Integrated Analysis of Metabolome and Transcriptome Reveals the Difference in Flavonoid Biosynthesis between the Red- and White-Sarcocarp Pomelo Fruits
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. UPLC and Electrospray Ionization-Triple Quadrupole Linear Ion Trap (ESI-Q TRAP)-MS/MS
2.3. RNA-Seq Analysis
2.4. Gene Expression Analysis by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.5. Statistical Analysis
3. Results
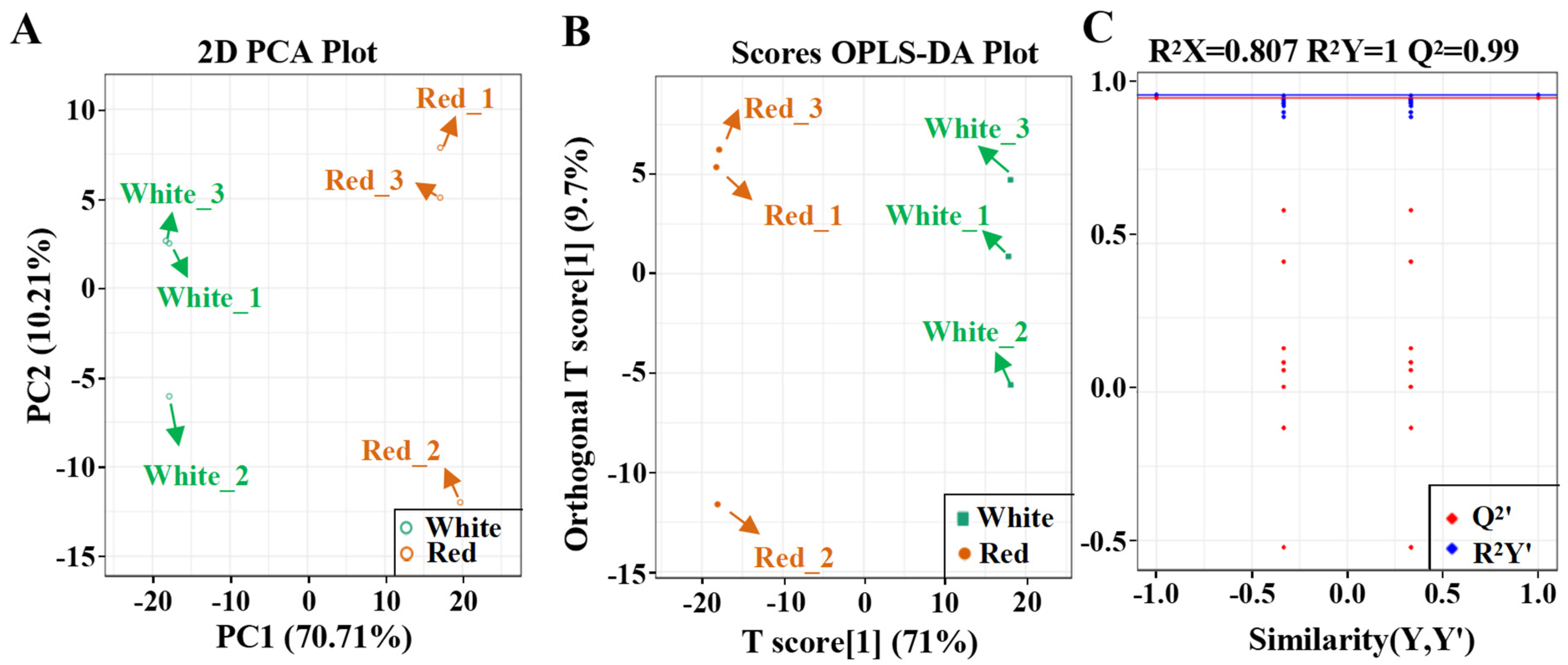
3.1. PCA and OPLS–DA Analysis in the Fruit of White and Red Pomelo Cultivars
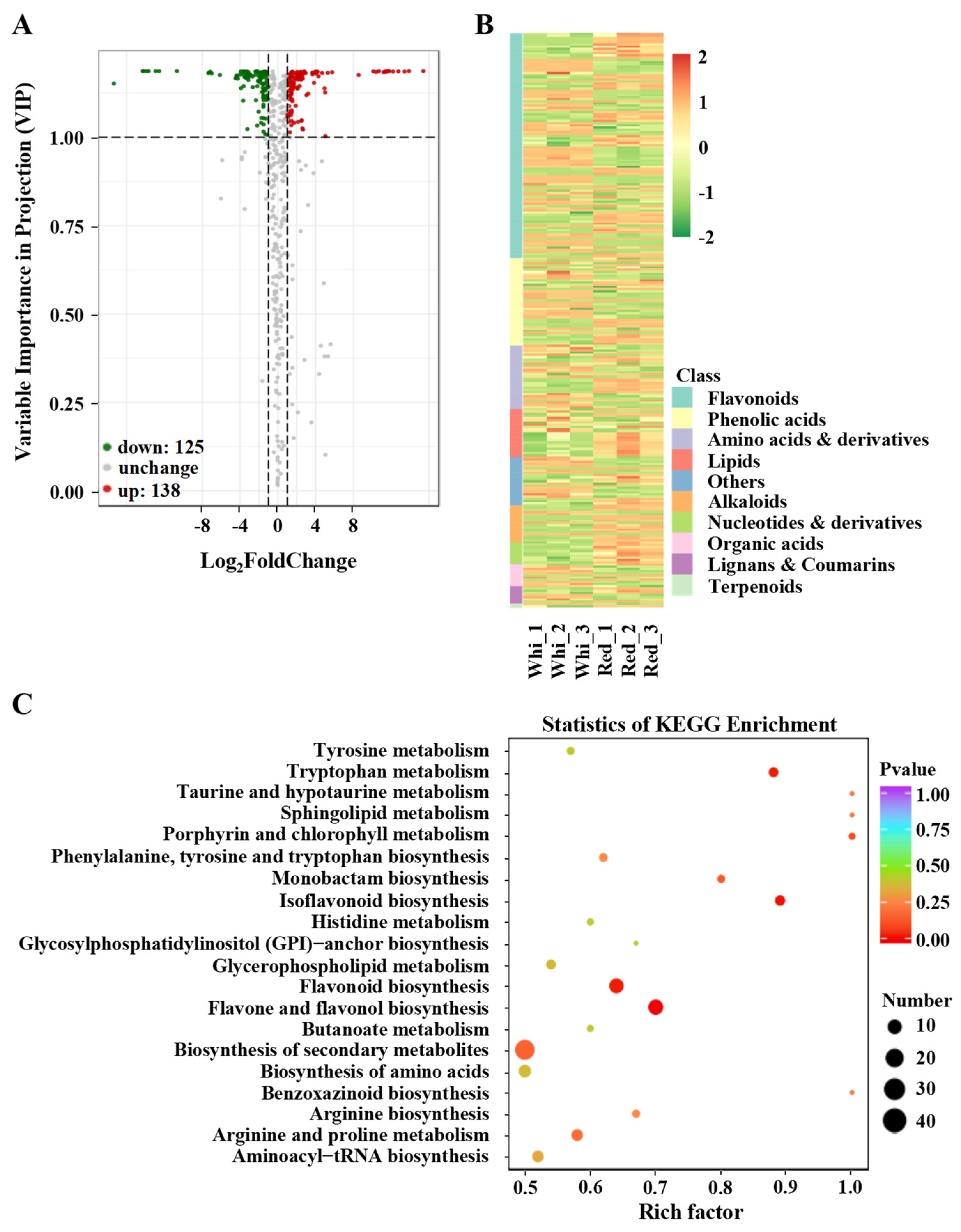
3.2. Screening of Significantly Changed Flavonoids
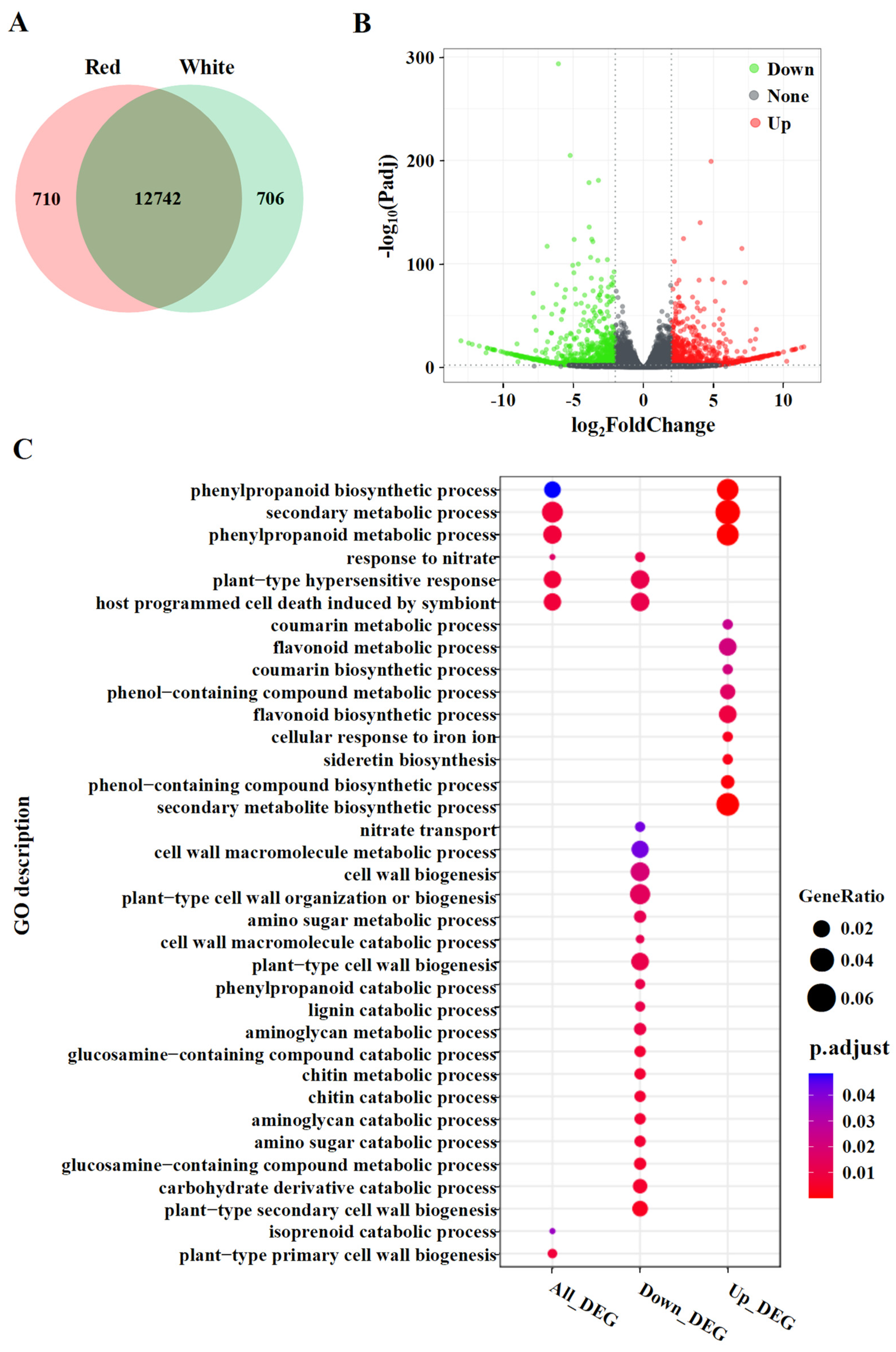
3.3. Analysis of DEGs and Gene Ontology (GO) Enrichment in Red- and White-Sarcocarp Pomelo
3.4. Correlation Network Analysis of DEGs and SCMs Related to Flavonoid Biosynthesis
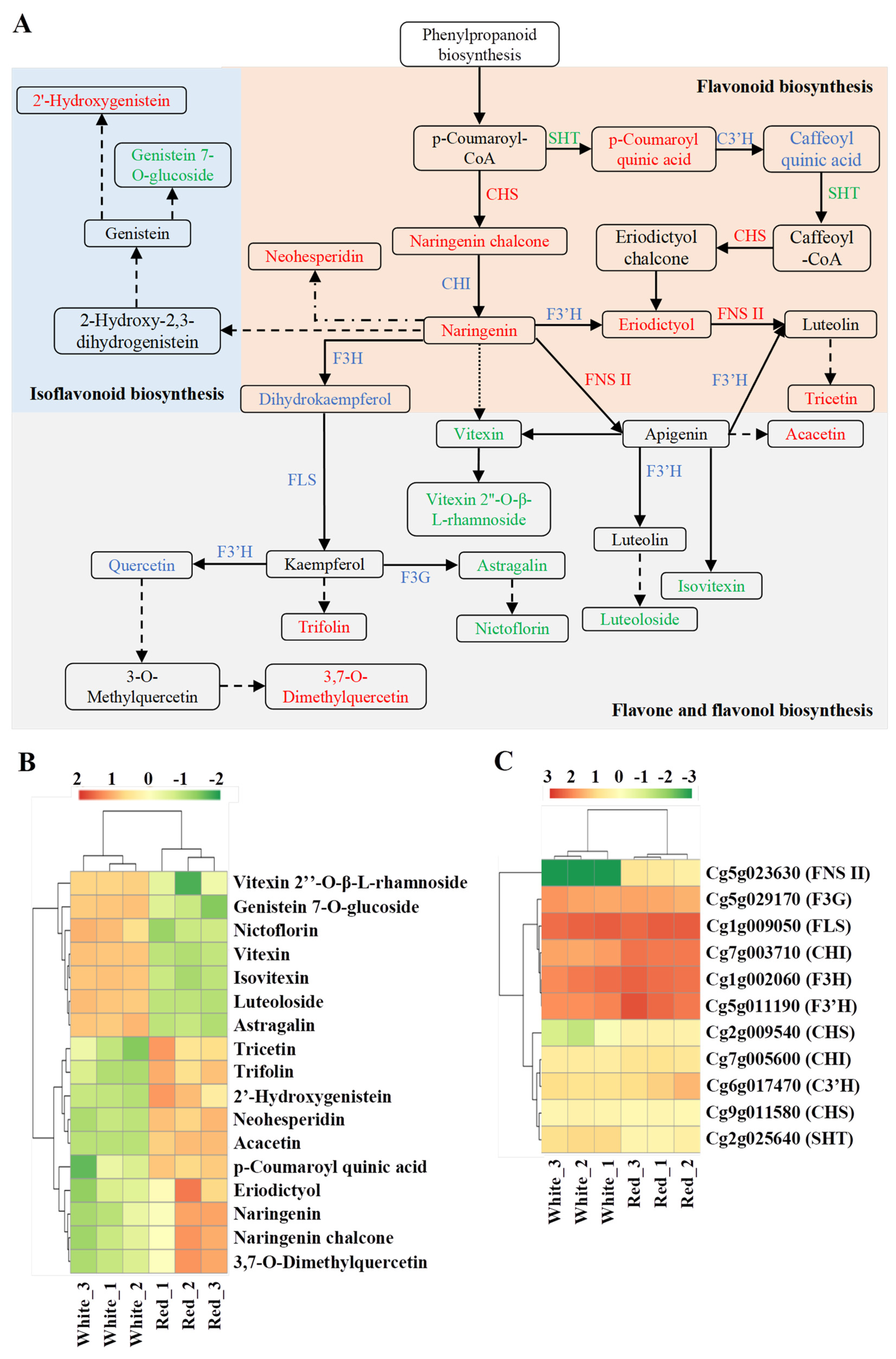
3.5. Analysis of Flavonoid Biosynthesis Network
3.6. qRT-PCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brunetti, C.; Fini, A.; Sebastiani, F.; Gori, A.; Tattini, M. Modulation of Phytohormone Signaling: A Primary Function of Flavonoids in Plant-Environment Interactions. Front. Plant Sci. 2018, 9, 1042. [Google Scholar] [CrossRef]
- Li, J.; Hossain, M.S.; Ma, H.; Yang, Q.; Gong, X.; Yang, P.; Feng, B. Comparative metabolomics reveals differences in flavonoid metabolites among different coloured buckwheat flowers. J. Food Compos. Anal. 2020, 85, 103335. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, J.; Liu, Z.; Wang, J.; Yang, B.; Chen, W.; Ou, L.; Dai, X.; Zhang, Z.; Zou, X. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.). Food Chem. 2020, 306, 125629. [Google Scholar] [CrossRef]
