Highly Efficient Biotransformation and Production of Selenium Nanoparticles and Polysaccharides Using Potential Probiotic Bacillus subtilis T5
Abstract
:1. Introduction
2. Materials and Methods
2.1. Supplies and Chemicals
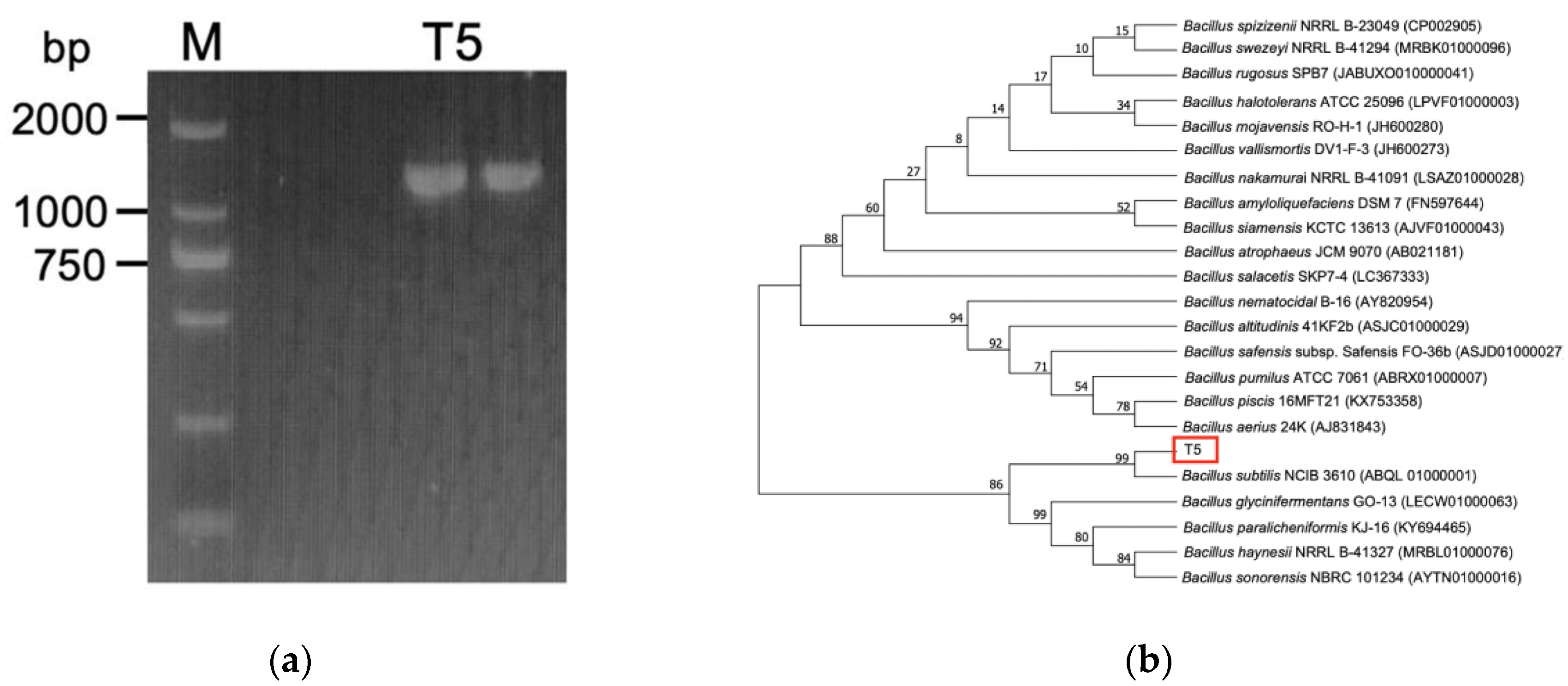
2.2. High Selenite Tolerance Strain Isolation and Identification
2.2.1. Screening of Selenite-Tolerant Strains
2.2.2. Selenite Tolerance Strain Identification
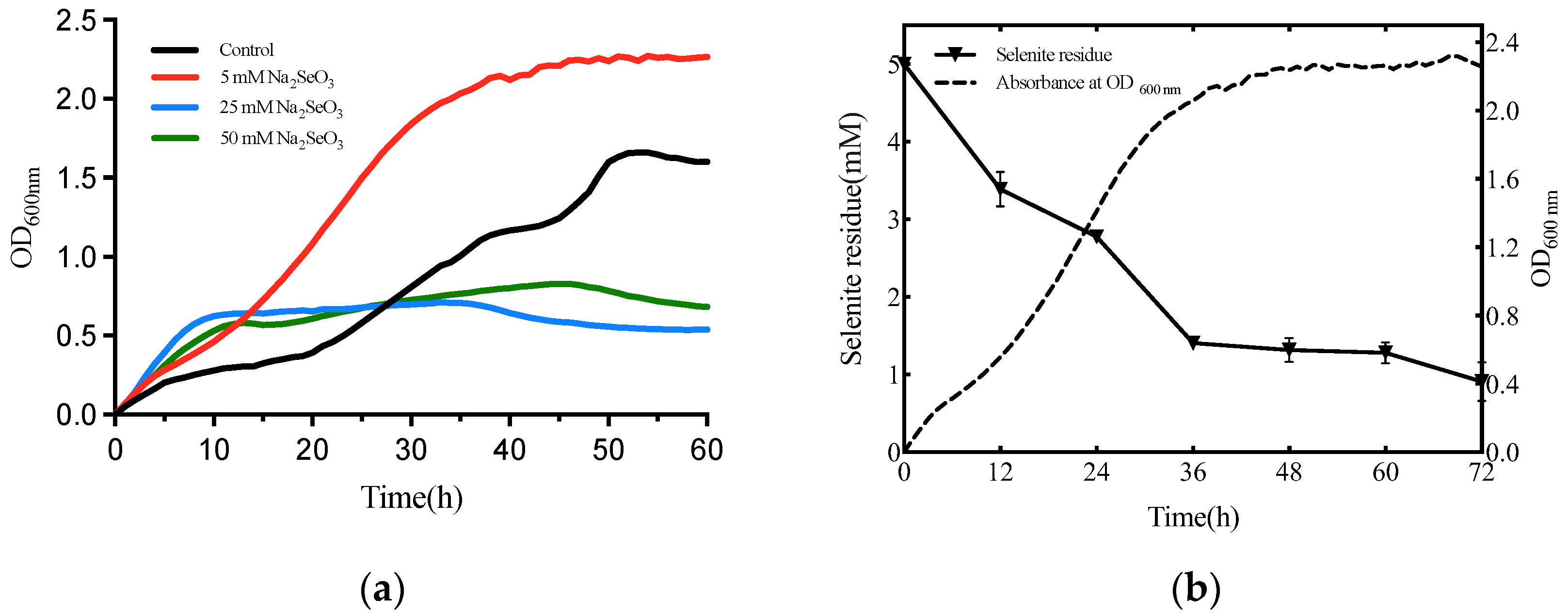
2.3. The Growth Dynamics and Selenite Reduction Activity of the Strain T5
2.4. Characterization of Selenium Nanoparticles by Strain T5
2.4.1. Scan Electron Microscopy
2.4.2. Preparation of SeNPs
2.5. Evaluate the Probiotic Properties and Safety Properties of Strain T5
2.5.1. Tolerance to the Low pH Condition
2.5.2. Bile Salts Tolerance
2.5.3. Cell Surface Hydrophobicity
2.5.4. Auto-Aggregation
2.5.5. Hemolytic Activity
2.6. Antioxidant Capacity of Strain T5
2.7. The α-Amylase Activity Produced by Strain T5
2.8. Production of Polysaccharides by Isolate T5
2.8.1. Extraction of Crude Polysaccharide
2.8.2. Determination of Se Content in the Polysaccharide
2.9. Statistical Analysis
3. Results
3.1. Identification of Selenite-Reducing Strain
3.2. Selenite Biotransformation Assays
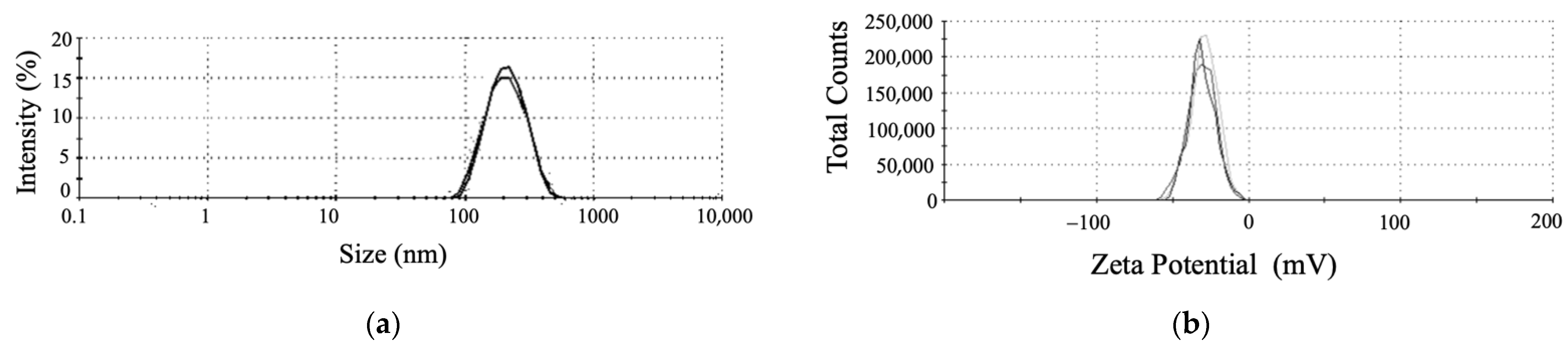
3.3. Characterization of SeNPs by B. subtilis T5
3.3.1. The DLS, Zeta Potential, and SEM Analyses Revealed the Shape and Stably of SeNPs
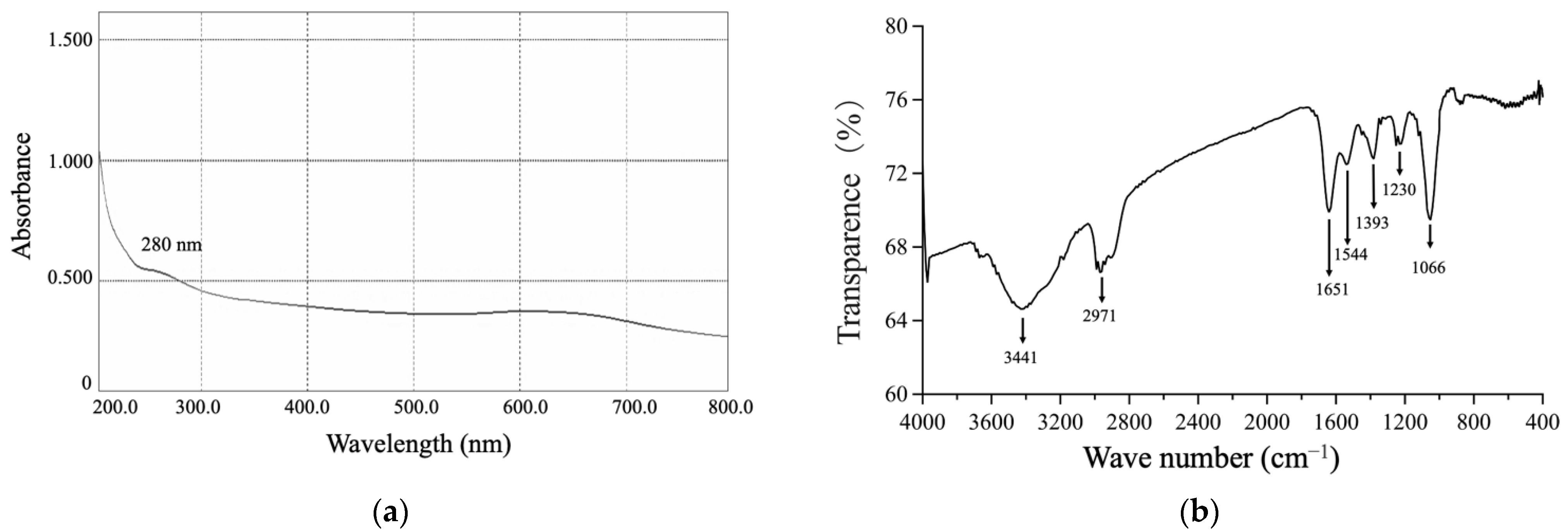
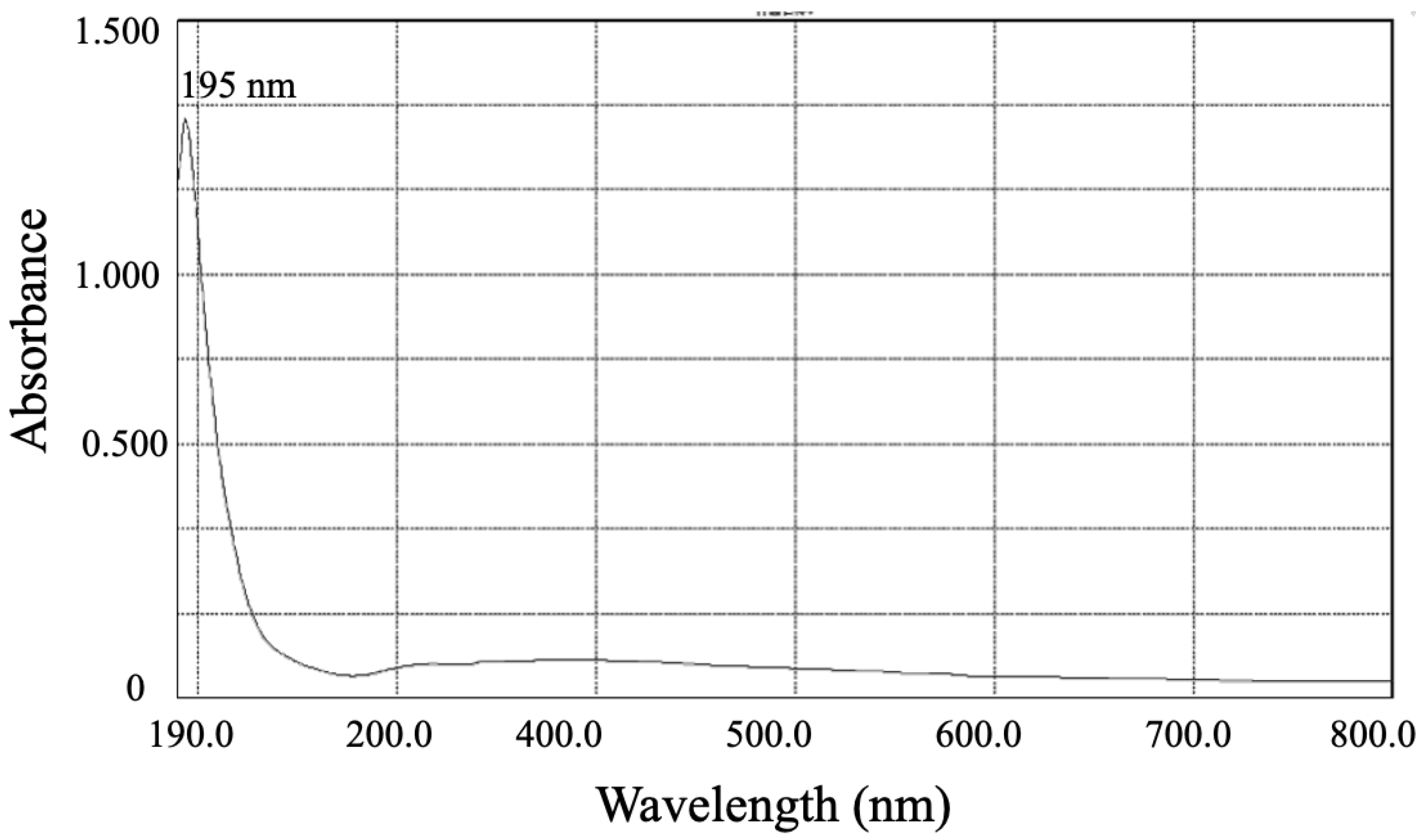
3.3.2. The UV-Vis and FTIR Spectrums of SeNPs
3.4. Probiotic Properties of B. subtilis T5
3.4.1. Tolerance to Low pH and Bile Salts
3.4.2. Hydrophobicity and Auto-Aggregation Properties
3.4.3. Hemolytic Activity
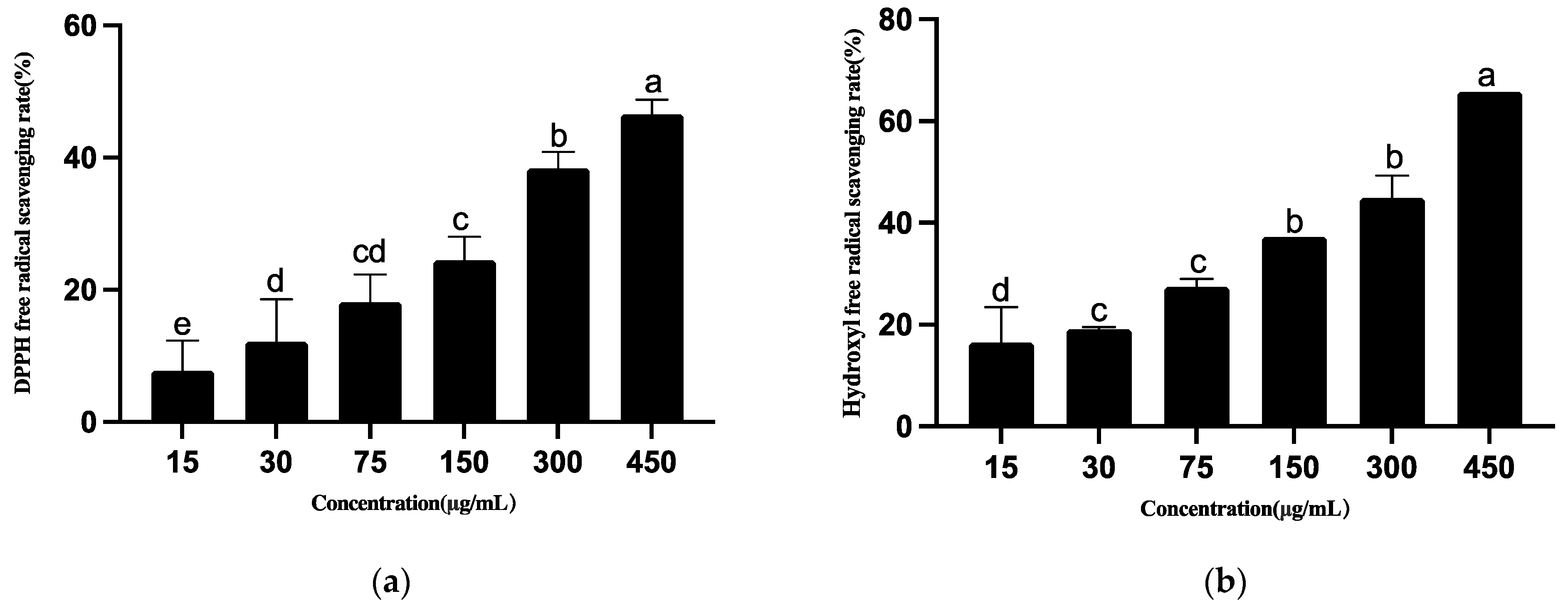
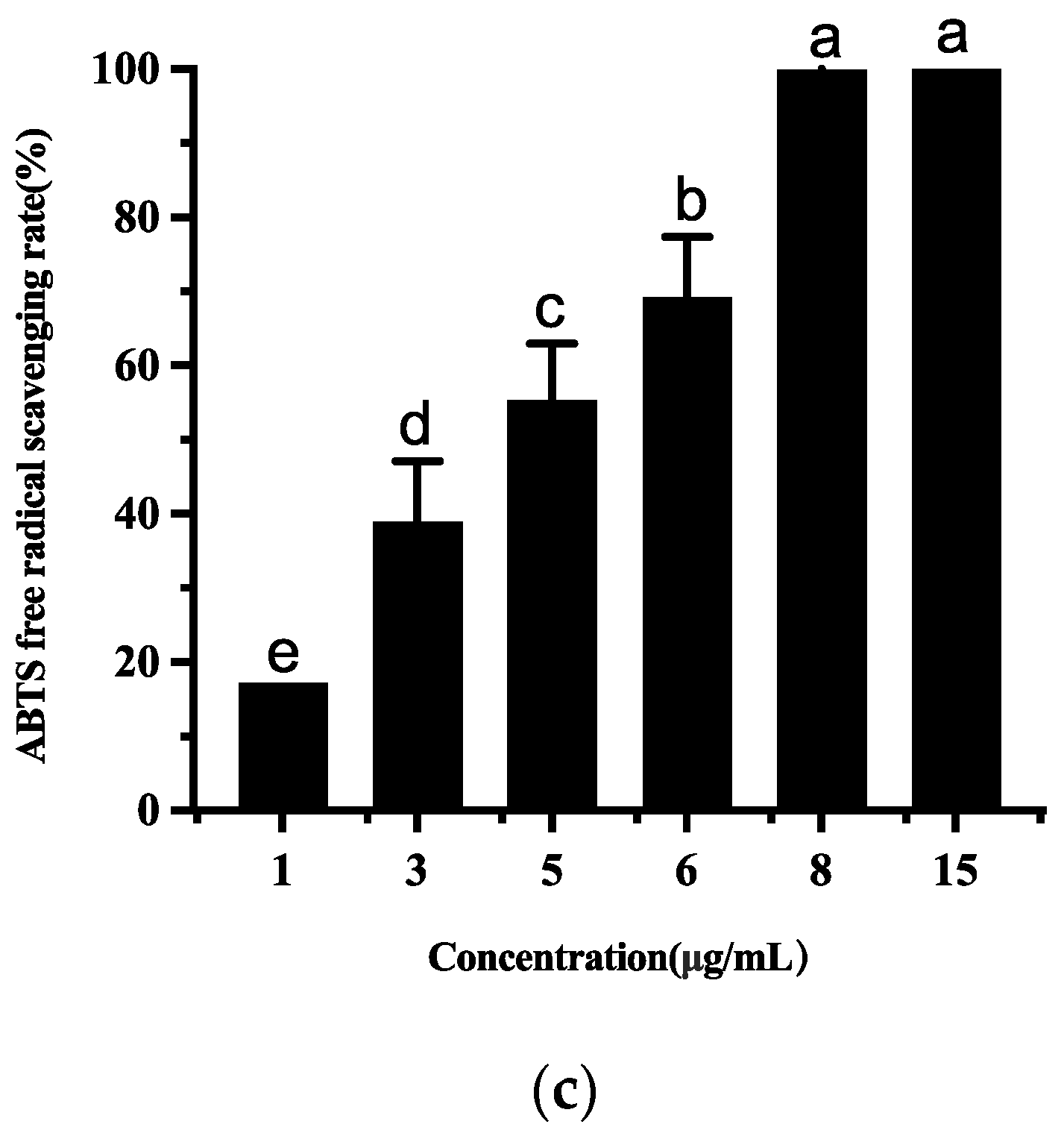
3.4.4. Antioxidant Activities of Ethyl Acetate Extract of T5
3.5. Functional Activity of B. subtilis T5
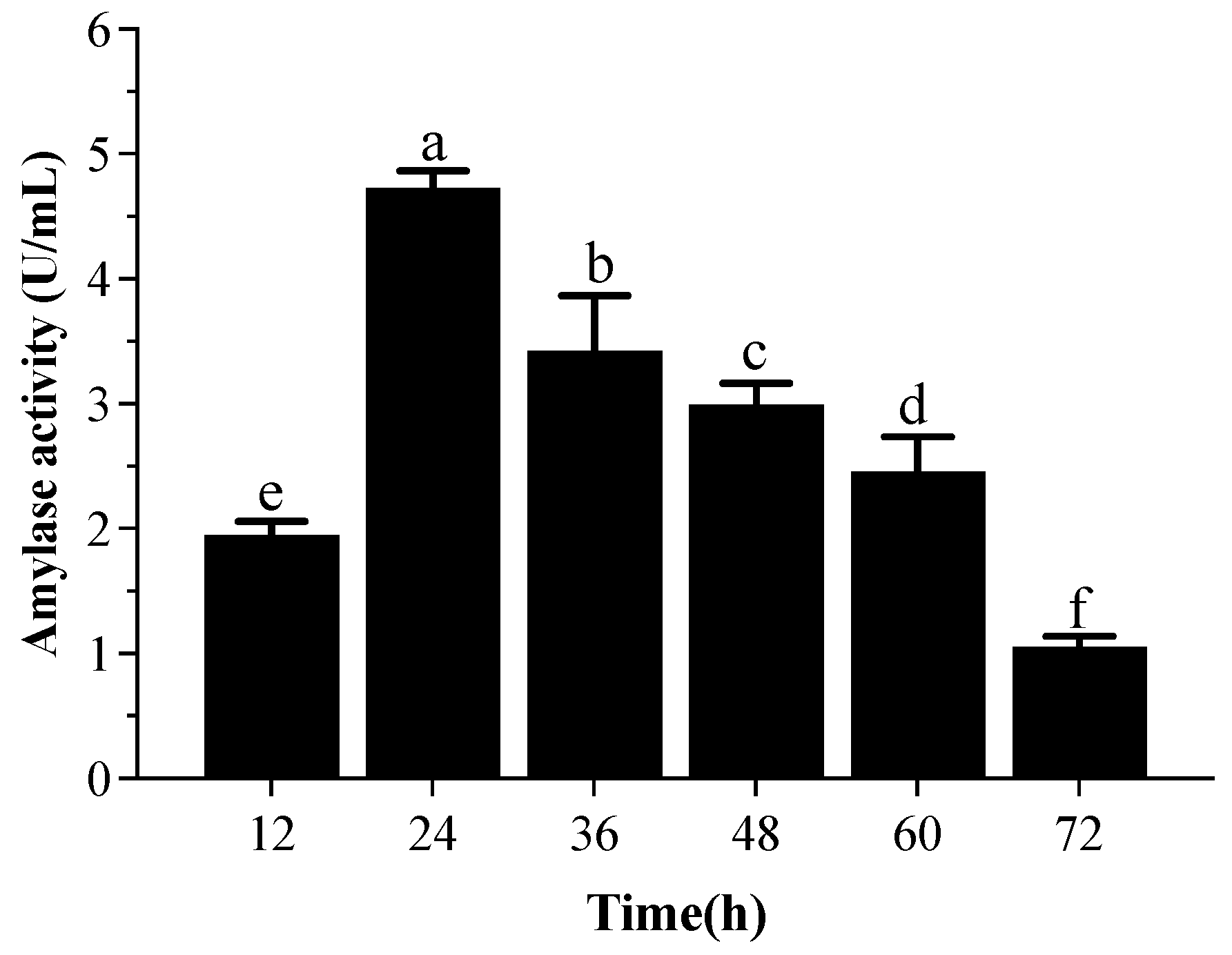
3.5.1. α-Amylase Activity
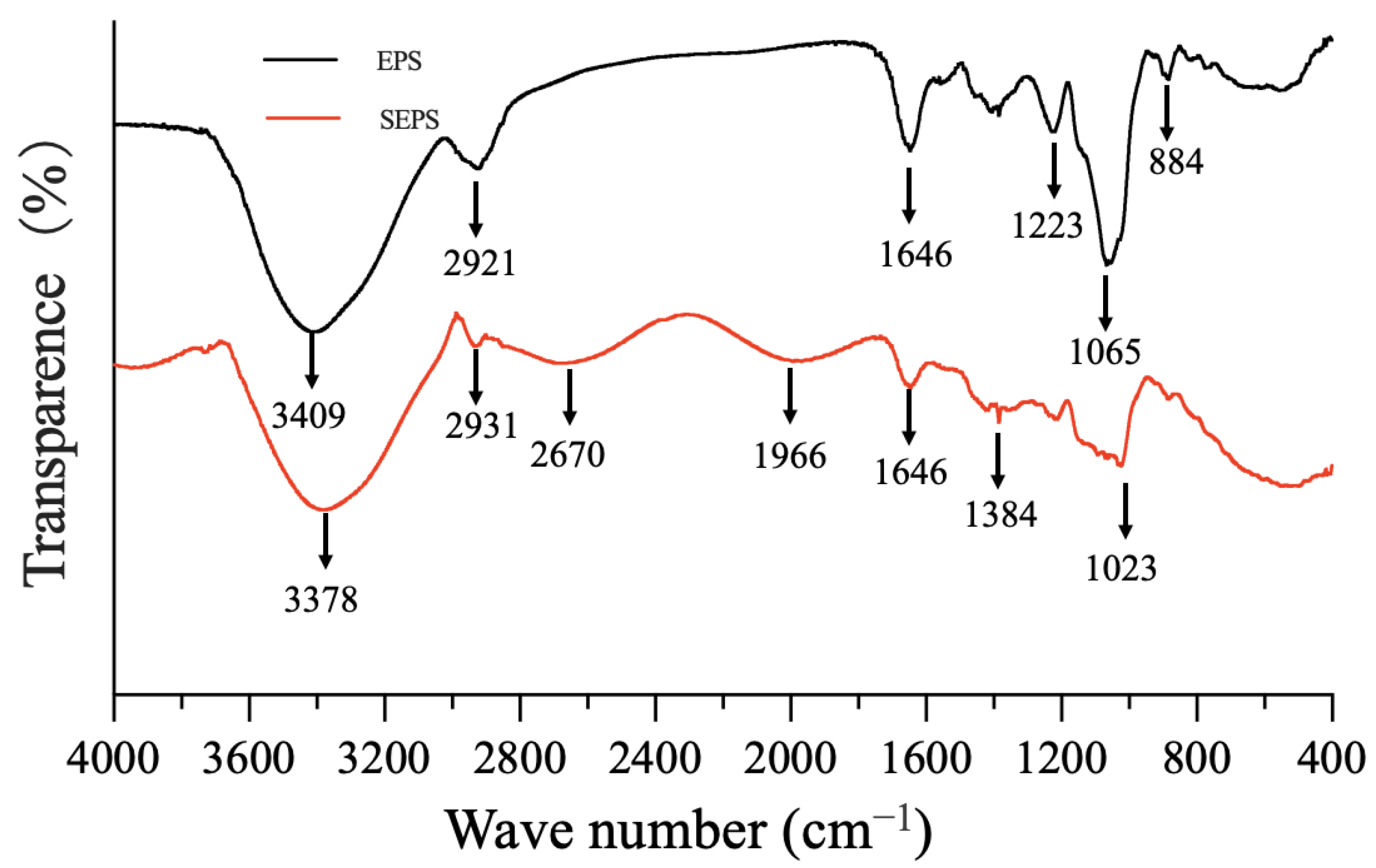
3.5.2. Assessment of Crude Polysaccharides
3.6. Effects of Selenite on Functional Activity by B. subtilis T5
3.6.1. Effects of Selenite on α-Amylase Activity by B. subtilis T5
3.6.2. Assessment of the Selenium Polysaccharides in the Presence of 5 mM Selenite
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Y.; Shi, Y.; Jia, H.; Ding, H.; Yue, T.; Yuan, Y. Biosynthesis of selenium nanoparticles of Monascus purpureus and their inhibition to Alicyclobacillus acidoterrestris. Food Control 2021, 130, 108366. [Google Scholar] [CrossRef]
