Dysbiosis of Gut Microbiota from the Perspective of the Gut–Brain Axis: Role in the Provocation of Neurological Disorders
Abstract
1. Introduction
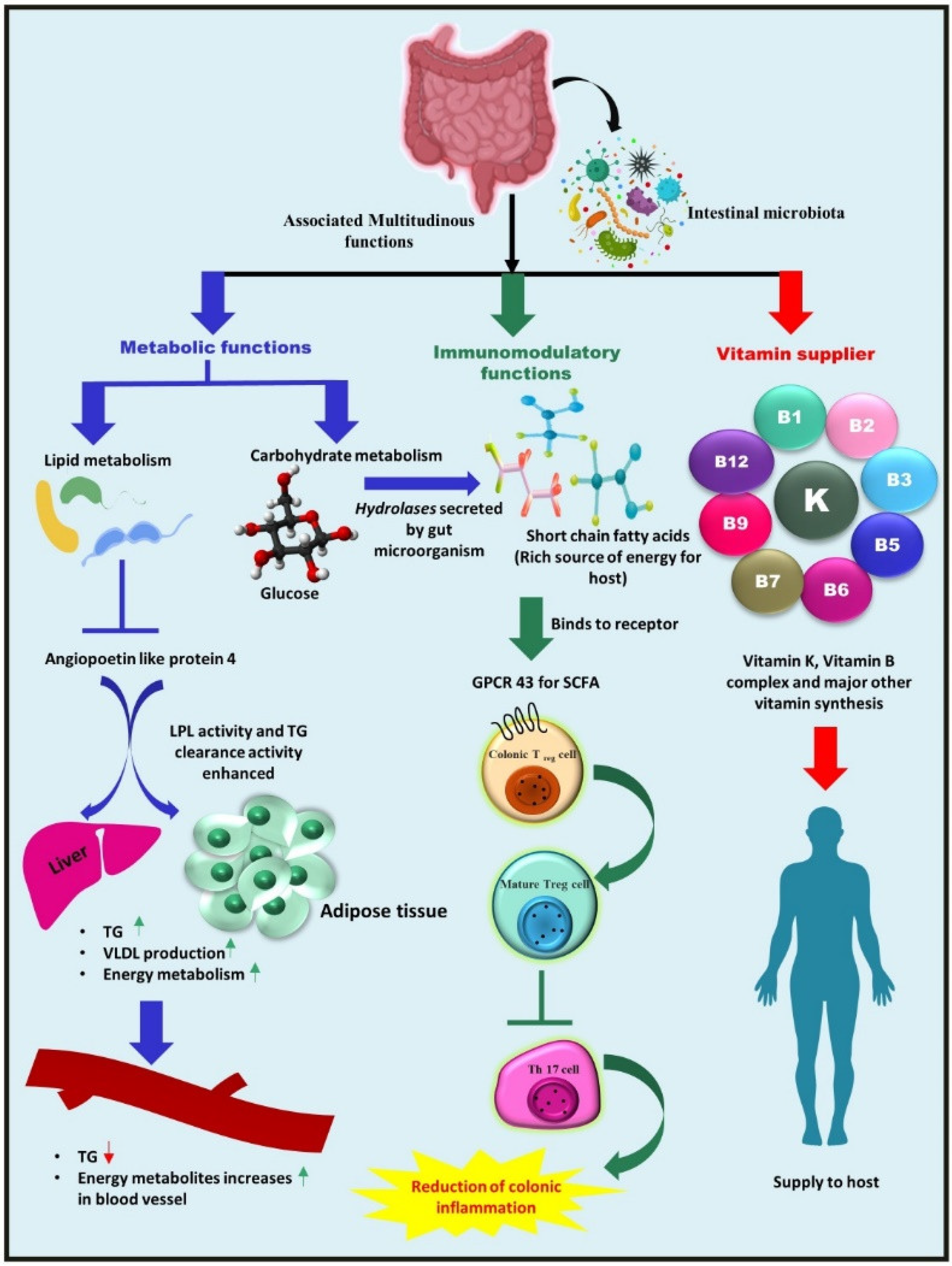
2. Composition of Gut Microbiota and Its Associated Multifarious Function in Host Heath
2.1. Metabolic Function—Nutrient and Other Dietary Component Metabolism
2.2. Vitamins Supplier—Bacteria as a Source of Vitamins for Their Hosts
2.3. Immunomodulatory Role of Gut Microbiota
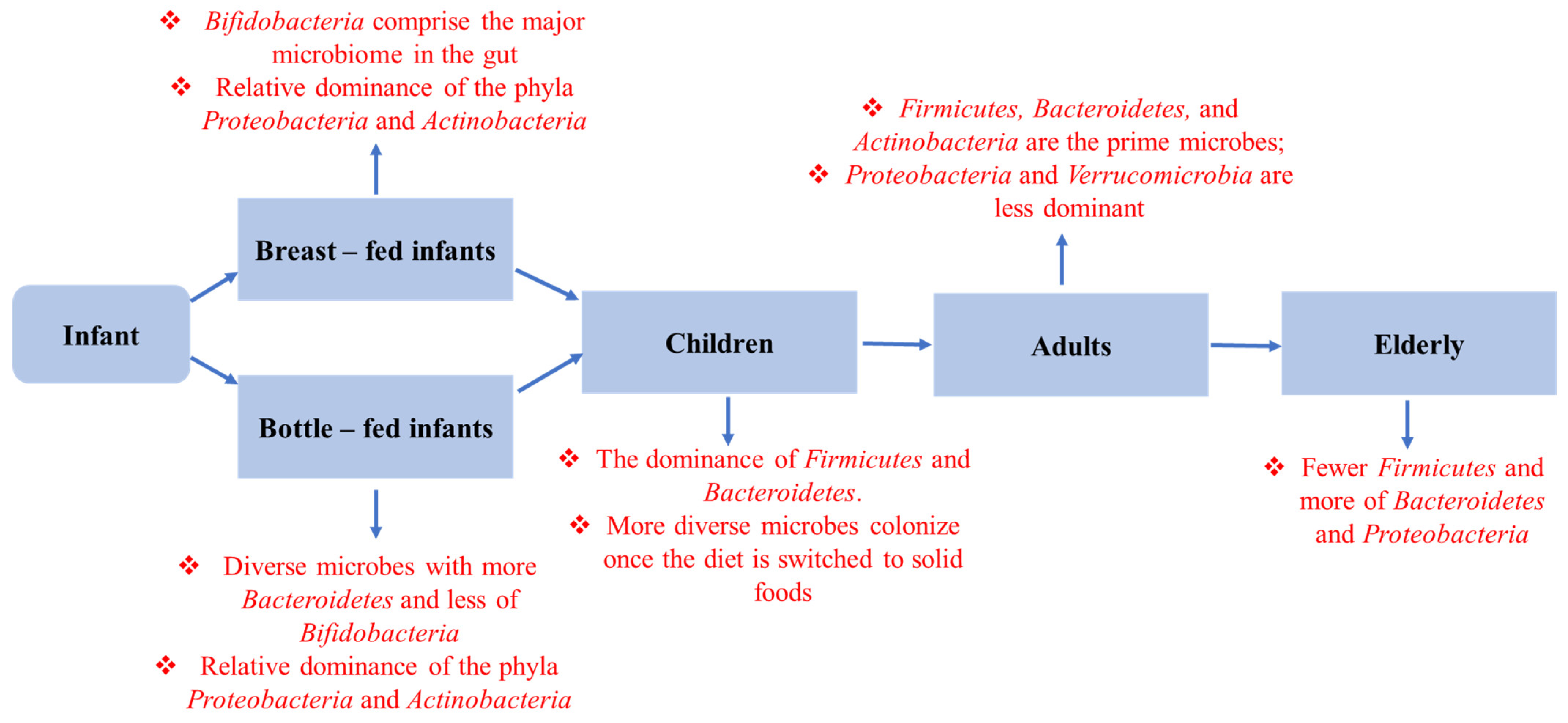
3. Host Factors Controlling Gut Microbiota
3.1. Delivery Pattern
3.2. Infant Feeding
3.3. Medication: Antibiotics
3.4. Genetics: mi-RNAs
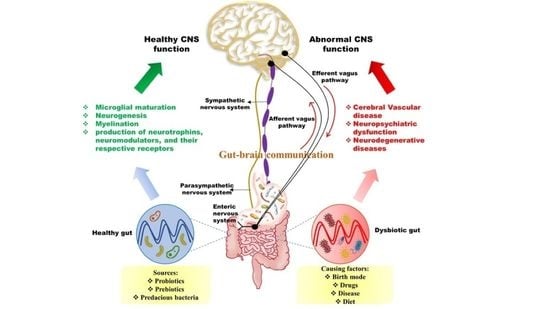
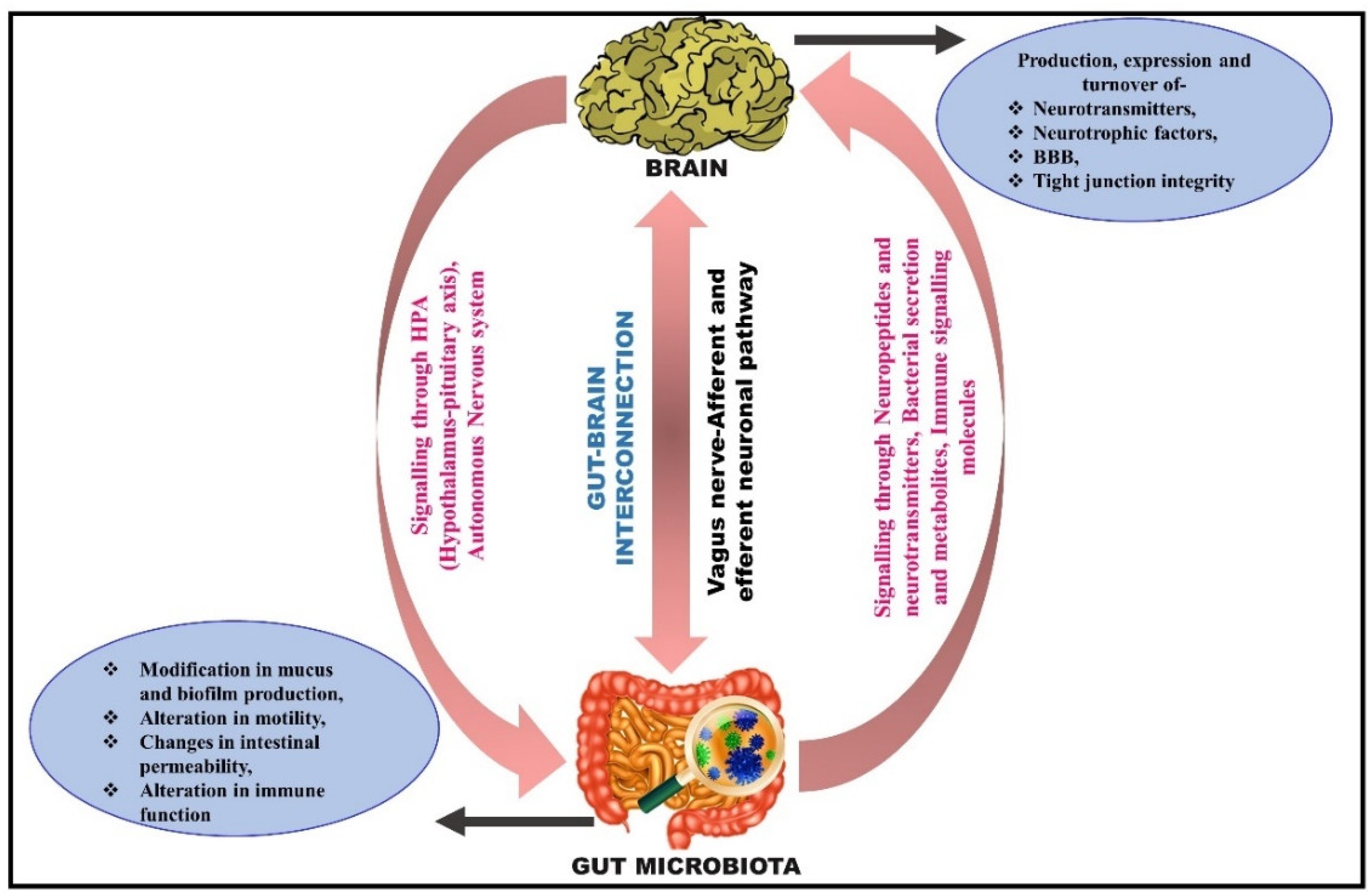
4. Microbiota–Gut Brain Interconnection
5. Routeways for the Bidirectional Communication between the Gut and Brain
5.1. Neuronal Pathway: Activation of the Vagus Nerve
5.2. Microbial Signaling Molecules as a Pathway of Communication
5.3. Immune Signaling Pathway
6. Correlation between the Gut Microbiota and Cerebral Function
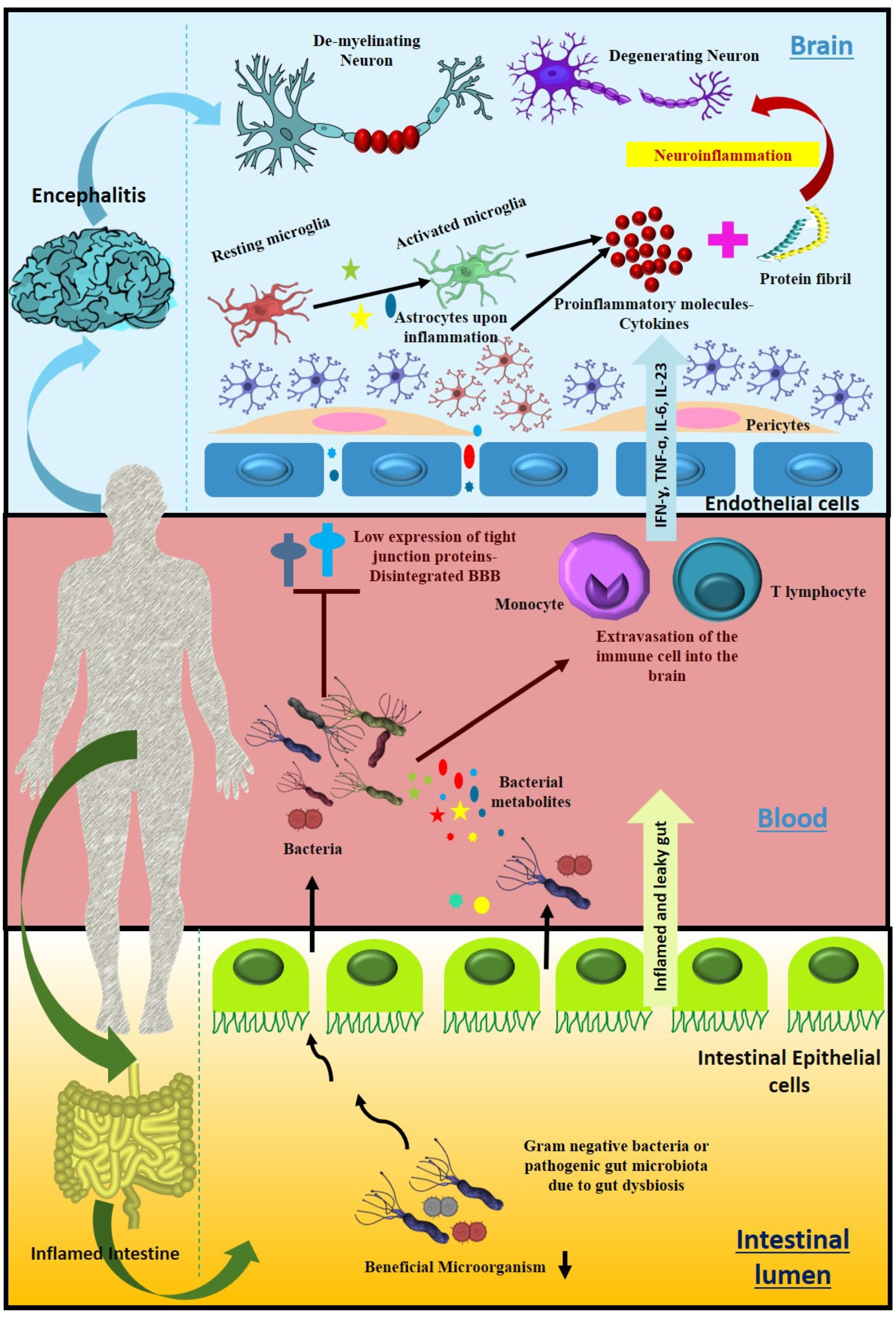
7. Gut Dysbacteriosis: Consequences, Diagnostic and Therapeutic Options
7.1. Microbial Imbalance Leads to Several Neurological Disorders
7.2. Strategies to Prevent Dysbiosis of Microbiota
7.3. Potential Biomarker for Dysbiosis and Its Implications
8. Effect of Intestinal Microbiota-Derived Metabolites on Neurological Disorders
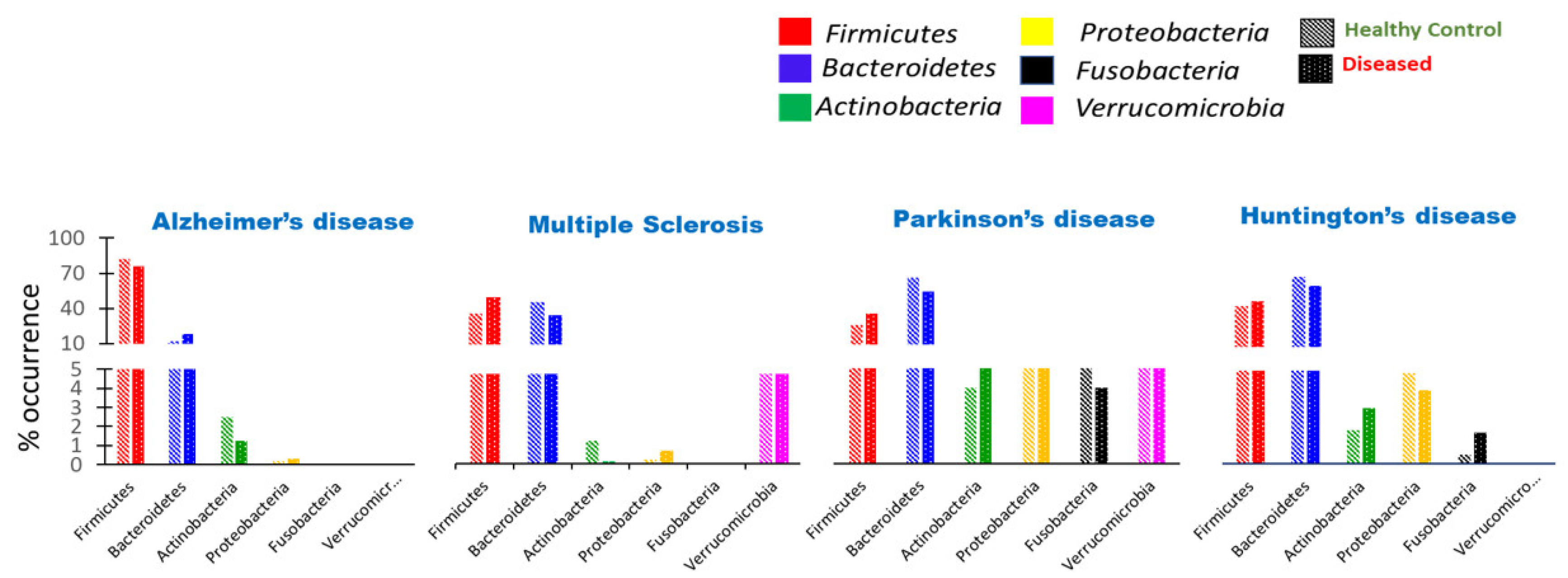
8.1. Aggregate-Forming Tendency of Gut Bacterial Proteins in Alzheimer’s Disease
8.2. Impact of Microbiota on the Induction of Parkinson’s Disease
8.3. Huntington’s Disease Association with Gut Dysbiosis
8.4. Substantial Alteration of Human Gut Microbiota in Multiple Sclerosis
9. Altered Bile Acid Profile Associates with Neurological Dysfunction
10. The Role of Dysbiosis in the Aging Process
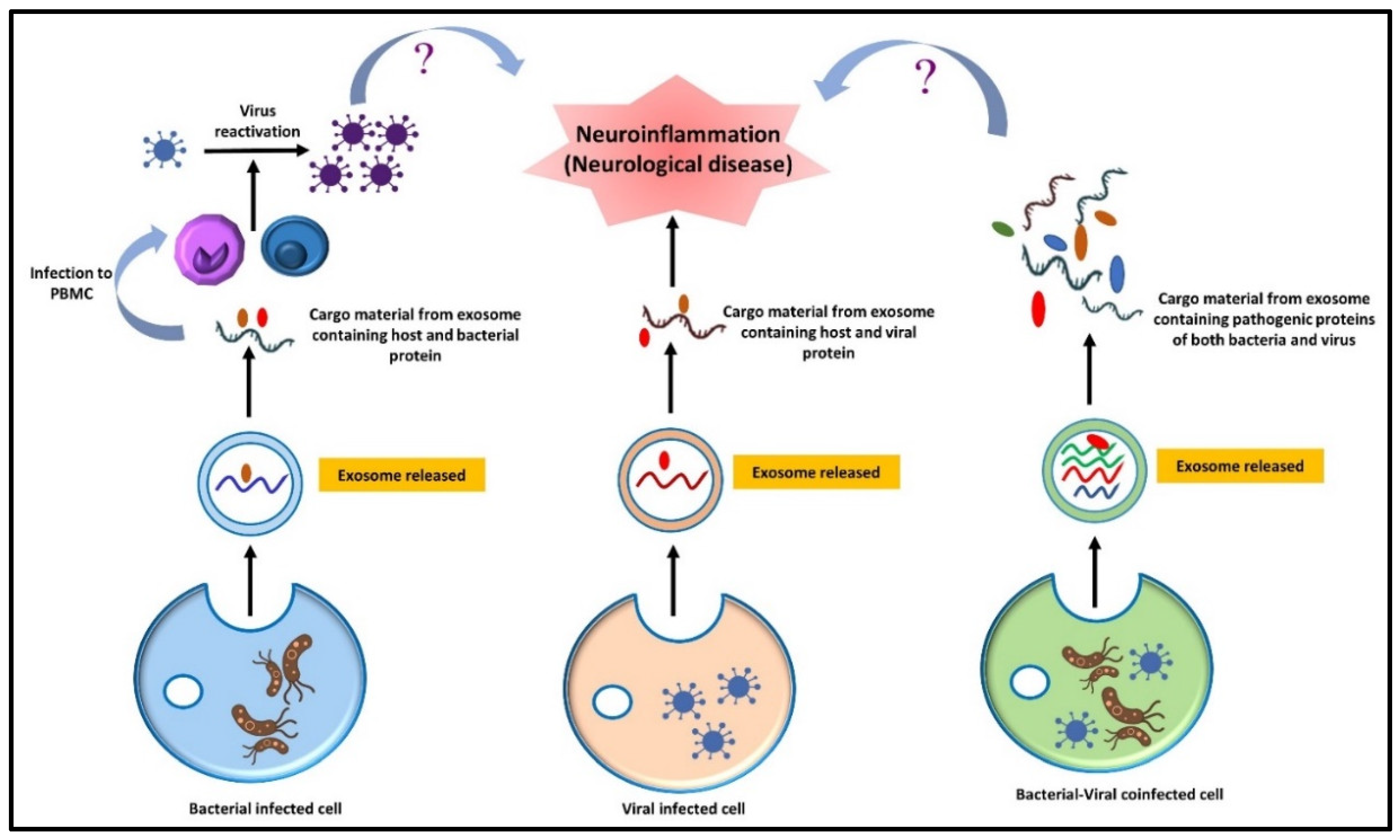
11. Synergetic Effect of Co-Infection and Microbial Interaction on a Neurological Disorder
12. Modulation of Gut Microbiota for Neurological Disorders in the Perspective of Host-Directed Therapy: Microbiota-Targeted Technique “Fecal Microbiota Transplantation”
13. Function of Modulated Microbial Communities in Healthy Aging and Rejuvenation
14. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Matijašić, M.; Meštrović, T.; Paljetak, H.; Perić, M.; Barešić, A.; Verbanac, D. Gut Microbiota beyond Bacteria—Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int. J. Mol. Sci. 2020, 21, 82668. [Google Scholar] [CrossRef] [PubMed]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2015, 65, 225–237. [Google Scholar] [CrossRef]
- Liu, S.; Gao, J.; Zhu, M.; Liu, K.; Zhang, H.-L. Gut Microbiota and Dysbiosis in Alzheimer’s Disease: Implications for Pathogenesis and Treatment. Mol. Neurobiol. 2020, 57, 5026–5043. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, Y.; Chaudhuri, K.R.; Reynolds, R.; Tan, E.-K.; Pettersson, S. The role of gut dysbiosis in Parkinson’s disease: Mechanistic insights and therapeutic options. Brain 2021, 144, 2571–2593. [Google Scholar] [CrossRef]
- Gkolfakis, P.; Dimitriadis, G.; Triantafyllou, K. Gut microbiota and non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 572–581. [Google Scholar] [CrossRef]
- Rodarte-Acosta, J.S.; Morales-Sánchez, M.A. El rol de la microbiota intestinal en la dermatitis atópica. Rev. Del Cent. Derm. Pascua 2021, 30, 76–83. [Google Scholar] [CrossRef]
- Yu, C.; Su, Z.; Li, Y.; Li, Y.; Liu, K.; Chu, F.; Liu, T.; Chen, R.; Ding, X. Dysbiosis of gut microbiota is associated with gastric carcinogenesis in rats. Biomed. Pharmacother. 2020, 126, 110036. [Google Scholar] [CrossRef]
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020, 202, 2147–2167. [Google Scholar] [CrossRef]
- The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef]
- Galland, L. The Gut Microbiome and the Brain. J. Med. Food 2014, 17, 1261–1272. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- Arnold, J.W.; Roach, J.; Azcarate-Peril, M.A. Emerging Technologies for Gut Microbiome Research. Trends Microbiol. 2016, 24, 887–901. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Z.; Yu, Y.-J.; Adeli, K. Role of Gut Microbiota in Neuroendocrine Regulation of Carbohydrate and Lipid Metabolism via the Microbiota-Gut-Brain-Liver Axis. Microorganisms 2020, 8, 527. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, C.H. Regulation of common neurological disorders by gut microbial metabolites. Exp. Mol. Med. 2021, 53, 1821–1833. [Google Scholar] [CrossRef] [PubMed]
- Korf, J.M.; Ganesh, B.P.; McCullough, L.D. Gut dysbiosis and age-related neurological diseases in females. Neurobiol. Dis. 2022, 168, 105695. [Google Scholar] [CrossRef] [PubMed]
- Baizabal-Carvallo, J.F.; Alonso-Juarez, M. The Link between Gut Dysbiosis and Neuroinflammation in Parkinson’s Disease. Neuroscience 2020, 432, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Wasser, C.I.; Mercieca, E.-C.; Kong, G.; Hannan, A.J.; McKeown, S.J.; Glikmann-Johnston, Y.; Stout, J.C. Gut dysbiosis in Huntington’s disease: Associations among gut microbiota, cognitive performance and clinical outcomes. Brain Commun. 2020, 2, 110. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Anwar, H.; Irfan, S.; Hussain, G.; Naeem Faisal, M.; Muzaffar, H.; Mustafa, I.; Mukhtar, I.; Malik, S.; Irfan Ullah, M. Gut Microbiome: A New Organ System in Body. In Parasitology and Microbiology Research; Antonio Bastidas Pacheco, G., Ali Kamboh, A., Eds.; IntechOpen: London, UK, 2020; ISBN 978-1-78985-901-0. [Google Scholar]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Boziki, M.K.; Mentis, A.-F.A.; Shumilina, M.; Makshakov, G.; Evdoshenko, E.; Grigoriadis, N. COVID-19 Immunopathology and the Central Nervous System: Implication for Multiple Sclerosis and Other Autoimmune Diseases with Associated Demyelination. Brain Sci. 2020, 10, 345. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Johnson, E.L.; Heaver, S.L.; Walters, W.A.; Ley, R.E. Microbiome and metabolic disease: Revisiting the bacterial phylum Bacteroidetes. Klin. Wochenschr. 2016, 95, 1–8. [Google Scholar] [CrossRef]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimaraes, V.D.; Sokol, H.; Dore, J.; Corthier, G.; Furet, J.-P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Devaraj, S.; Hemarajata, P.; Versalovic, J. The Human Gut Microbiome and Body Metabolism: Implications for Obesity and Diabetes. Clin. Chem. 2013, 59, 617–628. [Google Scholar] [CrossRef]
- Deleu, S.; Machiels, K.; Raes, J.; Verbeke, K.; Vermeire, S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? eBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Lombard, V.; Henrissat, B. Complex Carbohydrate Utilization by the Healthy Human Microbiome. PLoS ONE 2012, 7, e28742. [Google Scholar] [CrossRef]
- Velagapudi, V.R.; Hezaveh, R.; Reigstad, C.S.; Gopalacharyulu, P.; Yetukuri, L.; Islam, S.; Felin, J.; Perkins, R.; Borén, J.; Orešič, M.; et al. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 2010, 51, 1101–1112. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Sidhu, H.; Hoppe, B.; Hesse, A.; Tenbrock, K.; Bromme, S.; Rietschel, E.; Peck, A.B. Absence of Oxalobacter formigenes in cystic fibrosis patients: A risk factor for hyperoxaluria. Lancet 1998, 352, 1026–1029. [Google Scholar] [CrossRef]