- Zhu, H.H.; Yang, J.X.; Xiao, C.H.; Mao, T.Y.; Zhang, J.; Zhang, H.Y. Differences in flavonoid pathway metabolites and transcripts affect yellow petal colouration in the aquatic plant Nelumbo nucifera. BMC Plant Biol. 2019, 19, 277. [Google Scholar] [CrossRef] [PubMed]
- Ohkatsu, Y.; Sakurai, T.; Sato, T. Relationship between Chemical Structure and Antioxidant Function of Flavonoids. J. Jpn. Pet. Inst. 2010, 53, 213–221. [Google Scholar] [CrossRef]
- Fardoun, M.M.; Maaliki, D.; Halabi, N.; Iratni, R.; Bitto, A.; Baydoun, E.; Eid, A.H. Flavonoids in adipose tissue inflammation and atherosclerosis: One arrow, two targets. Clin. Sci. 2020, 134, 1403–1432. [Google Scholar] [CrossRef] [PubMed]
- Jian, T.; Chen, J.; Ding, X.; Lv, H.; Li, J.; Wu, Y.; Ren, B.; Tong, B.; Zuo, Y.; Su, K. Flavonoids isolated from loquat (Eriobotrya japonica) leaves inhibit oxidative stress and inflammation induced by cigarette smoke in COPD mice: The role of TRPV1 signaling pathways. Food Funct. 2020, 11, 3516–3526. [Google Scholar] [CrossRef] [PubMed]
- Millar, C.L.; Duclos, Q.; Blesso, C.N. Effects of dietary flavonoids on reverse cholesterol transport, HDL metabolism, and HDL function. Adv. Nutr. 2017, 8, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, F.; Lian, Y.; Xiao, H.; Zheng, J. Biosynthesis of citrus flavonoids and their health effects. Crit. Rev. Food Sci. Nutr. 2020, 60, 566–583. [Google Scholar] [CrossRef]
