Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial
Abstract
:1. Introduction
2. Methods
2.1. Study Population
2.2. Metabolomic Profiling
2.3. Covariate Assessment
2.4. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- American Cancer Society. Cancer Facts & Figures 2022; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Hancock, S.B.; Georgiades, C.S. Kidney cancer. Cancer J. 2016, 22, 387–392. [Google Scholar] [CrossRef] [PubMed]
- The World Cancer Research Fund/American Institute for Cancer Research. Diet, Nutrition, Physical Activity and Cancer: A Global Perspective; Continuous Update Project Expert Report Update; The World Cancer Research Fund/American Institute for Cancer Research: Washington, DC, USA, 2018. [Google Scholar]
- Hidayat, K.; Du, X.; Zou, S.Y.; Shi, B.M. Blood pressure and kidney cancer risk: Meta-analysis of prospective studies. J. Hypertens. 2017, 35, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Macleod, L.C.; Hotaling, J.M.; Wright, J.L.; Davenport, M.T.; Gore, J.L.; Harper, J.; White, E. Risk factors for renal cell carcinoma in the VITAL study. J. Urol. 2013, 190, 1657–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaelin, W.G., Jr. Von hippel-lindau disease. Annu. Rev. Pathol. Mech. Dis. 2007, 2, 145–173. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Linehan, W.M.; Srinivasan, R.; Schmidt, L.S. The genetic basis of kidney cancer: A metabolic disease. Nat. Rev. Urol. 2010, 7, 277–285. [Google Scholar] [CrossRef]
- Lucarelli, G.; Loizzo, D.; Franzin, R.; Battaglia, S.; Ferro, M.; Cantiello, F.; Castellano, G.; Bettocchi, C.; Ditonno, P.; Battaglia, M. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev. Mol. Diagn. 2019, 19, 397–407. [Google Scholar] [CrossRef]
- Lucarelli, G.; Rutigliano, M.; Sallustio, F.; Ribatti, D.; Giglio, A.; Signorile, M.L.; Grossi, V.; Sanese, P.; Napoli, A.; Maiorano, E.; et al. Integrated multi-omics characterization reveals a distinctive metabolic signature and the role of NDUFA4L2 in promoting angiogenesis, chemoresistance, and mitochondrial dysfunction in clear cell renal cell carcinoma. Aging 2018, 10, 3957. [Google Scholar] [CrossRef]
- Bombelli, S.; Torsello, B.; De Marco, S.; Lucarelli, G.; Cifola, I.; Grasselli, C.; Strada, G.; Bovo, G.; Perego, R.A.; Bianchi, C. 36-kDa annexin A3 isoform negatively modulates lipid storage in clear cell renal cell carcinoma cells. Am. J. Pathol. 2020, 190, 2317–2326. [Google Scholar] [CrossRef]