- Hashem, A.H.; Khalil, A.M.A.; Reyad, A.M.; Salem, S.S. Biomedical Applications of Mycosynthesized Selenium Nanoparticles Using Penicillium expansum ATTC 36200. Biol. Trace Elem. Res. 2021, 199, 3998–4008. [Google Scholar] [CrossRef] [PubMed]
- Pieczynska, J.; Grajeta, H. The role of selenium in human conception and pregnancy. J. Trace Elem. Med. Biol. 2015, 29, 31–38. [Google Scholar] [CrossRef]
- Xie, M.; Sun, X.; Li, P.; Shen, X.; Fang, Y. Selenium in cereals: Insight into species of the element from total amount. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2914–2940. [Google Scholar] [CrossRef]
- Liu, L.; He, Y.; Xiao, Z.; Tao, W.; Zhu, J.; Wang, B.; Liu, Z.; Wang, M. Effects of Selenium Nanoparticles on Reproductive Performance of Male Sprague-Dawley Rats at Supranutritional and Nonlethal Levels. Biol. Trace Elem. Res. 2017, 180, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, H.; Yan, X.; Zhang, L. Comparison of short-term toxicity between Nano-Se and selenite in mice. Life Sci. 2005, 76, 1099–1109. [Google Scholar] [CrossRef]
- Yazdi, M.H.; Mahdavi, M.; Varastehmoradi, B.; Faramarzi, M.A.; Shahverdi, A.R. The Immunostimulatory Effect of Biogenic Selenium Nanoparticles on the 4T1 Breast Cancer Model: An In Vivo Study. Biol. Trace Elem. Res. 2012, 149, 22–28. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef]
- Borah, S.N.; Goswami, L.; Sen, S.; Sachan, D.; Sarma, H.; Montes, M.; Peralta-Videa, J.R.; Pakshirajan, K.; Narayan, M. Selenite bioreduction and biosynthesis of selenium nanoparticles by Bacillus paramycoides SP3 isolated from coal mine overburden leachate. Environ. Pollut. 2021, 285, 117519. [Google Scholar] [CrossRef]
- Ashengroph, M.; Hosseini, S.R. A newly isolated Bacillus amyloliquefaciens SRB04 for the synthesis of selenium nanoparticles with potential antibacterial properties. Int. Microbiol. 2021, 24, 103–114. [Google Scholar] [CrossRef]
- Huang, S.; Wang, Y.; Tang, C.; Jia, H.; Wu, L. Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16-A novel mechanism of selenite reduction based on proteomic analysis. J. Hazard. Mater. 2021, 406, 124690. [Google Scholar] [CrossRef]
- Ullah, A.; Yin, X.; Wang, F.; Xu, B.; Mirani, Z.A.; Xu, B.; Chan, M.W.H.; Ali, A.; Usman, M.; Ali, N.; et al. Biosynthesis of Selenium Nanoparticles (via Bacillus subtilis BSN313), and Their Isolation, Characterization, and Bioactivities. Molecules 2021, 26, 5559. [Google Scholar] [CrossRef]
