; Fahmi, T.; Pyrek, S.J.; Yao-Borengasser, A.; Borowa-Mazgaj, B.; Bratton, S.M.; Kadlubar, S.A.; Mackenzie, P.I.; Haun, R.S.; Radominska-Pandya, A. Human UDP-Glucuronosyltransferases: Effects of altered expression in breast and pancreatic cancer cell lines. Cancer Biol. Ther. 2015, 16, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Bozza, P.T.; Bakker-Abreu, I.; Navarro-Xavier, R.A.; Bandeira-Melo, C. Lipid body function in eicosanoid synthesis: An update. Prostaglandins Leukot. Essent. Fat. Acids 2011, 85, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Herms, A.; Bosch, M.; Ariotti, N.; Reddy, B.J.; Fajardo, A.; Fernandez-Vidal, A.; Alvarez-Guaita, A.; Fernandez-Rojo, M.A.; Rentero, C.; Tebar, F.; et al. Cell-to-cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr. Biol. 2013, 23, 1489–1496. [Google Scholar] [CrossRef]
- Nguyen, N.; Bonzo, J.A.; Chen, S.; Chouinard, S.; Kelner, M.J.; Hardiman, G.; Belanger, A.; Tukey, R.H. Disruption of the ugt1 locus in mice resembles human Crigler-Najjar type I disease. J. Biol. Chem. 2008, 283, 7901–7911. [Google Scholar] [CrossRef] [PubMed]
- Bunik, V.; Mkrtchyan, G.; Grabarska, A.; Oppermann, H.; Daloso, D.; Araujo, W.L.; Juszczak, M.; Rzeski, W.; Bettendorff, L.; Fernie, A.R.; et al. Inhibition of mitochondrial 2-oxoglutarate dehydrogenase impairs viability of cancer cells in a cell-specific metabolism-dependent manner. Oncotarget 2016, 7, 26400–26421. [Google Scholar] [CrossRef] [PubMed]
- Schell, J.C.; Olson, K.A.; Jiang, L.; Hawkins, A.J.; Van Vranken, J.G.; Xie, J.; Egnatchik, R.A.; Earl, E.G.; DeBerardinis, R.J.; Rutter, J. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol. Cell 2014, 56, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Sen, T.; Sen, N.; Noordhuis, M.G.; Ravi, R.; Wu, T.C.; Ha, P.K.; Sidransky, D.; Hoque, M.O. OGDHL is a modifier of AKT-dependent signaling and NF-kappaB function. PLoS ONE 2012, 7, e48770. [Google Scholar] [CrossRef] [PubMed]
- Vincent, E.E.; Sergushichev, A.; Griss, T.; Gingras, M.C.; Samborska, B.; Ntimbane, T.; Coelho, P.P.; Blagih, J.; Raissi, T.C.; Choiniere, L.; et al. Mitochondrial Phosphoenolpyruvate Carboxykinase Regulates Metabolic Adaptation and Enables Glucose-Independent Tumor Growth. Mol. Cell 2015, 60, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ko, B.; Hensley, C.T.; Jiang, L.; Wasti, A.T.; Kim, J.; Sudderth, J.; Calvaruso, M.A.; Lumata, L.; Mitsche, M.; et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 2014, 56, 414–424. [Google Scholar] [CrossRef]
- Bellemare, J.; Rouleau, M.; Harvey, M.; Popa, I.; Pelletier, G.; Tetu, B.; Guillemette, C. Immunohistochemical expression of conjugating UGT1A-derived isoforms in normal and tumoral drug-metabolizing tissues in humans. J. Pathol. 2011, 223, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Bellemare, J.; Rouleau, M.; Harvey, M.; Têtu, B.; Guillemette, C. Alternative-splicing forms of the major phase II conjugating UGT1A gene negatively regulate glucuronidation in human carcinoma cell lines. Pharm. J. 2010, 10, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Vitting-Seerup, K.; Sandelin, A. The Landscape of Isoform Switches in Human Cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [Google Scholar] [CrossRef] [PubMed]