- Pfeiffer, P.; Hegedűs, A. Review of the molecular genetics of flavonoid biosynthesis in fruits. Acta Aliment. 2011, 40, 150–163. [Google Scholar] [CrossRef]
- Matsui, K.; Walker, A.R. Biosynthesis and regulation of flavonoids in buckwheat. Breed. Sci. 2020, 70, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Agati, G.; Biricolti, S.; Guidi, L.; Ferrini, F.; Fini, A.; Tattini, M. The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. J. Plant Physiol. 2011, 168, 204–212. [Google Scholar] [CrossRef]
- Guimarães, R.; Barros, L.; Barreira, J.C.; Sousa, M.J.; Carvalho, A.M.; Ferreira, I.C. Targeting excessive free radicals with peels and juices of citrus fruits: Grapefruit, lemon, lime and orange. Food Chem. Toxicol. 2010, 48, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, K.A.; Haldavanekar, P.C.; Parulekar, Y.R.; Haldankar, P.M. Survey and characterization of pummelo genotypes (Citrus grandis L. Osbeck) grown in coastal region of Maharashtra. Ecoscan 2015, 8, 371–380. [Google Scholar]
- Liu, Y.; Heying, E.; Tanumihardjo, S.A. History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Food Saf. 2012, 11, 530–545. [Google Scholar] [CrossRef]
- Jiang, K.; Song, Q.; Wang, L.; Xie, T.; Wu, X.; Wang, P.; Yin, G.; Ye, W.; Wang, T. Antitussive, expectorant and anti-inflammatory activities of different extracts from Exocarpium Citri grandis. J. Ethnopharmacol. 2014, 156, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wu, H.; Su, W.; Shi, R.; Li, P.; Liao, Y.; Wang, Y.; Li, P. Effects of Total flavonoids from exocarpium citri grandis on air pollution particle-induced pulmonary inflammation and oxidative stress in mice. J. Food Sci. 2019, 84, 3843–3849. [Google Scholar] [CrossRef]