- Shakibaie, M.; Mohammadi-Khorsand, T.; Adeli-Sardou, M.; Jafari, M.; Amirpour-Rostami, S.; Ameri, A.; Forootanfar, H. Probiotic and antioxidant properties of selenium-enriched Lactobacillus brevis L. Se isolated from an Iranian traditional dairy product. J. Trace Elem. Med. Biol. 2017, 40, 1–9. [Google Scholar] [CrossRef]
- Xu, C.; Qiao, L.; Guo, Y.; Ma, L.; Cheng, Y. Preparation, characteristics and antioxidant activity of polysaccharides and proteins-capped selenium nanoparticles synthesized by Lactobacillus casei ATCC 393. Carbohydr. Polym. 2018, 195, 576–585. [Google Scholar] [CrossRef]
- Jin, W.; Yoon, C.; Johnston, T.V.; Ku, S.; Ji, G.E. Production of Selenomethionine-Enriched Bifidobacterium bifidum BGN4 via Sodium Selenite Biocatalysis. Molecules 2018, 23, 2860. [Google Scholar] [CrossRef] [Green Version]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Tamtaji, O.R.; Heidari-Soureshjani, R.; Mirhosseini, N.; Kouchaki, E.; Bahmani, F.; Aghadavod, E.; Tajabadi-Ebrahimi, M.; Asemi, Z. Probiotic and selenium co-supplementation, and the effects on clinical, metabolic and genetic status in Alzheimer’s disease: A randomized, double-blind, controlled trial. Clin. Nutr. 2019, 38, 2569–2575. [Google Scholar] [CrossRef]
- Jamilian, M.; Mansury, S.; Bahmani, F.; Heidar, Z.; Amirani, E.; Asemi, Z. The effects of probiotic and selenium co-supplementation on parameters of mental health, hormonal profiles, and biomarkers of inflammation and oxidative stress in women with polycystic ovary syndrome. J. Ovarian Res. 2018, 11, 80. [Google Scholar] [CrossRef] [Green Version]
- Simon, W.V.O.; Scharek, L. Micro-Organisms as Feed Additives—Probiotics. Adv. Pork Prod. 2005, 16, 161. [Google Scholar]
- Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Jujjavarapu, S.E.; Dhagat, S. Evolutionary Trends in Industrial Production of α-amylase. Recent Pat. Biotechnol. 2019, 13, 4–18. [Google Scholar] [CrossRef]
- Contesini, F.J.; Melo, R.R.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef]
- Dhruw, C.; Husain, K.; Kumar, V.; Sonawane, V.C. Novel xylanase producing Bacillus strain X2: Molecular phylogenetic analysis and its application for production of xylooligosaccharides. 3 Biotech 2020, 10, 328. [Google Scholar] [CrossRef]