- Gonzalez-Regueiro, J.A.; Moreno-Castaneda, L.; Uribe, M.; Chavez-Tapia, N.C. The Role of Bile Acids in Glucose Metabolism and Their Relation with Diabetes. Ann. Hepatol. 2017, 16, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.F. The Continuing Importance of Bile Acids in Liver and Intestinal Disease. Arch. Intern. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Muto, A.; Takei, H.; Nittono, H.; Murai, T.; Kurosawa, T.; Hofmann, A.F.; Pandak, W.M.; Bajaj, J.S. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: Validation by GC-MS and LC-MS. J. Lipid Res. 2014, 55, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Quinn, R.A.; Melnik, A.V.; Vrbanac, A.; Fu, T.; Patras, K.A.; Christy, M.P.; Bodai, Z.; Belda-Ferre, P.; Tripathi, A.; Chung, L.K.; et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 2020, 579, 123–129. [Google Scholar] [CrossRef]
- Gu, Q.; Li, P. Biosynthesis of Vitamins by Probiotic Bacteria. In Probiotics and Prebiotics in Human Nutrition and Health; Rao, V., Rao, L.G., Eds.; IntechOpen: London, UK, 2016; ISBN 978-953-51-2475-7. [Google Scholar]
- Engevik, M.; Morra, C.N.; Röth, D.; Engevik, K.; Spinler, J.K.; Devaraj, S.; Crawford, S.E.; Estes, M.K.; Kalkum, M.; Versalovic, J. Microbial Metabolic Capacity for Intestinal Folate Production and Modulation of Host Folate Receptors. Front. Microbiol. 2019, 10, 2305. [Google Scholar] [CrossRef]
- Magnusdottir, S.; Ravcheev, D.; De Crécy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef]
- De Assis, D.A.; Matte, C.; Aschidamini, B.; Rodrigues, E.; Ayub, M.A.Z. Biosynthesis of vitamin B12 by Propionibacterium freudenreichii subsp. shermanii ATCC 13673 using liquid acid protein residue of soybean as culture medium. Biotechnol. Prog. 2020, 36, e3011. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2012, 24, 160–168. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Shrestha, U.K. Immunology of the gut and oral tolerance. J. Adv. Intern. Med. 2015, 4, 16–24. [Google Scholar] [CrossRef]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.; Ng, D.C.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal Bacterial Colonization Induces Mutualistic Regulatory T Cell Responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef]
- Van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- White, J.R.; Dauros-Singorenko, P.; Hong, J.; Vanholsbeeck, F.; Phillips, A.; Swift, S. The role of host molecules in communication with the resident and pathogenic microbiota: A review. Med. Microecol. 2020, 4, 100005. [Google Scholar] [CrossRef]
- Reyman, M.; Van Houten, M.A.; Van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef]
- Neu, J.; Rushing, J. Cesarean Versus Vaginal Delivery: Long-term Infant Outcomes and the Hygiene Hypothesis. Clin. Perinatol. 2011, 38, 321–331. [Google Scholar] [CrossRef]
- Grölund, M.-M.; Lehtonen, O.-P.; Eerola, E.; Kero, P. Fecal Microflora in Healthy Infants Born by Different Methods of Delivery: Permanent Changes in Intestinal Flora After Cesarean Delivery. J. Pediatr. Gastroenterol. Nutr. 1999, 28, 19–25. [Google Scholar] [CrossRef]
- Coelho, G.D.P.; Ayres, L.F.A.; Barreto, D.S.; Henriques, B.D.; Prado, M.R.M.C.; Dos Passos, C.M. Acquisition of microbiota according to the type of birth: An integrative review. Rev. Lat.-Am. Enferm. 2021, 29, e3446. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M. The Origin of Human Milk Bacteria: Is There a Bacterial Entero-Mammary Pathway during Late Pregnancy and Lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Marin-Gómez, W.; Grande, M.J.; Pérez-Pulido, R.; Galvez, A.; Lucas, R. Changes in the Bacterial Diversity of Human Milk during Late Lactation Period (Weeks 21 to 48). Foods 2020, 9, 1184. [Google Scholar] [CrossRef] [PubMed]
- Obermajer, T.; Pogačić, T. Commentary: Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front. Microbiol. 2016, 7, 1281. [Google Scholar] [CrossRef]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial Changes during Pregnancy, Birth, and Infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef]
- Toscano, M.; De Grandi, R.; Grossi, E.; Drago, L. Role of the Human Breast Milk-Associated Microbiota on the Newborns’ Immune System: A Mini Review. Front. Microbiol. 2017, 8, 2100. [Google Scholar] [CrossRef]
- Panagos, P.G.; Vishwanathan, R.; Penfield-Cyr, A.; Matthan, N.R.; Shivappa, N.; Wirth, M.D.; Hebert, J.R.; Sen, S. Breastmilk from obese mothers has pro-inflammatory properties and decreased neuroprotective factors. J. Perinatol. 2016, 36, 284–290. [Google Scholar] [CrossRef]
- Wen, L.; Duffy, A. Factors Influencing the Gut Microbiota, Inflammation, and Type 2 Diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef]
- Klingensmith, N.J.; Coopersmith, C.M. The Gut as the Motor of Multiple Organ Dysfunction in Critical Illness. Crit. Care Clin. 2016, 32, 203–212. [Google Scholar] [CrossRef]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar] [CrossRef]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Hehemann, J.-H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef]
- Li, M.; Chen, W.-D.; Wang, Y.-D. The roles of the gut microbiota–miRNA interaction in the host pathophysiology. Mol. Med. 2020, 26, 101. [Google Scholar] [CrossRef]
- Djuranovic, S.; Nahvi, A.; Green, R. miRNA-Mediated Gene Silencing by Translational Repression Followed by mRNA Deadenylation and Decay. Science 2012, 336, 237–240. [Google Scholar] [CrossRef]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta 2010, 1803, 1231–1243. [Google Scholar] [CrossRef]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef]
- Link, A.; Becker, V.; Goel, A.; Wex, T.; Malfertheiner, P. Feasibility of Fecal MicroRNAs as Novel Biomarkers for Pancreatic Cancer. PLoS ONE 2012, 7, e42933. [Google Scholar] [CrossRef]
- Wortelboer, K.; Bakker, G.J.; Winkelmeijer, M.; van Riel, N.; Levin, E.; Nieuwdorp, M.; Herrema, H.; Davids, M. Fecal microbiota transplantation as tool to study the interrelation between microbiota composition and miRNA expression. Microbiol. Res. 2022, 257, 126972. [Google Scholar] [CrossRef]
- Viennois, E.; Chassaing, B.; Tahsin, A.; Pujada, A.; Wang, L.; Gewirtz, A.T.; Merlin, D. Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation. Theranostics 2019, 9, 4542–4557. [Google Scholar] [CrossRef] [PubMed]
- Schächtle, M.A.; Rosshart, S.P. The Microbiota-Gut-Brain Axis in Health and Disease and Its Implications for Translational Research. Front. Cell. Neurosci. 2021, 15, 698172. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Taki, Y.; Sato, K.; Kinomura, S.; Goto, R.; Okada, K.; Kawashima, R.; He, Y.; Evans, A.C.; Fukuda, H. Age-related changes in topological organization of structural brain networks in healthy individuals. Hum. Brain Mapp. 2011, 33, 552–568. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Nunez, C.; Meneguetti, B.T.; Franco, O.L.; Lu, T.K. Neuromicrobiology: How Microbes Influence the Brain. ACS Chem. Neurosci. 2017, 9, 141–150. [Google Scholar] [CrossRef]
- Ignatova, V. Influence of Gut Microbiota on Behavior and Its Disturbances. In Behavioral Neuroscience; Palermo, S., Morese, R., Eds.; IntechOpen: London, UK, 2019; ISBN 978-1-78984-051-3. [Google Scholar]