- Wettersten, H.I.; Hakimi, A.A.; Morin, D.; Bianchi, C.; Johnstone, M.E.; Donohoe, D.R.; Trott, J.F.; Aboud, O.A.; Stirdivant, S.; Neri, B.; et al. Grade-Dependent Metabolic Reprogramming in Kidney Cancer Revealed by Combined Proteomics and Metabolomics AnalysisCombined Metabolomics and Proteomics Analysis of RCC. Cancer Res. 2015, 75, 2541–2552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balabanov, S.; Zimmermann, U.; Protzel, C.; Scharf, C.; Klebingat, K.-J.; Walther, R. Tumour-related enzyme alterations in the clear cell type of human renal cell carcinoma identified by two-dimensional gel electrophoresis. Eur. J. Biochem. 2001, 268, 5977–5980. [Google Scholar] [CrossRef] [PubMed]
- Zira, A.N.; Theocharis, S.E.; Mitropoulos, D.; Migdalis, V.; Mikros, E. 1H NMR metabonomic analysis in renal cell carcinoma: A possible diagnostic tool. J. Proteome Res. 2010, 9, 4038–4044. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Huang, Z.; Gao, Y.; Yan, X.; Xing, J.; Hang, W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res. 2011, 10, 1396–1405. [Google Scholar] [CrossRef]
- Lin, L.; Yu, Q.; Yan, X.; Hang, W.; Zheng, J.; Xing, J.; Huang, B. Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst 2010, 135, 2970–2978. [Google Scholar] [CrossRef]
- Gao, H.; Dong, B.; Liu, X.; Xuan, H.; Huang, Y.; Lin, D. Metabonomic profiling of renal cell carcinoma: High-resolution proton nuclear magnetic resonance spectroscopy of human serum with multivariate data analysis. Anal. Chim. Acta 2008, 624, 269–277. [Google Scholar] [CrossRef]
- Ganti, S.; Taylor, S.; Kim, K.; Hoppel, C.L.; Guo, L.; Yang, J.; Evans, C.; Weiss, R.H. Urinary acylcarnitines are altered in human kidney cancer. Int. J. Cancer 2012, 130, 2791–2800. [Google Scholar] [CrossRef]
- Kordalewska, M.; Wawrzyniak, R.; Jacyna, J.; Godzień, J.; Gonzálves, L.; Raczak-Gutknecht, J.; Markuszewski, M.; Gutknecht, P.; Siebert, J.; Barbas, C.; et al. Molecular signature of renal cell carcinoma by means of a multiplatform metabolomics analysis. Biochem. Biophys. Rep. 2022, 31, 101318. [Google Scholar] [CrossRef]
- Kim, K.; Taylor, S.; Ganti, S.; Guo, L.; Osier, M.V.; Weiss, R.H. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. Omics J. Integr. Biol. 2011, 15, 293–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, M.S.; Barros, A.S.; Pinto, J.; Carvalho, M.; Pires-Luis, A.S.; Henrique, R.; Jeronimo, C.; Bastos, M.D.L.; Gil, A.M.; Guedes de Pinho, P. Nuclear Magnetic Resonance metabolomics reveals an excretory metabolic signature of renal cell carcinoma. Sci. Rep. 2016, 6, 37275. [Google Scholar] [CrossRef]