- Bello-Morales, R.; Andreu, S.; Ruiz-Carpio, V.; Ripa, I.; López-Guerrero, J.A. Extracellular Polymeric Substances: Still Promising Antivirals. Viruses 2022, 14, 1337. [Google Scholar] [CrossRef]
- Duan, W.-X.; Yang, X.-H.; Zhang, H.-F.; Feng, J.; Zhang, M.-Y. Chemical Structure, Hypoglycemic Activity, and Mechanism of Action of Selenium Polysaccharides. Biol. Trace Elem. Res. 2021, 200, 4404–4418. [Google Scholar] [CrossRef]
- Górska-Jakubowska, S.; Klimaszewska, M.; Podsadni, P.; Kaleta, B.; Zagożdżon, R.; Górska, S.; Gamian, A.; Strączek, T.; Kapusta, C.; Cieślak, M.; et al. Selenium-Containing Exopolysaccharides Isolated from the Culture Medium of Lentinula edodes: Structure and Biological Activity. Int. J. Mol. Sci. 2021, 22, 13039. [Google Scholar] [CrossRef]
- Ye, S.; Zhang, J.; Liu, Z.; Zhang, Y.; Li, J.; Li, Y.O. Biosynthesis of selenium rich exopolysaccharide (Se-EPS) by Pseudomonas PT-8 and characterization of its antioxidant activities. Carbohydr. Polym. 2016, 142, 230–239. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, D.; Li, H.; Sun, Y.; Zeng, X.; Yan, B. Antioxidant and immunomodulatory activity of selenium exopolysaccharide produced by Lactococcus lactis subsp. lactis. Food Chem. 2013, 138, 84–89. [Google Scholar] [CrossRef]
- Xu, C.L.; Wang, Y.Z.; Jin, M.L.; Yang, X.Q. Preparation, characterization and immunomodulatory activity of selenium-enriched exopolysaccharide produced by bacterium Enterobacter cloacae Z0206. Bioresour. Technol. 2009, 100, 2095–2097. [Google Scholar] [CrossRef]
- Li, J.; Shen, B.; Nie, S.; Duan, Z.; Chen, K. A combination of selenium and polysaccharides: Promising therapeutic potential. Carbohydr. Polym. 2019, 206, 163–173. [Google Scholar] [CrossRef]
- Jhon, H.G.; Krieg, N.R.; Petyer, H.A.; Sneath, J.T.S.; Stanle, T. Williams, Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams and Wilkins: Baltimore, MD, USA, 1994; pp. 255–260. [Google Scholar]
- Janda, J.M.; Sharon, L.A. 16S rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Ashe, S.; Maji, U.J.; Sen, R.; Mohanty, S.; Maiti, N.K. Specific oligonucleotide primers for detection of endoglucanase positive Bacillus subtilis by PCR. 3 Biotech 2014, 4, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Avendaño, N.C.R. Production of selenium nanoparticles in Pseudomonas putida KT2440. Sci. Rep. 2016, 6, 37155. [Google Scholar] [CrossRef] [Green Version]