- Al-Asmakh, M.; Anuar, F.; Zadjali, F.; Rafter, J.; Pettersson, S. Gut microbial communities modulating brain development and function. Gut Microbes 2012, 3, 366–373. [Google Scholar] [CrossRef]
- Ogbonnaya, E.S.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; O’Leary, O.F. Adult Hippocampal Neurogenesis Is Regulated by the Microbiome. Biol. Psychiatry 2015, 78, e7–e9. [Google Scholar] [CrossRef]
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef]
- Indari, O.; Chandramohanadas, R.; Jha, H.C. Epstein–Barr virus infection modulates blood–brain barrier cells and its co-infection with Plasmodium falciparum induces RBC adhesion. Pathog. Dis. 2020, 79, ftaa080. [Google Scholar] [CrossRef]
- Kashyap, D.; Baral, B.; Jakhmola, S.; Singh, A.K.; Jha, H.C. Helicobacter pylori and Epstein-Barr Virus Coinfection Stimulates Aggressiveness in Gastric Cancer through the Regulation of Gankyrin. mSphere 2021, 6, e00751-21. [Google Scholar] [CrossRef]
- Singh, S.; Jha, H.C. Status of Epstein-Barr Virus Coinfection with Helicobacter pylori in Gastric Cancer. J. Oncol. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Montiel-Castro, A.J.; González-Cervantes, R.M.; Ebravo-Ruiseco, G.; Epacheco-Lopez, G. The microbiota-gut-brain axis: Neurobehavioral correlates, health and sociality. Front. Integr. Neurosci. 2013, 7, 70. [Google Scholar] [CrossRef]
- Mazzoli, R.; Pessione, E. The Neuro-endocrinological Role of Microbial Glutamate and GABA Signaling. Front. Microbiol. 2016, 7, 1934. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef]
- Caspani, G.; Swann, J. Small talk: Microbial metabolites involved in the signaling from microbiota to brain. Curr. Opin. Pharmacol. 2019, 48, 99–106. [Google Scholar] [CrossRef]
- Breit, S.; Kupferberg, A.; Rogler, G.; Hasler, G. Vagus Nerve as Modulator of the Brain–Gut Axis in Psychiatric and Inflammatory Disorders. Front. Psychiatry 2018, 9, 44. [Google Scholar] [CrossRef]
- McCorry, L.K. Physiology of the Autonomic Nervous System. Am. J. Pharm. Educ. 2007, 71, 78. [Google Scholar] [CrossRef]
- Cunningham, D.J.C.; Robbins, P.A.; Wolff, C.B. Integration of Respiratory Responses to Changes in Alveolar Partial Pressures of CO2 and O2 and in Arterial pH. In Comprehensive Physiology; Terjung, R., Ed.; Wiley: Hoboken, NJ, USA, 1986; pp. 475–528. ISBN 978-0-470-65071-4. [Google Scholar]
- Costa, M.; Brookes, S.J.H.; Hennig, G.W. Anatomy and physiology of the enteric nervous system. Gut 2000, 47, 15iv–19. [Google Scholar] [CrossRef]
- Fülling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagus…. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Rangon, C.-M.; Niezgoda, A. Understanding the Pivotal Role of the Vagus Nerve in Health from Pandemics. Bioengineering 2022, 9, 352. [Google Scholar] [CrossRef] [PubMed]
- Bonaz, B.; Bazin, T.; Pellissier, S. The Vagus Nerve at the Interface of the Microbiota-Gut-Brain Axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef]
- Yu, K.B.; Hsiao, E.Y. Roles for the gut microbiota in regulating neuronal feeding circuits. J. Clin. Investig. 2021, 131, e143772. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, P.; Bienenstock, J.; Kunze, W.A. Vagal Pathways for Microbiome-Brain-Gut Axis Communication. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease; Advances in Experimental Medicine and Biology; Lyte, M., Cryan, J.F., Eds.; Springer: New York, NY, USA, 2014; Volume 817, pp. 115–133. ISBN 978-1-4939-0896-7. [Google Scholar]
- Goehler, L.E.; Gaykema, R.P.; Opitz, N.; Reddaway, R.; Badr, N.; Lyte, M. Activation in vagal afferents and central autonomic pathways: Early responses to intestinal infection with Campylobacter jejuni. Brain Behav. Immun. 2005, 19, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Bercik, P.; Park, A.J.; Sinclair, D.; Khoshdel, A.; Lu, J.; Huang, X.; Deng, Y.; Blennerhassett, P.A.; Fahnestock, M.; Moine, D.; et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol. Motil. 2011, 23, 1132–1139. [Google Scholar] [CrossRef]
- Alim, E.; Gülekon, I.N.; Atalar, K.; Bahçelioğlu, M. Enteric nervous system, gut-brain connection and related neurodevelopmental disorders. Anatomy 2020, 14, 61–67. [Google Scholar] [CrossRef]
- Goehler, L.E.; Park, S.M.; Opitz, N.; Lyte, M.; Gaykema, R.P. Campylobacter jejuni infection increases anxiety-like behavior in the holeboard: Possible anatomical substrates for viscerosensory modulation of exploratory behavior. Brain Behav. Immun. 2008, 22, 354–366. [Google Scholar] [CrossRef]
- Browning, K.N.; Verheijden, S.; Boeckxstaens, G.E. The Vagus Nerve in Appetite Regulation, Mood, and Intestinal Inflammation. Gastroenterology 2017, 152, 730–744. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. (Lausanne) 2020, 11, 25. [Google Scholar] [CrossRef]
- Ma, Q.; Xing, C.; Long, W.; Wang, H.Y.; Liu, Q.; Wang, R.-F. Impact of microbiota on central nervous system and neurological diseases: The gut-brain axis. J. Neuroinflamm. 2019, 16, 53. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef]
- Sarubbo, F.; Cavallucci, V.; Pani, G. The Influence of Gut Microbiota on Neurogenesis: Evidence and Hopes. Cells 2022, 11, 382. [Google Scholar] [CrossRef]
- Ahmed, H.; Leyrolle, Q.; Koistinen, V.; Kärkkäinen, O.; Layé, S.; Delzenne, N.; Hanhineva, K. Microbiota-derived metabolites as drivers of gut–brain communication. Gut Microbes 2022, 14, 2102878. [Google Scholar] [CrossRef]
- Yarandi, S.S.; Peterson, D.A.; Treisman, G.J.; Moran, T.H.; Pasricha, P.J. Modulatory Effects of Gut Microbiota on the Central Nervous System: How Gut Could Play a Role in Neuropsychiatric Health and Diseases. J. Neurogastroenterol. Motil. 2016, 22, 201–212. [Google Scholar] [CrossRef]
- Rhee, S.H.; Pothoulakis, C.; Mayer, E.A. Principles and clinical implications of the brain–gut–enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 306–314. [Google Scholar] [CrossRef]
- Talham, G.L.; Jiang, H.-Q.; Bos, N.A.; Cebra, J.J. Segmented Filamentous Bacteria Are Potent Stimuli of a Physiologically Normal State of the Murine Gut Mucosal Immune System. Infect. Immun. 1999, 67, 1992–2000. [Google Scholar] [CrossRef]
- McKernan, D.P. Toll-like receptors and immune cell crosstalk in the intestinal epithelium. AIMS Allergy Immunol. 2019, 3, 13–31. [Google Scholar] [CrossRef]
- Vijay, K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int. Immunopharmacol. 2018, 59, 391–412. [Google Scholar] [CrossRef]
- Frosali, S.; Pagliari, D.; Gambassi, G.; Landolfi, R.; Pandolfi, F.; Cianci, R. How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology. J. Immunol. Res. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Alhasson, F.; Das, S.; Seth, R.; Dattaroy, D.; Chandrashekaran, V.; Ryan, C.N.; Chan, L.S.; Testerman, T.; Burch, J.; Hofseth, L.J.; et al. Altered gut microbiome in a mouse model of Gulf War Illness causes neuroinflammation and intestinal injury via leaky gut and TLR4 activation. PLoS ONE 2017, 12, e0172914. [Google Scholar] [CrossRef]