- Dietmair, S.; Hodson, M.P.; Quek, L.-E.; Timmins, N.E.; Gray, P.; Nielsen, L.K. A Multi-Omics Analysis of Recombinant Protein Production in Hek293 Cells. PLoS ONE 2012, 7, e43394. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Nakajima, M.; Yamanaka, H.; Fujiwara, R.; Yokoi, T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab. Dispos. 2008, 36, 1461–1464. [Google Scholar] [CrossRef] [PubMed]
- Radominska-Pandya, A.; Ouzzine, M.; Fournel-Gigleux, S.; Magdalou, J. Structure of UDP-glucuronosyltransferases in membranes. Methods Enzymol. 2005, 400, 116–147. [Google Scholar] [PubMed]



| UGT1A1 | |||
|---|---|---|---|
| Number of Metabolites 1 | Enzyme | Alternative Isoform | Enz. + Alt. |
| Total | 276 | 207 | 241 |
| Up|Down | 176|100 | 97|110 | 155|86 |
| UGT2B7 | |||
| Number of Metabolites 1 | Enzyme | Alternative Isoform | Enz. + Alt. |
| Total | 345 | 292 | 280 |
| Up|Down | 228|117 | 184|108 | 149|131 |
| Cell Line | Pathway | Metabolite | Fold Change | p-Value |
|---|---|---|---|---|
| UGT1A1 | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | Isobar: fructose 1,6-diphosphate, glucose 1,6-diphosphate, myo-inositol 1,4 or 1,3-diphosphate | −12.5 | 4.4 × 10−8 |
| Fructose, Mannose and Galactose Metabolism | mannose-6-phosphate | −9.1 | 8.6 × 10−5 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | glucose 6-phosphate | −5.3 | 9.4 × 10−5 | |
| Pentose Phosphate Pathway | 6-phosphogluconate | −5.3 | 5.8 × 10−5 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | 3-phosphoglycerate | −5.0 | 2.0 × 10−6 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | dihydroxyacetone phosphate (DHAP) | −5.0 | 5.8 × 10−8 | |
| Glycerolipid Metabolism | glycerophosphoglycerol | 5.4 | 1.4 × 10−10 | |
| Polyunsaturated Fatty Acid (n3 and n6) | arachidonate (20:4n6) | 5.4 | 2.7 × 10−8 | |
| Monoacylglycerol | 1-arachidonylglycerol (20:4) | 5.5 | 5.9 × 10−8 | |
| Purine Metabolism, Adenine containing | adenine | 5.5 | 2.4 × 10−10 | |
| Pyrimidine Metabolism, Cytidine containing | 2′-deoxycytidine | 5.6 | 9.8 × 10−6 | |
| Polyamine Metabolism | N1,N12-diacetylspermine | 5.9 | 1.0 × 10−4 | |
| UGT2B7 | Fructose, Mannose and Galactose Metabolism | mannose-6-phosphate | −20.0 | 5.9 × 10−11 |
| Pyrimidine Metabolism, Cytidine containing | 2′-deoxycytidine | −20.0 | 5.1 × 10−10 | |
| Polyamine Metabolism | N1,N12-diacetylspermine | −14.3 | 2.2 × 10−6 | |
| Polyamine Metabolism | N(1)-acetylspermine | −9.1 | 1.1 × 10−7 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | glucose 6-phosphate | −6.3 | 3.9 × 10−7 | |
| Nicotinate and Nicotinamide Metabolism | adenosine 5′-diphosphoribose (ADP-ribose) | −6.3 | 4.1 × 10−6 | |
| Polyunsaturated Fatty Acid (n3 and n6) | arachidonate (20:4n6) | 13.3 | 6.5 × 10−9 | |
| Glutathione Metabolism | cysteine-glutathione disulfide | 14.5 | 8.3 × 10−7 | |
| Pyrimidine Metabolism, Thymine containing | 5,6-dihydrothymine | 19.5 | 3.6 × 10−9 | |
| Methionine, Cysteine, SAM and Taurine Metabolism | cystine | 21.0 | 1.4 × 10−9 | |
| Pyrimidine Metabolism, Thymine containing | thymine | 26.1 | 7.3 × 10−9 | |