- Nizioł, J.; Bonifay, V.; Ossoliński, K.; Ossoliński, T.; Ossolińska, A.; Sunner, J.; Beech, I.; Arendowski, A.; Ruman, T. Metabolomic study of human tissue and urine in clear cell renal carcinoma by LC-HRMS and PLS-DA. Anal. Bioanal. Chem. 2018, 410, 3859–3869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakimi, A.A.; Reznik, E.D.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganti, S.; Taylor, S.L.; Abu Aboud, O.; Yang, J.; Evans, C.; Osier, M.V.; Alexander, D.C.; Kim, K.; Weiss, R.H. Kidney Tumor Biomarkers Revealed by Simultaneous Multiple Matrix Metabolomics AnalysisMultiple Matrix Metabolomics Analysis of Kidney Cancer. Cancer Res. 2012, 72, 3471–3479. [Google Scholar] [CrossRef] [Green Version]
- Guida, F.; Tan, V.Y.; Corbin, L.J.; Smith-Byrne, K.; Alcala, K.; Langenberg, C.; Stewart, I.D.; Butterworth, A.S.; Surendran, P.; Achaintre, D.; et al. The blood metabolome of incident kidney cancer: A case–control study nested within the MetKid consortium. PLoS Med. 2021, 18, e1003786. [Google Scholar] [CrossRef] [PubMed]
- Dobrian, A.D.; Davies, M.J.; Schriver, S.D.; Lauterio, T.J.; Prewitt, R.L. Oxidative stress in a rat model of obesity-induced hypertension. Hypertension 2001, 37, 554–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.E.; Crook, E.D.; Jones, D.W.; Wofford, M.R.; Dubbert, P.M. Mechanisms of obesity-associated cardiovascular and renal disease. Am. J. Med. Sci. 2002, 324, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Bagby, S.P. Obesity-initiated metabolic syndrome and the kidney: A recipe for chronic kidney disease? J. Am. Soc. Nephrol. 2004, 15, 2775–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chade, A.R.; Lerman, A.; Lerman, L.O. Kidney in early atherosclerosis. Hypertension 2005, 45, 1042–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kooijman, R. Regulation of apoptosis by insulin-like growth factor (IGF)-I. Cytokine Growth Factor Rev. 2006, 17, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Fan, Z.; Masui, H.; Rosen, N.; Mendelsohn, J. Apoptosis induced by an anti-epidermal growth factor receptor monoclonal antibody in a human colorectal carcinoma cell line and its delay by insulin. J. Clin. Investig. 1995, 95, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.M.; Weinstein, S.J.; Pollak, M.; Li, Z.; Virtamo, J.; Albanes, D.; Chow, W.H.; Purdue, M.P. Prediagnostic circulating adipokine concentrations and risk of renal cell carcinoma in male smokers. Carcinogenesis 2013, 34, 109–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bråkenhielm, E.; Veitonmäki, N.; Cao, R.; Kihara, S.; Matsuzawa, Y.; Zhivotovsky, B.; Funahashi, T.; Cao, Y. Adiponectin-induced antiangiogenesis and antitumor activity involve caspase-mediated endothelial cell apoptosis. Proc. Natl. Acad. Sci. USA 2004, 101, 2476–2481. [Google Scholar] [CrossRef] [Green Version]