- Holzer, P.; Farzi, A. Neuropeptides and the Microbiota-Gut-Brain Axis. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease; Advances in Experimental Medicine and Biology; Lyte, M., Cryan, J.F., Eds.; Springer: New York, NY, USA, 2014; Volume 817, pp. 195–219. ISBN 978-1-4939-0896-7. [Google Scholar]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of Gut Bacteria on Human Health and Diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Sampson, T.R.; Geschwind, D.H.; Mazmanian, S.K. The Central Nervous System and the Gut Microbiome. Cell 2016, 167, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Faruqui, N.A.; Prium, D.H.; Mowna, S.A.; Ullah, A.; Araf, Y.; Sarkar, B.; Zohora, U.S.; Rahman, M.S. Gut microorganisms and neurological disease perspectives. Futur. Neurol. 2021, 16, FNL53. [Google Scholar] [CrossRef]
- Lehner, C.; Gehwolf, R.; Tempfer, H.; Krizbai, I.; Hennig, B.; Bauer, H.-C.; Bauer, H. Oxidative Stress and Blood–Brain Barrier Dysfunction Under Particular Consideration of Matrix Metalloproteinases. Antioxid. Redox Signal. 2011, 15, 1305–1323. [Google Scholar] [CrossRef]
- Liebner, S.; Dijkhuizen, R.M.; Reiss, Y.; Plate, K.H.; Agalliu, D.; Constantin, G. Functional morphology of the blood–brain barrier in health and disease. Acta Neuropathol. 2018, 135, 311–336. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Zhu, W. Gut Microbiota: The Brain Peacekeeper. Front. Microbiol. 2016, 7, 345. [Google Scholar] [CrossRef]
- Jena, A.; Montoya, C.A.; Mullaney, J.A.; Dilger, R.N.; Young, W.; McNabb, W.C.; Roy, N.C. Gut-Brain Axis in the Early Postnatal Years of Life: A Developmental Perspective. Front. Integr. Neurosci. 2020, 14, 44. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef]
- Parker, A.; Fonseca, S.; Carding, S.R. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes 2019, 11, 135–157. [Google Scholar] [CrossRef]
- Liu, W.-Y.; Wang, Z.-B.; Zhang, L.-C.; Wei, X.; Li, L. Tight Junction in Blood-Brain Barrier: An Overview of Structure, Regulation, and Regulator Substances. CNS Neurosci. Ther. 2012, 18, 609–615. [Google Scholar] [CrossRef]
- Tang, W.; Zhu, H.; Feng, Y.; Guo, R.; Wan, D. The Impact of Gut Microbiota Disorders on the Blood–Brain Barrier. Infect. Drug Resist. 2020, 13, 3351–3363. [Google Scholar] [CrossRef]
- Kabouridis, P.S.; Lasrado, R.; McCallum, S.; Chng, S.H.; Snippert, H.J.; Clevers, H.; Pettersson, S.; Pachnis, V. The gut microbiota keeps enteric glial cells on the move; prospective roles of the gut epithelium and immune system. Gut Microbes 2015, 6, 398–403. [Google Scholar] [CrossRef]
- Kabouridis, P.S.; Lasrado, R.; McCallum, S.; Chng, S.H.; Snippert, H.J.; Clevers, H.; Pettersson, S.; Pachnis, V. Microbiota Controls the Homeostasis of Glial Cells in the Gut Lamina Propria. Neuron 2015, 85, 289–295. [Google Scholar] [CrossRef]
- De Vadder, F.; Grasset, E.; Holm, L.M.; Karsenty, G.; MacPherson, A.J.; Olofsson, L.E.; Bäckhed, F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. USA 2018, 115, 6458–6463. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The Human Microbiome: Our Second Genome. Annu. Rev. Genom. Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef]
- Clarke, S.; Murphy, E.F.; Nilaweera, K.; Ross, R.; Shanahan, F.; O’Toole, P.W.; Cotter, P.D. The gut microbiota and its relationship to diet and obesity. Gut Microbes 2012, 3, 186–202. [Google Scholar] [CrossRef]
- Dhikav, V.; Anand, K.S. Hippocampus in health and disease: An overview. Ann. Indian Acad. Neurol. 2012, 15, 239–246. [Google Scholar] [CrossRef]
- Luczynski, P.; Whelan, S.O.; O’Sullivan, C.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Adult microbiota-deficient mice have distinct dendritic morphological changes: Differential effects in the amygdala and hippocampus. Eur. J. Neurosci. 2016, 44, 2654–2666. [Google Scholar] [CrossRef]
- Kawase, T.; Nagasawa, M.; Ikeda, H.; Yasuo, S.; Koga, Y.; Furuse, M. Gut microbiota of mice putatively modifies amino acid metabolism in the host brain. Br. J. Nutr. 2017, 117, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.; Garcia-Perez, I.; Braniste, V.; Wilson, I.; Sidaway, J.; Nicholson, J.; Pettersson, S.; Holmes, E. Application of 1 H NMR spectroscopy to the metabolic phenotyping of rodent brain extracts: A metabonomic study of gut microbial influence on host brain metabolism. J. Pharm. Biomed. Anal. 2017, 143, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Meng, Z.; Li, N.; Liu, Y.; Li, L.; Chen, D.; Yang, Y. Roles of Gut Microbiota in the Regulation of Hippocampal Plasticity, Inflammation, and Hippocampus-Dependent Behaviors. Front. Cell. Infect. Microbiol. 2021, 10, 611014. [Google Scholar] [CrossRef] [PubMed]
- Luck, B.; Engevik, M.A.; Ganesh, B.; Lackey, E.P.; Lin, T.; Balderas, M.; Major, A.; Runge, J.; Luna, R.A.; Sillitoe, R.V.; et al. Bifidobacteria shape host neural circuits during postnatal development by promoting synapse formation and microglial function. Sci. Rep. 2020, 10, 7737. [Google Scholar] [CrossRef] [PubMed]
- Björkholm, C.; Monteggia, L.M. BDNF—A Key Transducer of Antidepressant Effects. Neuropharmacology 2015, 102, 72–79. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, A.; Singh, D. Dietary Flavonoids Interaction with CREB-BDNF Pathway: An Unconventional Approach for Comprehensive Management of Epilepsy. Curr. Neuropharmacol. 2019, 17, 1158–1175. [Google Scholar] [CrossRef]
- Lv, M.; Yang, S.; Cai, L.; Qin, L.-Q.; Li, B.-Y.; Wan, Z. Effects of Quercetin Intervention on Cognition Function in APP/PS1 Mice was Affected by Vitamin D Status. Mol. Nutr. Food Res. 2018, 62, e1800621. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut Microbiota and Metabolome Alterations Associated with Parkinson’s Disease. mSystems 2020, 5, e00561-20. [Google Scholar] [CrossRef] [PubMed]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Zhu, F.; Hart, J.; Roalstad, S.; Graves, J.; Lynch, S.; Waubant, E.; Centers, T.U.N.O.P.M. Gut microbiota in early pediatric multiple sclerosis: A case−control study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Mazmanian, S.K. Control of Brain Development, Function, and Behavior by the Microbiome. Cell Host Microbe 2015, 17, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhao, D.; Shah, S.Z.A.; Wu, W.; Lai, M.; Zhang, X.; Li, J.; Guan, Z.; Zhao, H.; Li, W.; et al. The Role of the Gut Microbiota in the Pathogenesis of Parkinson’s Disease. Front. Neurol. 2019, 10, 1155. [Google Scholar] [CrossRef]
- Park, J.C.; Im, S.-H. The gut-immune-brain axis in neurodevelopment and neurological disorders. Microbiome Res. Rep. 2022, 1, 23. [Google Scholar] [CrossRef]
- Sarkar, S.R.; Banerjee, S. Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 2019, 328, 98–104. [Google Scholar] [CrossRef]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Mirzaei, R.; Bouzari, B.; Hosseini-Fard, S.R.; Mazaheri, M.; Ahmadyousefi, Y.; Abdi, M.; Jalalifar, S.; Karimitabar, Z.; Teimoori, A.; Keyvani, H.; et al. Role of microbiota-derived short-chain fatty acids in nervous system disorders. Biomed. Pharmacother. 2021, 139, 111661. [Google Scholar] [CrossRef]
- Tran, S.M.-S.; Mohajeri, M.H. The Role of Gut Bacterial Metabolites in Brain Development, Aging and Disease. Nutrients 2021, 13, 732. [Google Scholar] [CrossRef]
- Vogt, N.M.; Romano, K.A.; Darst, B.F.; Engelman, C.D.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Blennow, K.; Zetterberg, H.; Bendlin, B.B.; et al. The gut microbiota-derived metabolite trimethylamine N-oxide is elevated in Alzheimer’s disease. Alzheimer’s Res. Ther. 2018, 10, 124. [Google Scholar] [CrossRef]