- Loeschner, K.; Hadrup, N.; Hansen, M.; Pereira, S.A.; Gammelgaard, B.; Moller, L.H.; Mortensen, A.; Lam, H.R.; Larsen, E.H. Absorption, distribution, metabolism and excretion of selenium following oral administration of elemental selenium nanoparticles or selenite in rats. Metallomics 2014, 6, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shu, X.; Zhou, Q.; Fan, T.; Wang, T.; Chen, X.; Li, M.; Ma, Y.; Ni, J.; Hou, J.; et al. Selenite Reduction and the Biogenesis of Selenium Nanoparticles by Alcaligenes faecalis Se03 Isolated from the Gut of Monochamus alternatus (Coleoptera: Cerambycidae). Int. J. Mol. Sci. 2018, 19, 2799. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Li, Q.; Jin, Z.; Yu, X.; Ding, M.; Ju, Y. Establishment and application of a new serum sodium candidate reference method. Clin. Chim. Acta 2020, 508, 249–253. [Google Scholar] [CrossRef]
- Khoei, N.S.; Lampis, S.; Zonaro, E.; Yrjälä, K.; Bernardi, P.; Vallini, G. Insights into selenite reduction and biogenesis of elemental selenium nanoparticles by two environmental isolates of Burkholderia fungorum. New Biotechnol. 2017, 34, 1–11. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Duan, Y.; Wang, Q.; Cong, X.; He, Y.; Gao, C.; Hafeez, M.; Jan, S.; Rasheed, S.M.; et al. Enhancing the Activity of Carboxymethyl Cellulase Enzyme Using Highly Stable Selenium Nanoparticles Biosynthesized by Bacillus paralicheniformis Y4. Molecules 2022, 27, 4585. [Google Scholar] [CrossRef]
- Joint FAO/WHO Working Group. Guidelines for the Evaluation of Probiotics in Food; Report; Food and Agriculture Organization of the United Nations: Rome, Italy, 2002. [Google Scholar]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Guo, C.F.; Zhang, S.; Yuan, Y.H.; Yue, T.L.; Li, J.Y. Comparison of lactobacilli isolated from Chinese suan-tsai and koumiss for their probiotic and functional properties. J. Funct. Foods 2015, 12, 294–302. [Google Scholar] [CrossRef]
- Vinderola, C.G.; Reinheimer, J.A. Lactic acid starter and probiotic bacteria: A comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 2008, 226, 1065–1073. [Google Scholar] [CrossRef]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; de Cadiñanos, L.P.G.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Sharma, A.; Lavania, M.; Singh, R.; Lal, B. Identification and probiotic potential of lactic acid bacteria from camel milk. Saudi J. Biol. Sci. 2021, 28, 1622–1632. [Google Scholar] [CrossRef]