- Khandekar, M.J.; Cohen, P.; Spiegelman, B.M. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 2011, 11, 886–895. [Google Scholar] [CrossRef] [PubMed]
- McClain, K.M.; Moore, S.C.; Sampson, J.N.; Henderson, T.R.; Gebauer, S.K.; Newman, J.W.; Ross, S.; Pedersen, T.L.; Baer, D.J.; A Zanetti, K. Preanalytical Sample Handling Conditions and Their Effects on the Human Serum Metabolome in Epidemiologic Studies. Am. J. Epidemiol. 2021, 190, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Townsend, M.K.; Clish, C.B.; Kraft, P.; Wu, C.; Souza, A.L.; A Deik, A.; Tworoger, S.S.; Wolpin, B.M. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin. Chem. 2013, 59, 1657–1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimberly, W.T.; O’Sullivan, J.; Nath, A.K.; Keyes, M.; Shi, X.; Larson, M.G.; Yang, Q.; Long, M.T.; Vasan, R.; Peterson, R.T.; et al. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight 2017, 2, e92989. [Google Scholar] [CrossRef] [Green Version]
- Moore, S.C.; Mazzilli, K.M.; Sampson, J.N.; Matthews, C.E.; Carter, B.D.; Playdon, M.C.; Wang, Y.; Stevens, V.L. A metabolomics analysis of postmenopausal breast cancer risk in the cancer prevention study II. Metabolites 2021, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.C.; Playdon, M.C.; Sampson, J.N.; Hoover, R.N.; Trabert, B.; Matthews, C.E.; Ziegler, R.G. A Metabolomics Analysis of Body Mass Index and Postmenopausal Breast Cancer Risk. J. Natl. Cancer Inst. 2018, 110, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Playdon, M.C.; Ziegler, R.G.; Sampson, J.N.; Stolzenberg-Solomon, R.; Thompson, H.J.; Irwin, M.L.; Mayne, S.T.; Hoover, R.N.; Moore, S. Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 2017, 106, 637–649. [Google Scholar] [CrossRef] [Green Version]
- VanderWeele, T.J.; Vansteelandt, S. Odds ratios for mediation analysis for a dichotomous outcome. Am. J. Epidemiol. 2010, 172, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, R.P.; Vaidyanathan, N.; Rengasamy, M.; Oommen, A.M.; Somaiya, N.; Jagannath, M.R. Sphingolipid metabolic pathway: An overview of major roles played in human diseases. J. Lipids 2013, 2013, 178910. [Google Scholar]