- Koszewiczz, M.; Jaroch, J.; Brzecka, A.; Ejma, M.; Budrewicz, S.; Mikhaleva, L.M.; Muresanu, C.; Schield, P.; Somasundaram, S.G.; Kirkland, C.E.; et al. Dysbiosis is one of the risk factor for stroke and cognitive impairment and potential target for treatment. Pharmacol. Res. 2020, 164, 105277. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.A.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Chen, C.; Ahn, E.H.; Kang, S.S.; Liu, X.; Alam, A.; Ye, K. Gut dysbiosis contributes to amyloid pathology, associated with C/EBPβ/AEP signaling activation in Alzheimer’s disease mouse model. Sci. Adv. 2020, 6, eaba0466. [Google Scholar] [CrossRef]
- Gagliardi, A.; Totino, V.; Cacciotti, F.; Iebba, V.; Neroni, B.; Bonfiglio, G.; Trancassini, M.; Passariello, C.; Pantanella, F.; Schippa, S. Rebuilding the Gut Microbiota Ecosystem. Int. J. Environ. Res. Public Health 2018, 15, 1679. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Begum-Haque, S.; Dasgupta, S.; Kasper, D.L.; Kasper, L.H. Central Nervous System Demyelinating Disease Protection by the Human Commensal Bacteroides fragilis Depends on Polysaccharide A Expression. J. Immunol. 2010, 185, 4101–4108. [Google Scholar] [CrossRef]
- Huang, H.; Xu, H.; Luo, Q.; He, J.; Li, M.; Chen, H.; Tang, W.; Nie, Y.; Zhou, Y. Fecal microbiota transplantation to treat Parkinson’s disease with constipation. Medicine 2019, 98, e16163. [Google Scholar] [CrossRef]
- König, R.S.; Albrich, W.C.; Kahlert, C.R.; Bahr, L.S.; Löber, U.; Vernazza, P.; Scheibenbogen, C.; Forslund, S.K. The Gut Microbiome in Myalgic Encephalomyelitis (ME)/Chronic Fatigue Syndrome (CFS). Front. Immunol. 2022, 12, 628741. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Azm, S.A.N.; Djazayeri, A.; Safa, M.; Azami, K.; Ahmadvand, B.; Sabbaghziarani, F.; Sharifzadeh, M.; Vafa, M. Lactobacilli and bifidobacteria ameliorate memory and learning deficits and oxidative stress in β-amyloid (1–42) injected rats. Appl. Physiol. Nutr. Metab. 2018, 43, 718–726. [Google Scholar] [CrossRef]
- Jing, Y.; Yu, Y.; Bai, F.; Wang, L.; Yang, D.; Zhang, C.; Qin, C.; Yang, M.; Zhang, D.; Zhu, Y.; et al. Effect of fecal microbiota transplantation on neurological restoration in a spinal cord injury mouse model: Involvement of brain-gut axis. Microbiome 2021, 9, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-S.; Cha, L.; Sim, M.; Jung, S.; Chun, W.Y.; Baik, H.W.; Shin, D.-M. Probiotic Supplementation Improves Cognitive Function and Mood with Changes in Gut Microbiota in Community-Dwelling Older Adults: A Randomized, Double-Blind, Placebo-Controlled, Multicenter Trial. J. Gerontol. Ser. A 2020, 76, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Desbonnet, L.; Garrett, L.; Clarke, G.; Kiely, B.; Cryan, J.F.; Dinan, T.G. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 2010, 170, 1179–1188. [Google Scholar] [CrossRef]
- Du, D.; Tang, W.; Zhou, C.; Sun, X.; Wei, Z.; Zhong, J.; Huang, Z. Fecal Microbiota Transplantation Is a Promising Method to Restore Gut Microbiota Dysbiosis and Relieve Neurological Deficits after Traumatic Brain Injury. Oxidative Med. Cell. Longev. 2021, 2021, 1–21. [Google Scholar] [CrossRef]
- Bonfili, L.; Cecarini, V.; Gogoi, O.; Berardi, S.; Scarpona, S.; Angeletti, M.; Rossi, G.; Eleuteri, A.M. Gut microbiota manipulation through probiotics oral administration restores glucose homeostasis in a mouse model of Alzheimer’s disease. Neurobiol. Aging 2019, 87, 35–43. [Google Scholar] [CrossRef]
- Kechagia, M.; Basoulis, D.; Konstantopoulou, S.; Dimitriadi, D.; Gyftopoulou, K.; Skarmoutsou, N.; Fakiri, E.M. Health Benefits of Probiotics: A Review. Int. Sch. Res. Not. 2013, 2013, 1–7. [Google Scholar] [CrossRef]
- Aponte, M.; Murru, N.; Shoukat, M. Therapeutic, Prophylactic, and Functional Use of Probiotics: A Current Perspective. Front. Microbiol. 2020, 11, 562048. [Google Scholar] [CrossRef]
- Berg, R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996, 4, 430–435. [Google Scholar] [CrossRef]
- Liao, S.F.; Nyachoti, M. Using Probiotics to Improve Swine Gut Health and Nutrient Utilization. Anim. Nutr. 2017, 3, 331–343. [Google Scholar] [CrossRef]
- Maldonado Galdeano, C.; Cazorla, S.I.; Lemme Dumit, J.M.; Vélez, E.; Perdigón, G. Beneficial Effects of Probiotic Consumption on the Immune System. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar] [CrossRef]
- Fijan, S. Microorganisms with Claimed Probiotic Properties: An Overview of Recent Literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef]
- Mani-López, E.; Palou, E.; López-Malo, A. Probiotic viability and storage stability of yogurts and fermented milks prepared with several mixtures of lactic acid bacteria. J. Dairy Sci. 2014, 97, 2578–2590. [Google Scholar] [CrossRef]
- Amara, A.; Shibl, A. Role of Probiotics in health improvement, infection control and disease treatment and management. Saudi Pharm. J. 2013, 23, 107–114. [Google Scholar] [CrossRef]
- Stavropoulou, E.; Bezirtzoglou, E. Probiotics in Medicine: A Long Debate. Front. Immunol. 2020, 11, 2192. [Google Scholar] [CrossRef]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2012, 6, 39–51. [Google Scholar] [CrossRef]
- Neef, A.; Sanz, Y. Future for probiotic science in functional food and dietary supplement development. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 679–687. [Google Scholar] [CrossRef]
- Khangwal, I.; Shukla, P. Prospecting prebiotics, innovative evaluation methods, and their health applications: A review. 3 Biotech 2019, 9, 187. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, P.; Chen, M.; Luo, Y.; Prabhakar, M.; Zheng, H.; He, Y.; Qi, Q.; Long, H.; Zhang, Y.; et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) Increase Bifidobacterium but Reduce Butyrate Producing Bacteria with Adverse Glycemic Metabolism in healthy young population. Sci. Rep. 2017, 7, 11789. [Google Scholar] [CrossRef] [PubMed]
- Michel, M.R.; Rodríguez-Jasso, R.M.; Aguilar, C.N.; Gonzalez-Herrera, S.M.; Flores-Gallegos, A.C.; Rodríguez-Herrera, R. Fructosyltransferase Sources, Production, and Applications for Prebiotics Production. In Probiotics and Prebiotics in Human Nutrition and Health; Rao, V., Rao, L.G., Eds.; IntechOpen: London, UK, 2016; ISBN 978-953-51-2475-7. [Google Scholar]
- Savignac, H.M.; Corona, G.; Mills, H.; Chen, L.; Spencer, J.P.; Tzortzis, G.; Burnet, P.W. Prebiotic feeding elevates central brain derived neurotrophic factor, N-methyl-d-aspartate receptor subunits and d-serine. Neurochem. Int. 2013, 63, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Rossen, N.G.; Fuentes, S.; van der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.A.; Duflou, A.; Löwenberg, M.; Van Den Brink, G.R.; Mathus-Vliegen, E.M.H.; de Vos, W.M.; et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 2015, 149, 110–118.e4. [Google Scholar] [CrossRef]
- Chen, H.; Athar, R.; Zheng, G.; Williams, H.N. Prey bacteria shape the community structure of their predators. ISME J. 2011, 5, 1314–1322. [Google Scholar] [CrossRef]
- Atterbury, R.J.; Hobley, L.; Till, R.; Lambert, C.; Capeness, M.J.; Lerner, T.R.; Fenton, A.K.; Barrow, P.; Sockett, R.E. Effects of Orally Administered Bdellovibrio bacteriovorus on the Well-Being and Salmonella Colonization of Young Chicks. Appl. Environ. Microbiol. 2011, 77, 5794–5803. [Google Scholar] [CrossRef]