| Pyrimidine Metabolism, Uracil containing | uracil | 28.3 | 1.9 × 10−8 | |
| alt. UGT1A1 | Plasmalogen | 1-(1-enyl-stearoyl)-2-linoleoyl-GPE (P-18:0/18:2) | −6.3 | 1.1 × 10−6 |
| Polyamine Metabolism | putrescine | −4.3 | 8.8 × 10−8 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | Isobar: fructose 1,6-diphosphate, glucose 1,6-diphosphate, myo-inositol 1,4 or 1,3-diphosphate | −4.0 | 7.4 × 10−5 | |
| Pentose Phosphate Pathway | 6-phosphogluconate | −3.6 | 8.0 × 10−4 | |
| Polyamine Metabolism | N-acetylputrescine | −3.6 | 9.1 × 10−7 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | dihydroxyacetone phosphate (DHAP) | −3.4 | 1.7 × 10−6 | |
| Leucine, Isoleucine and Valine Metabolism | 3-methyl-2-oxovalerate | 5.5 | 7.3 × 10−7 | |
| Leucine, Isoleucine and Valine Metabolism | 4-methyl-2-oxopentanoate | 5.8 | 2.1 × 10−6 | |
| Leucine, Isoleucine and Valine Metabolism | 3-methyl-2-oxobutyrate | 5.9 | 1.6 × 10−6 | |
| Diacylglycerol | palmitoleoyl-oleoyl-glycerol (16:1/18:1) [1] | 6.9 | 4.9 × 10−5 | |
| Diacylglycerol | diacylglycerol (12:0/18:1, 14:0/16:1, 16:0/14:1) [1] | 8.0 | 5.7 × 10−7 | |
| Pyrimidine Metabolism, Cytidine containing | 2′-deoxycytidine | 13.5 | 1.1 × 10−7 | |
| alt. UGT2B7 | Pyrimidine Metabolism, Cytidine containing | 2′-deoxycytidine | −100.0 | 1.0 × 10−13 |
| Diacylglycerol | diacylglycerol (12:0/18:1, 14:0/16:1, 16:0/14:1) [1] | −7.7 | 2.2 × 10−3 | |
| Pyrimidine Metabolism, Cytidine containing | cytidine | −7.1 | 5.7 × 10−7 | |
| Pyrimidine Metabolism, Cytidine containing | 5-methylcytidine | −7.1 | 9.8 × 10−6 | |
| Polyamine Metabolism | N1,N12-diacetylspermine | −6.7 | 2.0 × 10−4 | |
| Endocannabinoid | N-oleoyltaurine | −3.8 | 7.9 × 10−6 | |
| Polyunsaturated Fatty Acid (n3 and n6) | mead acid (20:3n9) | 4.5 | 6.4 × 10−6 | |
| Pentose Phosphate Pathway | 6-phosphogluconate | 4.6 | 1.8 × 10−7 | |
| Pyrimidine Metabolism, Thymine containing | 3-aminoisobutyrate | 4.8 | 1.1 × 10−12 | |
| Pyrimidine Metabolism, Uracil containing | uracil | 5.5 | 3.0 × 10−4 | |
| Plasmalogen | 1-(1-enyl-stearoyl)-2-linoleoyl-GPE (P-18:0/18:2) | 6.1 | 1.8 × 10−7 | |
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | glucose | 18.2 | 5.7 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Audet-Delage, Y.; Rouleau, M.; Villeneuve, L.; Guillemette, C. The Glycosyltransferase Pathway: An Integrated Analysis of the Cell Metabolome. Metabolites 2022, 12, 1006. https://doi.org/10.3390/metabo12101006
Audet-Delage Y, Rouleau M, Villeneuve L, Guillemette C. The Glycosyltransferase Pathway: An Integrated Analysis of the Cell Metabolome. Metabolites. 2022; 12(10):1006. https://doi.org/10.3390/metabo12101006
Chicago/Turabian StyleAudet-Delage, Yannick, Michèle Rouleau, Lyne Villeneuve, and Chantal Guillemette. 2022. "The Glycosyltransferase Pathway: An Integrated Analysis of the Cell Metabolome" Metabolites 12, no. 10: 1006. https://doi.org/10.3390/metabo12101006
APA StyleAudet-Delage, Y., Rouleau, M., Villeneuve, L., & Guillemette, C. (2022). The Glycosyltransferase Pathway: An Integrated Analysis of the Cell Metabolome. Metabolites, 12(10), 1006. https://doi.org/10.3390/metabo12101006