- Feng, C.; Zhang, F.; Wang, B.; Zhang, L.; Dong, Y.; Shao, Y. Genome-wide analysis of fermentation and probiotic trait stability in Lactobacillus plantarum during continuous culture. J. Dairy Sci. 2020, 103, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.L.; Tang, Z.H.; Zhang, X.F.; Zhong, Y.H.; Yao, S.Z.; Wang, L.S.; Lin, C.W.; Luo, X. Chemical properties and antioxidant activity of a water-soluble polysaccharide from Dendrobium officinale. Int. J. Biol. Macromol. 2016, 89, 219–227. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Yassin, S.N.; Jiru, T.M.; Indracanti, M. Screening and Characterization of Thermostable Amylase-Producing Bacteria Isolated from Soil Samples of Afdera, Afar Region, and Molecular Detection of Amylase-Coding Gene. Int. J. Microbiol. 2021, 2021, 5592885. [Google Scholar] [CrossRef]
- Li, Q.; Wang, W.; Zhu, Y.; Chen, Y.; Zhang, W.; Yu, P.; Mao, G.; Zhao, T.; Feng, W.; Yang, L.; et al. Structural elucidation and antioxidant activity a novel Se-polysaccharide from Se-enriched Grifola frondosa. Carbohydr. Polym. 2017, 161, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Guo, Y.; Chen, X.; Cui, D.; Wang, M.; Yang, W.; Chen, F. Structural Characteristics, Antioxidant and Hypoglycemic Activities of Polysaccharide from Siraitia grosvenorii. Molecules 2022, 27, 4192. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Chen, J.; Li, L.; Zhao, Z.; Liu, X. Characteristics of Se in water-soil-plant system and threshold of soil Se in seleniferous areas in Enshi, China. Sci. Total Environ. 2022, 827, 154372. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, S.; Chuang, C.-y.; Miller, E.J.; Schwehr, K.A.; Santschi, P.H. Chemical composition and relative hydrophobicity of microbial exopolymeric substances (EPS) isolated by anion exchange chromatography and their actinide-binding affinities. Mar. Chem. 2011, 126, 27–36. [Google Scholar] [CrossRef]
- Wang, L.L.; Wang, L.F.; Ren, X.M.; Ye, X.D.; Li, W.W.; Yuan, S.J.; Sun, M.; Sheng, G.P.; Yu, H.Q.; Wang, X.K. pH dependence of structure and surface properties of microbial EPS. Environ. Sci. Technol. 2012, 46, 737–744. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Hazard. Mater. 2017, 324, 3–14. [Google Scholar] [CrossRef]
- Jain, R.; Jordan, N.; Weiss, S.; Foerstendorf, H.; Heim, K.; Kacker, R.; Hübner, R.; Kramer, H.; van Hullebusch, E.D.; Farges, F.; et al. Extracellular polymeric substances govern the surface charge of biogenic elemental selenium nanoparticles. Environ. Sci. Technol. 2015, 49, 1713–1720. [Google Scholar] [CrossRef]