- Hajjo, R.; Sabbah, D.A.; Al Bawab, A.Q. Unlocking the Potential of the Human Microbiome for Identifying Disease Diagnostic Biomarkers. Diagnostics 2022, 12, 1742. [Google Scholar] [CrossRef]
- Zhou, Z.; Ge, S.; Li, Y.; Ma, W.; Liu, Y.; Hu, S.; Zhang, R.; Ma, Y.; Du, K.; Syed, A.; et al. Human Gut Microbiome-Based Knowledgebase as a Biomarker Screening Tool to Improve the Predicted Probability for Colorectal Cancer. Front. Microbiol. 2020, 11, 596027. [Google Scholar] [CrossRef]
- Romo-Vaquero, M.; Fernández-Villalba, E.; Gil-Martinez, A.-L.; Cuenca-Bermejo, L.; Espín, J.C.; Herrero, M.T.; Selma, M.V. Urolithins: Potential biomarkers of gut dysbiosis and disease stage in Parkinson’s patients. Food Funct. 2022, 13, 6306–6316. [Google Scholar] [CrossRef]
- Schwiertz, A.; Spiegel, J.; Dillmann, U.; Grundmann, D.; Bürmann, J.; Faßbender, K.; Schäfer, K.-H.; Unger, M.M. Fecal markers of intestinal inflammation and intestinal permeability are elevated in Parkinson’s disease. Park. Relat. Disord. 2018, 50, 104–107. [Google Scholar] [CrossRef]
- Rüb, A.M.; Tsakmaklis, A.; Gräfe, S.K.; Simon, M.-C.; Vehreschild, M.J.; Wuethrich, I. Biomarkers of human gut microbiota diversity and dysbiosis. Biomark. Med. 2021, 15, 139–150. [Google Scholar] [CrossRef]
- Brydges, C.R.; Fiehn, O.; Mayberg, H.S.; Schreiber, H.; Dehkordi, S.M.; Bhattacharyya, S.; Cha, J.; Choi, K.S.; Craighead, W.E.; Krishnan, R.R.; et al. Indoxyl Sulfate, a Gut Microbiome-Derived Uremic Toxin, Is Associated with Psychic Anxiety and Its Functional Magnetic Resonance Imaging-Based Neurologic Signature. Neuroscience 2020, 11, 21011. [Google Scholar] [CrossRef]
- Ling, Y.; Gong, T.; Zhang, J.; Gu, Q.; Gao, X.; Weng, X.; Liu, J.; Sun, J. Gut Microbiome Signatures Are Biomarkers for Cognitive Impairment in Patients With Ischemic Stroke. Front. Aging Neurosci. 2020, 12, 511562. [Google Scholar] [CrossRef]
- De Ture, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological Alterations in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef]
- Murphy, M.P.; LeVine, H., 3rd. Alzheimer’s Disease and the Amyloid-β Peptide. J. Alzheimer’s Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, S.; Shin, S.J.; Park, Y.H.; Nam, Y.; Kim, C.W.; Lee, K.W.; Kim, S.-M.; Jung, I.D.; Yang, H.D.; et al. Gram-negative bacteria and their lipopolysaccharides in Alzheimer’s disease: Pathologic roles and therapeutic implications. Transl. Neurodegener. 2021, 10, 49. [Google Scholar] [CrossRef]
- Zhan, X.; Stamova, B.; Sharp, F.R. Lipopolysaccharide Associates with Amyloid Plaques, Neurons and Oligodendrocytes in Alzheimer’s Disease Brain: A Review. Front. Aging Neurosci. 2018, 10, 42. [Google Scholar] [CrossRef]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef]
- Chapman, M.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli Curli Operons in Directing Amyloid Fiber Formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef]
- Friedland, R.P.; Chapman, M.R. The role of microbial amyloid in neurodegeneration. PLoS Pathog. 2017, 13, e1006654. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, D.; Singh, V.K.; Baral, B.; Pathak, D.K.; Jayabalan, J.; Kumar, R.; Tapryal, S.; Jha, H.C. Indication of Neurodegenerative Cascade Initiation by Amyloid-like Aggregate-Forming EBV Proteins and Peptide in Alzheimer’s Disease. ACS Chem. Neurosci. 2021, 12, 3957–3967. [Google Scholar] [CrossRef] [PubMed]
- Alexander, G.E. Biology of Parkinson’s disease: Pathogenesis and pathophysiology of a multisystem neurodegenerative disorder. Dialog- Clin. Neurosci. 2004, 6, 259–280. [Google Scholar] [CrossRef]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. npj Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Zapała, B.; Stefura, T.; Wójcik-Pędziwiatr, M.; Kabut, R.; Bałajewicz-Nowak, M.; Milewicz, T.; Dudek, A.; Stój, A.; Rudzińska-Bar, M. Differences in the Composition of Gut Microbiota between Patients with Parkinson’s Disease and Healthy Controls: A Cohort Study. J. Clin. Med. 2021, 10, 5698. [Google Scholar] [CrossRef]
- Shen, T.; Yue, Y.; He, T.; Huang, C.; Qu, B.; Lv, W.; Lai, H.-Y. The Association Between the Gut Microbiota and Parkinson’s Disease, a Meta-Analysis. Front. Aging Neurosci. 2021, 13, 636545. [Google Scholar] [CrossRef]
- Huang, Y.; Liao, J.; Liu, X.; Zhong, Y.; Cai, X.; Long, L. Review: The Role of Intestinal Dysbiosis in Parkinson’s Disease. Front. Cell. Infect. Microbiol. 2021, 11, 615075. [Google Scholar] [CrossRef]
- Caspani, G.; Kennedy, S.; Foster, J.A.; Swann, J. Gut microbial metabolites in depression: Understanding the biochemical mechanisms. Microb. Cell 2019, 6, 454–481. [Google Scholar] [CrossRef]
- Van Ijzendoorn, S.C.D.; Derkinderen, P. The Intestinal Barrier in Parkinson’s Disease: Current State of Knowledge. J. Park. Dis. 2019, 9, S323–S329. [Google Scholar] [CrossRef]
- Mandel, I.; Paperna, T.; Glass-Marmor, L.; Volkowich, A.; Badarny, S.; Schwartz, I.; Vardi, P.; Koren, I.; Miller, A. Tight junction proteins expression and modulation in immune cells and multiple sclerosis. J. Cell. Mol. Med. 2011, 16, 765–775. [Google Scholar] [CrossRef]
- Gorlé, N.; Bauwens, E.; Haesebrouck, F.; Smet, A.; Vandenbroucke, R.E. Helicobacter and the Potential Role in Neurological Disorders: There Is More Than Helicobacter pylori. Front. Immunol. 2021, 11, 584165. [Google Scholar] [CrossRef]
- Goldenberg, M.M. Medical management of Parkinson’s disease. Pharm. Ther. 2008, 33, 590–606. [Google Scholar]
- Nyholm, D.; Hellström, P.M. Effects of Helicobacter pylori on Levodopa Pharmacokinetics. J. Park. Dis. 2021, 11, 61–69. [Google Scholar] [CrossRef]
- Lolekha, P.; Sriphanom, T.; Vilaichone, R.-K. Helicobacter pylori eradication improves motor fluctuations in advanced Parkinson’s disease patients: A prospective cohort study (HP-PD trial). PLoS ONE 2021, 16, e0251042. [Google Scholar] [CrossRef]
- Niehues, M.; Hensel, A. In vitro interaction of L-Dopa with bacterial adhesins of Helicobacter pylori: An explanation for clinicial differences in bioavailability? Planta Med. 2009, 75, s-0029-1234811. [Google Scholar] [CrossRef]
- Abdollahi, H.; Tadjrobehkar, O. The Role of Different Sugars, Amino Acids and Few Other Substances in Chemotaxis Directed Motility of Helicobacter Pylori. Iran. J. Basic Med. Sci. 2012, 15, 787–794. [Google Scholar] [CrossRef]
- Rekdal, V.M.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef]
- Zuccato, C.; Valenza, M.; Cattaneo, E. Molecular Mechanisms and Potential Therapeutical Targets in Huntington’s Disease. Physiol. Rev. 2010, 90, 905–981. [Google Scholar] [CrossRef]
- Kong, G.; Cao, K.-A.L.; Judd, L.M.; Li, S.; Renoir, T.; Hannan, A.J. Microbiome profiling reveals gut dysbiosis in a transgenic mouse model of Huntington’s disease. Neurobiol. Dis. 2020, 135, 104268. [Google Scholar] [CrossRef]
- Du, G.; Dong, W.; Yang, Q.; Yu, X.; Ma, J.; Gu, W.; Huang, Y. Altered Gut Microbiota Related to Inflammatory Responses in Patients With Huntington’s Disease. Front. Immunol. 2021, 11, 603594. [Google Scholar] [CrossRef]