- Batchuluun, B.; Al Rijjal, D.; Prentice, K.J.; Eversley, J.A.; Burdett, E.; Mohan, H.; Bhattacharjee, A.; Gunderson, E.P.; Liu, Y.; Wheeler, M.B. Elevated medium-chain acylcarnitines are associated with gestational diabetes mellitus and early progression to type 2 diabetes and induce pancreatic β-cell dysfunction. Diabetes 2018, 67, 885–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.J.; Kwak, S.-Y.; Jo, G.; Song, T.-J.; Shin, M.-J. Serum metabolite profile associated with incident type 2 diabetes in Koreans: Findings from the Korean Genome and Epidemiology Study. Sci. Rep. 2018, 8, 8207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olshan, A.F.; Kuo, T.; Meyer, A.; Nielsen, M.E.; Purdue, M.P.; Rathmell, W.K. Racial difference in histologic subtype of renal cell carcinoma. Cancer Med. 2013, 2, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Mao, Y.; White, K.; Canadian Cancer Registries Epidemiology Research Group. Renal cell carcinoma and occupational exposure to chemicals in Canada. Occup. Med. 2002, 52, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Buhagen, M.; Grønskag, A.; Ragde, S.F.; Hilt, B. Association between kidney cancer and occupational exposure to trichloroethylene. J. Occup. Environ. Med. 2016, 58, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
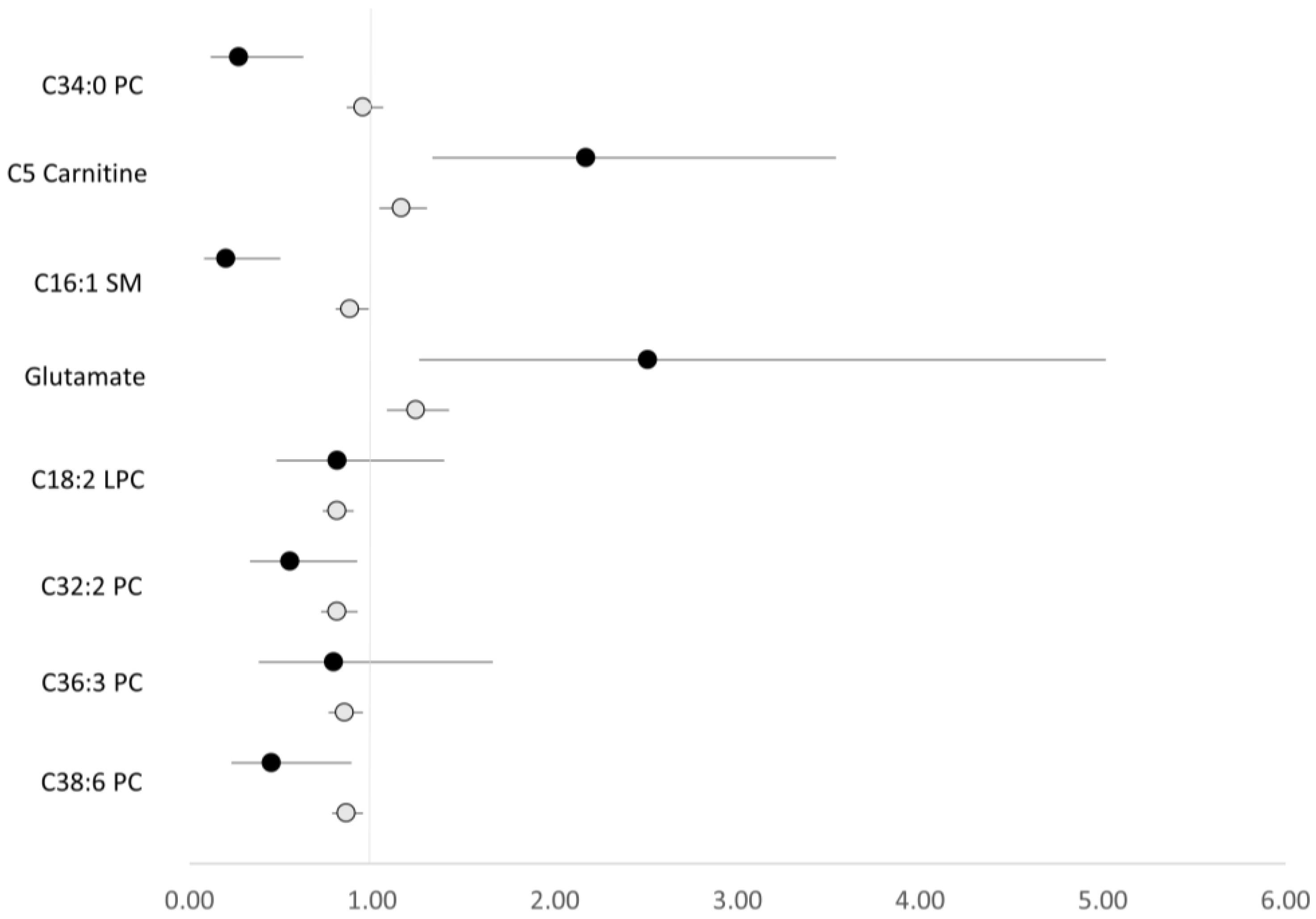
| Characteristic | Cases (n = 267) | Controls (n = 267) | p-Value |
|---|---|---|---|
| Age (years), mean ± SD | 63.0 ± 4.98 | 63.0 ± 4.97 | Matched |
| Sex, % | Matched | ||
| Male | 65.9 | 65.9 | |
| Female | 34.1 | 34.1 | |
| Race/ethnicity, % | 0.82 | ||
| White, non-Hispanic | 89.1 | 89.1 | |
| Black, non-Hispanic | 6.4 | 6.4 | |
| Other * | 4.5 | 4.5 | |
| Body mass index (kg/m2), mean ± SD | 28.9 ± 5.10 | 27.4 ± 4.20 | <0.0001 |
| Body mass index category, % | 0.001 | ||
| 0–18.5 kg/m2 | 0.8 | 0.8 | |
| 18.5–25 kg/m2 | 21.0 | 28.6 | |
| 25–30 kg/m2 | 41.2 | 48.5 | |
| 30+ kg/m2 | 37.1 | 22.2 | |
| Physical activity, % | 0.33 | ||
| None | 15.0 | 13.0 | |
| <1 h/week | 23.6 | 21.0 | |
| 1 h/week | 13.4 | 13.4 | |
| 2 h/week | 15.8 | 12.7 | |
| 3 h/week | 10.6 | 17.4 | |
| 4+ h/week | 21.7 | 22.5 | |
| History of diabetes, % | |||
| No | 88.8 | 90.9 | 0.41 |
| Yes | 11.2 | 9.1 | |
| History of hypertension, % | 0.08 | ||
| No | 54.5 | 62.0 | |
| Yes | 45.5 | 38.0 | |
| Cigarette smoking status, % | 0.94 | ||
| Never | 46.1 | 47.6 | |
| Former | 43.8 | 42.3 | |
| Current | 10.1 | 10.1 | |
| Alcohol consumption (g/day), mean ± SD | 9.2 ± 21.17 | 13.2 ± 29.32 | 0.07 |
| Family history of renal cancer, % | 0.46 | ||
| No | 93.2 | 95.5 | |
| Yes | 1.5 | 1.5 | |
| Unsure | 5.3 | 3.0 |
| Metabolite | Model 1 † OR (95% CI) | Model 2 ‡ OR (95% CI) |
|---|---|---|
| Glycerophospholipids | ||
| C38:4 PI | 0.35 (0.21–0.61) | 0.32 (0.18–0.58) |
| C34:0 PC | 0.43 (0.26–0.72) | 0.43 (0.26–0.74) |
| Fatty acyls (acylcarnitines) | ||
| C3-DC-CH3 Carnitine | 2.83 (1.73–4.64) | 2.61 (1.53–4.47) |