- Xu, G.; Liu, D.; Chen, J.; Ye, X.; Ma, Y.; Shi, J. Juice components and antioxidant capacity of citrus varieties cultivated in China. Food Chem. 2008, 106, 545–551. [Google Scholar] [CrossRef]
- Zhang, M.; Duan, C.; Zang, Y.; Huang, Z.; Liu, G. The flavonoid composition of flavedo and juice from the pummelo cultivar (Citrus grandis (L.) Osbeck) and the grapefruit cultivar (Citrus paradisi) from China. Food Chem. 2011, 129, 1530–1536. [Google Scholar] [CrossRef]
- Wang, F.; Chen, L.; Chen, H.; Chen, S.; Liu, Y. Analysis of flavonoid metabolites in citrus peels (Citrus reticulata “Dahongpao”) using UPLC-ESI-MS/MS. Molecules 2019, 24, 2680. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Song, J. Analysis of widely targeted metabolites of the euhalophyte Suaeda salsa under saline conditions provides new insights into salt tolerance and nutritional value in halophytic species. BMC Plant Biol. 2019, 19, 388. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sun, J.; Yang, Z.; Zhao, C.; Zhu, M.; Ma, D.; Dong, T.; Zhou, Z.; Liu, M.; Yang, D. Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative Ipomoea trifida. Gene 2019, 686, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, Y.; Shi, T.; Kou, M.; Sun, J.; Xu, T.; Li, Q.; Wu, S.; Cao, Q.; Hou, W. Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals the regulatory architecture of gene expression variation in the storage roots of sweet potato. Hortic. Res. 2020, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Dong, T.; Xu, T.; Kang, H. Identification, characterisation and expression analysis of MADS-box genes in sweetpotato wild relative Ipomoea trifida. Acta Physiol. Plant 2020, 42, 163. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Nawaz, G.; Zhao, C.; Li, Y.; Dong, T.; Zhu, M.; Du, X.; Zhang, L.; Li, Z. Exogenous melatonin attenuates post-harvest decay by increasing antioxidant activity in wax apple (Syzygium samarangense). Front. Plant Sci. 2020, 11, 569779. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, P.; Kang, H.; Liu, L.; Cao, Q.; Sun, J.; Dong, T.; Zhu, M.; Li, Z.; Xu, T. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genom. 2020, 21, 164. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Zhang, L.; Nawaz, G.; Zhao, C.; Zhang, J.; Cao, Q.; Dong, T.; Xu, T. Exogenous melatonin alleviates browning of fresh-cut sweetpotato by enhancing anti-oxidative process. Sci. Hortic. 2022, 297, 110937. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qiu, C.; Ding, Y.; Wang, Y.; Sun, L.; Fan, K.; Gai, Z.; Dong, G.; Wang, J.; Li, X. Fulvic acid ameliorates drought stress-induced damage in tea plants by regulating the ascorbate metabolism and flavonoids biosynthesis. BMC Genom. 2020, 21, 411. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Zhou, Z.; Ren, R.; Xu, M. Apigenin, flavonoid component isolated from Gentiana veitchiorum flower suppresses the oxidative stress through LDLR-LCAT signaling pathway. Biomed. Pharmacother. 2020, 128, 110298. [Google Scholar] [CrossRef]