- Gerhardt, S.; Mohajeri, M.H. Changes of Colonic Bacterial Composition in Parkinson’s Disease and Other Neurodegenerative Diseases. Nutrients 2018, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Engels, C.; Ruscheweyh, H.-J.; Beerenwinkel, N.; Lacroix, C.; Schwab, C. The Common Gut Microbe Eubacterium hallii also Contributes to Intestinal Propionate Formation. Front. Microbiol. 2016, 7, 713. [Google Scholar] [CrossRef] [PubMed]
- Pochard, P.; Gosset, P.; Grangette, C.; Andre, C.; Tonnel, A.-B.; Pestel, J.; Mercenier, A. Lactic acid bacteria inhibit TH2 cytokine production by mononuclear cells from allergic patients. J. Allergy Clin. Immunol. 2002, 110, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Bjorkqvist, M.; Wild, E.; Thiele, J.; Silvestroni, A.; Andre, R.; Lahiri, N.; Raibon, E.; Lee, R.V.; Benn, C.L.; Soulet, D.; et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J. Exp. Med. 2008, 205, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Gadani, S.P.; Cronk, J.C.; Norris, G.T.; Kipnis, J. IL-4 in the Brain: A Cytokine To Remember. J. Immunol. 2012, 189, 4213–4219. [Google Scholar] [CrossRef]
- Pirko, I.; Noseworthy, J.H. Demyelinating Disorders of the Central Nervous System. In Textbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1103–1133. ISBN 978-1-4160-3618-0. [Google Scholar]
- Wootla, B.; Eriguchi, M.; Rodriguez, M. Is Multiple Sclerosis an Autoimmune Disease? Autoimmune Dis. 2012, 2012, 969657. [Google Scholar] [CrossRef]
- Errede, M.; Girolamo, F.; Ferrara, G.; Strippoli, M.; Morando, S.; Boldrin, V.; Rizzi, M.; Uccelli, A.; Perris, R.; Bendotti, C.; et al. Blood-Brain Barrier Alterations in the Cerebral Cortex in Experimental Autoimmune Encephalomyelitis. J. Neuropathol. Exp. Neurol. 2012, 71, 840–854. [Google Scholar] [CrossRef]
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky Gut As a Danger Signal for Autoimmune Diseases. Front. Immunol. 2017, 8, 598. [Google Scholar] [CrossRef]
- Cox, L.M.; Maghzi, A.H.; Liu, S.; Tankou, S.K.; Dhang, F.H.; Willocq, V.; Song, A.; Wasén, C.; Tauhid, S.; Chu, R.; et al. Gut Microbiome in Progressive Multiple Sclerosis. Ann. Neurol. 2021, 89, 1195–1211. [Google Scholar] [CrossRef]
- Melbye, P.; Olsson, A.; Hansen, T.H.; Søndergaard, H.B.; Oturai, A.B. Short-chain fatty acids and gut microbiota in multiple sclerosis. Acta Neurol. Scand. 2018, 139, 208–219. [Google Scholar] [CrossRef]
- Liu, J.; Jin, Y.; Ye, Y.; Tang, Y.; Dai, S.; Li, M.; Zhao, G.; Hong, G.; Lu, Z.-Q. The Neuroprotective Effect of Short Chain Fatty Acids Against Sepsis-Associated Encephalopathy in Mice. Front. Immunol. 2021, 12, 626894. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Rumah, K.R.; Linden, J.; Fischetti, V.A.; Vartanian, T. Isolation of Clostridium perfringens Type B in an Individual at First Clinical Presentation of Multiple Sclerosis Provides Clues for Environmental Triggers of the Disease. PLoS ONE 2013, 8, e76359. [Google Scholar] [CrossRef]
- Tremlett, H.; Waubant, E. The multiple sclerosis microbiome? Ann. Transl. Med. 2017, 5, 53. [Google Scholar] [CrossRef]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef]
- Grant, S.M.; DeMorrow, S. Bile Acid Signaling in Neurodegenerative and Neurological Disorders. Int. J. Mol. Sci. 2020, 21, 75982. [Google Scholar] [CrossRef]
- Marksteiner, J.; Blasko, I.; Kemmler, G.; Koal, T.; Humpel, C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics 2017, 14, 1. [Google Scholar] [CrossRef]
- Graham, S.F.; Rey, N.L.; Ugur, Z.; Yilmaz, A.; Sherman, E.; Maddens, M.; Bahado-Singh, R.O.; Becker, K.; Schulz, E.; Meyerdirk, L.K.; et al. Metabolomic Profiling of Bile Acids in an Experimental Model of Prodromal Parkinson’s Disease. Metabolites 2018, 8, 71. [Google Scholar] [CrossRef]
- Ackerman, H.D.; Gerhard, G.S. Bile Acids in Neurodegenerative Disorders. Front. Aging Neurosci. 2016, 8, 263. [Google Scholar] [CrossRef]
- Nunes, A.F.; Amaral, J.D.; Lo, A.C.; Fonseca, M.B.; Viana, R.J.; Callaerts-Vegh, Z.; D’Hooge, R.; Rodrigues, C.M. TUDCA, a bile acid, attenuates amyloid precursor protein processing and amyloid-β deposition in APP/PS1 mice. Mol. Neurobiol. 2012, 45, 440–454. [Google Scholar] [CrossRef]
- Castro-Caldas, M.; Carvalho, A.N.; Rodrigues, E.; Henderson, C.; Wolf, C.R.; Rodrigues, C.; Gama, M.J. Tauroursodeoxycholic Acid Prevents MPTP-Induced Dopaminergic Cell Death in a Mouse Model of Parkinson’s Disease. Mol. Neurobiol. 2012, 46, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Health Aging 2018, 4, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Leite, G.; Pimentel, M.; Barlow, G.M.; Chang, C.; Hosseini, A.; Wang, J.; Parodi, G.; Sedighi, R.; Rezaie, A.; Mathur, R. Age and the aging process significantly alter the small bowel microbiome. Cell Rep. 2021, 36, 109765. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R. Emerging Interrelationship Between the Gut Microbiome and Cellular Senescence in the Context of Aging and Disease: Perspectives and Therapeutic Opportunities. Probiotics Antimicrob. Proteins 2022, 14, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Wilmanski, T.; Diener, C.; Rappaport, N.; Patwardhan, S.; Wiedrick, J.; Lapidus, J.; Earls, J.C.; Zimmer, A.; Glusman, G.; Robinson, M.; et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 2021, 3, 274–286. [Google Scholar] [CrossRef]
- Lee, H.-Y.; Lee, S.-H.; Lee, J.-H.; Lee, W.-J.; Min, K.-J. The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging 2019, 11, 4611–4640. [Google Scholar] [CrossRef]
- Smith, P.; Willemsen, D.; Popkes, M.; Metge, F.; Gandiwa, E.; Reichard, M.; Valenzano, D.R. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. eLife 2017, 6, e27014. [Google Scholar] [CrossRef]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Shah, Z.; Alsegiani, A. The influence of gut microbiota alteration on age-related neuroinflammation and cognitive decline. Neural Regen. Res. 2022, 17, 2407. [Google Scholar] [CrossRef]
- Zhan, X.; Hakoupian, M.; Jin, L.-W.; Sharp, F.R. Lipopolysaccharide, Identified Using an Antibody and by PAS Staining, Is Associated With Corpora amylacea and White Matter Injury in Alzheimer’s Disease and Aging Brain. Front. Aging Neurosci. 2021, 13, 705594. [Google Scholar] [CrossRef]
- Wu, M.-L.; Yang, X.-Q.; Xue, L.; Duan, W.; Du, J.-R. Age-related cognitive decline is associated with microbiota-gut-brain axis disorders and neuroinflammation in mice. Behav. Brain Res. 2021, 402, 113125. [Google Scholar] [CrossRef]
- Villarán, R.F.; Espinosa-Oliva, A.M.; Sarmiento, M.; De Pablos, R.M.; Argüelles, S.; Delgado-Cortés, M.J.; Sobrino, V.; Van Rooijen, N.; Venero, J.L.; Herrera, A.J.; et al. Ulcerative colitis exacerbates lipopolysaccharide-induced damage to the nigral dopaminergic system: Potential risk factor in Parkinson’s disease. J. Neurochem. 2010, 114, 1687–1700. [Google Scholar] [CrossRef]
- Devi, P.; Khan, A.; Chattopadhyay, P.; Mehta, P.; Sahni, S.; Sharma, S.; Pandey, R. Co-infections as Modulators of Disease Outcome: Minor Players or Major Players? Front. Microbiol. 2021, 12, 664386. [Google Scholar] [CrossRef]
- Traylen, C.M.; Patel, H.R.; Fondaw, W.; Mahatme, S.; Williams, J.F.; Walker, L.R.; Dyson, O.F.; Arce, S.; Akula, S.M. Virus reactivation: A panoramic view in human infections. Future Virol. 2011, 6, 451–463. [Google Scholar] [CrossRef]
- Li, N.; Ma, W.-T.; Pang, M.; Fan, Q.-L.; Hua, J.-L. The Commensal Microbiota and Viral Infection: A Comprehensive Review. Front. Immunol. 2019, 10, 1551. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef]