| C5 Carnitine | 2.88 (1.74–4.76) | 2.31 (1.36–3.93) |
| Sphingolipids | ||
| C14:0 SM | 0.45 (0.26–0.73) | 0.40 (0.24–0.68) |
| Organic nitrogen compounds | ||
| C16:1 SM | 0.40 (0.23–0.70) | 0.34 (0.19–0.63) |
| Metabolite | Chemical Class | Order Entered | Model Entry p * | Mutually Adjusted OR † (95% CI) | Mutually Adjusted p * |
|---|---|---|---|---|---|
| C38:4 PI | Glycerophospholipids | 1 | <0.0001 | 0.49 (0.26–0.95) | 0.03 |
| C3-DC-CH3 Carnitine | Fatty acyls (acylcarnitine) | 2 | 0.002 | 2.39 (1.39–4.13) | 0.002 |
| C14:0 SM | Sphingolipids | 3 | 0.02 | 0.52 (0.29–0.92) | 0.02 |
| Metabolite | BMI OR (95% CI) * | Attenuation of Log (OR) |
|---|---|---|
| None | 1.44 (1.19–1.74) | - |
| C38:4 PI | 1.51 (1.23–1.85) | −13.3% |
| C34:0 PC | 1.46 (1.20–1.77) | −3.8% |
| C3-DC-CH3 Carnitine | 1.34 (1.09–1.64) | 19.7% |
| C5 Carnitine | 1.33 (1.08–1.62) | 22.4% |
| C14:0 SM | 1.52 (1.24–1.86) | −15.7% |
| C16:1 SM | 1.52 (1.24–1.85) | −14.8% |
| Metabolite | Metabolite OR (95% CI) * Not BMI-Adjusted | Metabolite OR (95% CI) * BMI-Adjusted | Attenuation of Log (Metabolite OR) |
|---|---|---|---|
| C38:4 PI | 0.35 (0.21–0.61) | 0.31 (0.18–0.56) | −11.6% |
| C34:0 PC | 0.43 (0.26–0.72) | 0.41 (0.24–0.69) | −5.6% |
| C3-DC-CH3 Carnitine | 2.83 (1.73–4.64) | 2.49 (1.49–4.17) | 12.3% |
| C5 Carnitine | 2.88 (1.74–4.76) | 2.41 (1.44–4.05) | 16.8% |
| C14:0 SM | 0.45 (0.27–0.73) | 0.39 (0.23–0.65) | −14.7% |
| C16:1 SM | 0.40 (0.23–0.71) | 0.33 (0.18–0.60) | −21.0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McClain, K.M.; Sampson, J.N.; Petrick, J.L.; Mazzilli, K.M.; Gerszten, R.E.; Clish, C.B.; Purdue, M.P.; Lipworth, L.; Moore, S.C. Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Metabolites 2022, 12, 1189. https://doi.org/10.3390/metabo12121189
McClain KM, Sampson JN, Petrick JL, Mazzilli KM, Gerszten RE, Clish CB, Purdue MP, Lipworth L, Moore SC. Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Metabolites. 2022; 12(12):1189. https://doi.org/10.3390/metabo12121189
Chicago/Turabian StyleMcClain, Kathleen M., Joshua N. Sampson, Jessica L. Petrick, Kaitlyn M. Mazzilli, Robert E. Gerszten, Clary B. Clish, Mark P. Purdue, Loren Lipworth, and Steven C. Moore. 2022. "Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial" Metabolites 12, no. 12: 1189. https://doi.org/10.3390/metabo12121189
APA StyleMcClain, K. M., Sampson, J. N., Petrick, J. L., Mazzilli, K. M., Gerszten, R. E., Clish, C. B., Purdue, M. P., Lipworth, L., & Moore, S. C. (2022). Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Metabolites, 12(12), 1189. https://doi.org/10.3390/metabo12121189