- Hou, R.; Chen, J.; Yue, C.; Li, X.; Liu, J.; Gao, Z.; Liu, C.; Lu, Y.; Wang, D.; Li, H.; et al. Modification of lily polysaccharide by selenylation and the immune-enhancing activity. Carbohydr. Polym. 2016, 142, 73–81. [Google Scholar] [CrossRef]
- Zhu, Y.; Ren, B.; Li, H.; Lin, Z.; Bañuelos, G.; Li, L.; Zhao, G.; Guo, Y. Biosynthesis of selenium nanoparticles and effects of selenite, selenate, and selenomethionine on cell growth and morphology in Rahnella aquatilis HX2. Appl. Microbiol. Biotechnol. 2018, 102, 6191–6205. [Google Scholar] [CrossRef]
- Fernandez-Llamosas, H.; Castro, L.; Blazquez, M.L.; Diaz, E.; Carmona, M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef] [Green Version]
- Bebien, M.; Chauvin, J.P.; Adriano, J.M.; Grosse, S.; Verméglio, A. Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2001, 67, 4440–4447. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.S.; Ganthe, H.E.r. Acid-volatile selenium formation catalyzed by glutathione reductase. Biochemistry 1975, 14, 1632–1636. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Long, H.; Wang, C.; Yu, L.; Zhao, M.; Liu, X. Research progress on the biological activities of selenium polysaccharides. Food Funct. 2020, 11, 4834–4852. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Du, M.; Kai, L.; Du, S.; Hu, W.; Wang, Y.; Cheng, Y. Exopolymer-Functionalized Nanoselenium from Bacillus subtilis SR41: Characterization, Monosaccharide Analysis and Free Radical Scavenging Ability. Polymers 2022, 14, 3523. [Google Scholar] [CrossRef]
- Kashiwada, S. Distribution of nanoparticles in the see-through medaka (Oryzias latipes). Environ. Health Perspect. 2006, 114, 1697–1702. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Xu, H.; Huang, K.; Yang, X.; Xie, C.; Wu, J. Size effects of realgar particles on apoptosis in a human umbilical vein endothelial cell line: ECV-304. Pharm. Res. 2001, 44, 513–518. [Google Scholar] [CrossRef]
- Takeda, S.; Yamasaki, K.; Takeshita, M.; Kikuchi, Y.; Tsend-Ayush, C.; Dashnyam, B.; Ahhmed, A.M.; Kawahara, S.; Muguruma, M. The investigation of probiotic potential of lactic acid bacteria isolated from traditional Mongolian dairy products. Anim. Sci. J. 2011, 82, 571–579. [Google Scholar] [CrossRef]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [Green Version]