- Wink, M. Evolution of secondary metabolites in legumes (Fabaceae). S. Afr. J. Bot. 2013, 89, 164–175. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Li, S.; Xi, X.; Cao, D.; Wang, Z.; Wang, R.; Liu, B. Comprehensive influences of overexpression of a MYB transcriptor regulating anthocyanin biosynthesis on transcriptome and metabolome of tobacco leaves. Int. J. Mol. Sci. 2019, 20, 5123. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Shin, D.H.; Choi, S.; Park, Y. Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol. Cells 2012, 34, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Koskimäki, J.J.; Hokkanen, J.; Jaakola, L.; Suorsa, M.; Tolonen, A.; Mattila, S.; Pirttilä, A.M.; Hohtola, A. Flavonoid biosynthesis and degradation play a role in early defence responses of bilberry (Vaccinium myrtillus) against biotic stress. Eur. J. Plant Pathol. 2009, 125, 629–640. [Google Scholar] [CrossRef]
- Gharibi, S.; Tabatabaei, B.E.S.; Saeidi, G.; Talebi, M.; Matkowski, A. The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech. f. Phytochemistry 2019, 162, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Lazar, G.; Goodman, H.M. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 472–476. [Google Scholar] [CrossRef]
- Tan, G.; Ma, J.; Zhang, X.; Xu, Z.; Xiong, A. AgFNS overexpression increase apigenin and decrease anthocyanins in petioles of transgenic celery. Plant Sci. 2017, 263, 31–38. [Google Scholar] [CrossRef]