- Díaz, C.D.; García-Orozco, A.; Riera-Leal, A.; Padilla-Arellano, J.R.; Fafutis-Morris, M. Microbiota and Its Role on Viral Evasion: Is It With Us or Against Us? Front. Cell. Infect. Microbiol. 2019, 9, 256. [Google Scholar] [CrossRef]
- Follmer, C. Viral Infection-Induced Gut Dysbiosis, Neuroinflammation, and α-Synuclein Aggregation: Updates and Perspectives on COVID-19 and Neurodegenerative Disorders. ACS Chem. Neurosci. 2020, 11, 4012–4016. [Google Scholar] [CrossRef]
- Dutta, S.K.; Verma, S.; Jain, V.; Surapaneni, B.; Vinayek, R.; Phillips, L.; Nair, P.P. Parkinson’s Disease: The Emerging Role of Gut Dysbiosis, Antibiotics, Probiotics, and Fecal Microbiota Transplantation. J. Neurogastroenterol. Motil. 2019, 25, 363–376. [Google Scholar] [CrossRef]
- Nandwana, V.; Debbarma, S. Fecal Microbiota Transplantation: A Microbiome Modulation Technique for Alzheimer’s Disease. Cureus 2021, 13, e16503. [Google Scholar] [CrossRef]
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal microbiota transplantation: In perspective. Ther. Adv. Gastroenterol. 2015, 9, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Borody, T.; Brandt, L.J.; Paramsothy, S. Therapeutic faecal microbiota transplantation. Curr. Opin. Gastroenterol. 2014, 30, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Vendrik, K.E.W.; Ooijevaar, R.E.; De Jong, P.R.C.; Laman, J.D.; van Oosten, B.W.; Van Hilten, J.J.; Ducarmon, Q.R.; Keller, J.J.; Kuijper, E.J.; Contarino, M.F. Fecal Microbiota Transplantation in Neurological Disorders. Front. Cell. Infect. Microbiol. 2020, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, S.P.; Frye, A.K.; El-Gendy, A.O.; Castejon, M.; Keshavarzian, A.; van Dijk, G.; El Aidy, S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat. Commun. 2019, 10, 310–311. [Google Scholar] [CrossRef]
- Holmes, A.; Finger, C.; Morales-Scheihing, D.; Lee, J.; McCullough, L.D. Gut dysbiosis and age-related neurological diseases; an innovative approach for therapeutic interventions. Transl. Res. 2020, 226, 39–56. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Han, B.; Sivaramakrishnan, P.; Lin, C.-C.J.; Neve, I.A.; He, J.; Tay, L.W.R.; Sowa, J.N.; Sizovs, A.; Du, G.; Wang, J.; et al. Microbial Genetic Composition Tunes Host Longevity. Cell 2017, 169, 1249–1262.e13. [Google Scholar] [CrossRef]
- Shin, J.; Noh, J.-R.; Choe, D.; Lee, N.; Song, Y.; Cho, S.; Kang, E.-J.; Go, M.-J.; Ha, S.K.; Chang, D.-H.; et al. Ageing and rejuvenation models reveal changes in key microbial communities associated with healthy ageing. Microbiome 2021, 9, 240. [Google Scholar] [CrossRef]
- Wu, T.; Lyu, Y.; Li, X.; Wu, M.; Yu, K.; Li, S.; Ji, C.; Zhang, Q.; Zhang, Y.; Zhao, D.; et al. Impact of N-Acetylcysteine on the Gut Microbiota in the Piglets Infected With Porcine Epidemic Diarrhea Virus. Front. Veter.-Sci. 2021, 7, 582338. [Google Scholar] [CrossRef]
- Tenório, M.C.d.S.; Graciliano, N.G.; Moura, F.; de Oliveira, A.C.M.; Goulart, M.O.F. N-Acetylcysteine (NAC): Impacts on Human Health. Antioxidants 2021, 10, 967. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yuan, X.; Zhang, C.; Jia, P.; Jiao, S.; Zhao, X.; Yin, H.; Du, Y.; Liu, H. N -Acetylcysteine Alleviates Gut Dysbiosis and Glucose Metabolic Disorder in High-Fat Diet-Fed Mice. J. Diabetes 2018, 11, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Kang, K.S. N-acetylcysteine modulates lipopolysaccharide-induced intestinal dysfunction. Sci. Rep. 2019, 9, 1004. [Google Scholar] [CrossRef] [PubMed]
| GENUS | Alzheimer’s Disease | Parkinson’s Disease | Huntington’s Disease | Multiple Sclerosis |
|---|---|---|---|---|
| Bifidobacterium | ↓ | ↑ | ↑ | - |
| Clostridium | ↓ | ↓ | ↓ | ↑ |
| Dialister | ↓ | - | - | - |
| Turicibacter | ↓ | - | - | - |
| Bacteroides | ↑ | ↓ | ↓ | - |
| Blautia | ↑ | ↓ | - | ↑ |
| Bilophila | ↑ | - | - | - |
| Lactobacillus | - | ↓ | - | ↓ |
| Faecalibacterium | - | ↓ | ↑ | - |
| Coprococcus | - | ↓ | - | - |
| Prevotella | - | ↓ | ↑ | ↓ |
| Akkermansia | - | ↑ | - | ↑ |
| Methanobrevibacter | - | - | - | ↑ |
| Butyricimonas | - | - | - | ↓ |
| Collinsella | - | - | - | ↓ |
| Slackia | - | - | - | ↓ |
| Megamonas | - | - | ↓ | - |
| Gemmiger | - | - | ↑ | - |
| Allistipes | ↑ | - | ↓ | - |
| S. No. | Therapeutic Approach | Phylum/Genus or the Name of Particular Bacteria | Neurological Disorder | References |
|---|---|---|---|---|
| 1. | Recolonization of beneficial bacteria | Oral administration of Bacteroides fragilis | Experimental autoimmune encephalomyelitis | [174] |
| 2. | Fecal microbiota transplantation | Fecal microbiota suspension was injected through a TET tube. | Parkinson’s disease | [175] |
| 3. | Bacteriotherapy | Transcolonoscopic infusion of 13 non-pathogenic enteric bacteria | Chronic Fatigue Syndrome (CFS) | [176] |
| 4. | Administration of human commensal bacteria | Bacteroides fragilis | MIA mouse model of ASD (autism spectrum disorder) | [177] |
| 5. | Probiotic supplementation | Lactobacillus acidophilus, L. fermentum, Bifidobacterium lactis | Alzheimer’s disease | [178] |
| 6. | Fecal microbiota transplantation | FMT from healthy uninjured mice | Spinal cord injury (SCI) mice model | [179] |
| 7. | Probiotic supplementation | Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI | Alzheimer’s disease | [180] |
| 8. | Probiotic supplementation | Bifidobacterium infantis | Rat maternal separation (MS) model of depression | [181] |
| 9. | Fecal microbiota transplantation | Fecal suspension injected into the colon through the catheters | Traumatic brain injury (TBI) in male Sprague Dawley rats | [182] |
| 10. | Probiotic supplementation | Streptococcus thermophilus, Bifidobacterium lactis, Lactobacillus acidophilus, Lactobacillus helveticus, | Alzheimer’s disease triple-transgenic mice | [183] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kandpal, M.; Indari, O.; Baral, B.; Jakhmola, S.; Tiwari, D.; Bhandari, V.; Pandey, R.K.; Bala, K.; Sonawane, A.; Jha, H.C. Dysbiosis of Gut Microbiota from the Perspective of the Gut–Brain Axis: Role in the Provocation of Neurological Disorders. Metabolites 2022, 12, 1064. https://doi.org/10.3390/metabo12111064
Kandpal M, Indari O, Baral B, Jakhmola S, Tiwari D, Bhandari V, Pandey RK, Bala K, Sonawane A, Jha HC. Dysbiosis of Gut Microbiota from the Perspective of the Gut–Brain Axis: Role in the Provocation of Neurological Disorders. Metabolites. 2022; 12(11):1064. https://doi.org/10.3390/metabo12111064
Chicago/Turabian StyleKandpal, Meenakshi, Omkar Indari, Budhadev Baral, Shweta Jakhmola, Deeksha Tiwari, Vasundhra Bhandari, Rajan Kumar Pandey, Kiran Bala, Avinash Sonawane, and Hem Chandra Jha. 2022. "Dysbiosis of Gut Microbiota from the Perspective of the Gut–Brain Axis: Role in the Provocation of Neurological Disorders" Metabolites 12, no. 11: 1064. https://doi.org/10.3390/metabo12111064
APA StyleKandpal, M., Indari, O., Baral, B., Jakhmola, S., Tiwari, D., Bhandari, V., Pandey, R. K., Bala, K., Sonawane, A., & Jha, H. C. (2022). Dysbiosis of Gut Microbiota from the Perspective of the Gut–Brain Axis: Role in the Provocation of Neurological Disorders. Metabolites, 12(11), 1064. https://doi.org/10.3390/metabo12111064