- Ragul, K.; Kandasamy, S.; Devi, P.B.; Shetty, P.H. Evaluation of functional properties of potential probiotic isolates from fermented brine pickle. Food Chem. 2020, 311, 126057. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Alhuzani, M.R.; Kelany, M.S.; Hamed, M.M. Production and Partial Characterization of α-Amylase Enzyme from Marine Actinomycetes. BioMed Res. Int. 2021, 2021, 5289848. [Google Scholar] [CrossRef]
- Salman, T.; Kamal, M.; Ahmed, M.; Siddiqa, S.M.; Khan, R.A.; Hassan, A. Medium optimization for the production of amylase by Bacillus subtilis RM16 in Shake-flask fermentation. Pak. J. Pharm. Sci. 2016, 29, 439–444. [Google Scholar] [PubMed]
- Hough, D.W.; Danson, M.J. Extremozymes. Curr. Opin. Chem. Biol. 1999, 3, 39–46. [Google Scholar] [CrossRef] [PubMed]



| Probiotic Characteristics | Condition | Degree (%) |
|---|---|---|
| Survival tests: | ||
| Different pH levels (for 4 h) | pH 2.5 pH 3.5 | 62.75 ± 1.22 70.63 ± 2.07 |
| Different concentrations of bile salts (for 4 h) | 0.3% 0.6% | 51.12 ± 1.67 46.24 ± 2.58 |
| Auto-aggregation | 1 h 4 h 24 h | 5.49 ± 0.57 13.59 ± 1.37 80.47 ± 0.83 |
| Cell surface hydrophobicity | 0.5 h 1 h | 21.63 ± 3.44 37.99 ± 2.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Li, M.; Zhang, S.; Wang, Y.; Deng, J.; Wang, Q.; Yi, T.; Dong, X.; Cheng, S.; He, Y.; et al. Highly Efficient Biotransformation and Production of Selenium Nanoparticles and Polysaccharides Using Potential Probiotic Bacillus subtilis T5. Metabolites 2022, 12, 1204. https://doi.org/10.3390/metabo12121204
Duan Y, Li M, Zhang S, Wang Y, Deng J, Wang Q, Yi T, Dong X, Cheng S, He Y, et al. Highly Efficient Biotransformation and Production of Selenium Nanoparticles and Polysaccharides Using Potential Probiotic Bacillus subtilis T5. Metabolites. 2022; 12(12):1204. https://doi.org/10.3390/metabo12121204
Chicago/Turabian StyleDuan, Yuhua, Mengjun Li, Sishang Zhang, Yidan Wang, Jieya Deng, Qin Wang, Tian Yi, Xingxing Dong, Shuiyuan Cheng, Yi He, and et al. 2022. "Highly Efficient Biotransformation and Production of Selenium Nanoparticles and Polysaccharides Using Potential Probiotic Bacillus subtilis T5" Metabolites 12, no. 12: 1204. https://doi.org/10.3390/metabo12121204
APA StyleDuan, Y., Li, M., Zhang, S., Wang, Y., Deng, J., Wang, Q., Yi, T., Dong, X., Cheng, S., He, Y., Gao, C., & Wang, Z. (2022). Highly Efficient Biotransformation and Production of Selenium Nanoparticles and Polysaccharides Using Potential Probiotic Bacillus subtilis T5. Metabolites, 12(12), 1204. https://doi.org/10.3390/metabo12121204