- Righini, S.; Rodriguez, E.J.; Berosich, C.; Grotewold, E.; Casati, P.; Falcone Ferreyra, M.L. Apigenin produced by maize flavone synthase I and II protects plants against UV-B-induced damage. Plant, Cell Environ. 2019, 42, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Yazawa, T.; Kasuga, S.; Sawada, Y.; Kanamori, H.; Ogo, Y.; Hirai, M.Y.; Matsumoto, T.; Kawahigashi, H. Expression of flavone synthase II and flavonoid 3′-hydroxylase is associated with color variation in tan-colored injured leaves of sorghum. Front. Plant Sci. 2016, 7, 1718. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Wang, J.; Li, Y.; Zhang, L.; Nawaz, G.; Wu, S.; Xu, T. Integrated Analysis of Metabolome and Transcriptome Reveals the Difference in Flavonoid Biosynthesis between the Red- and White-Sarcocarp Pomelo Fruits. Metabolites 2022, 12, 1161. https://doi.org/10.3390/metabo12121161
Zhao C, Wang J, Li Y, Zhang L, Nawaz G, Wu S, Xu T. Integrated Analysis of Metabolome and Transcriptome Reveals the Difference in Flavonoid Biosynthesis between the Red- and White-Sarcocarp Pomelo Fruits. Metabolites. 2022; 12(12):1161. https://doi.org/10.3390/metabo12121161
Chicago/Turabian StyleZhao, Chenxu, Jiajia Wang, Yuxia Li, Lei Zhang, Ghazala Nawaz, Shaoyuan Wu, and Tao Xu. 2022. "Integrated Analysis of Metabolome and Transcriptome Reveals the Difference in Flavonoid Biosynthesis between the Red- and White-Sarcocarp Pomelo Fruits" Metabolites 12, no. 12: 1161. https://doi.org/10.3390/metabo12121161
APA StyleZhao, C., Wang, J., Li, Y., Zhang, L., Nawaz, G., Wu, S., & Xu, T. (2022). Integrated Analysis of Metabolome and Transcriptome Reveals the Difference in Flavonoid Biosynthesis between the Red- and White-Sarcocarp Pomelo Fruits. Metabolites, 12(12), 1161. https://doi.org/10.3390/metabo12121161







