Effects of Dietary Cottonseed Protein Concentrate Levels on Growth Performance, Health Status, Flesh Quality and Intestinal Microbiota of Grass Carp (Ctenopharyngodon idellus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Diets
2.2. Fish and Feeding Trial
2.3. Sample Collection
2.4. Sample Analysis
2.4.1. Determination of Serum Biochemical Indices
2.4.2. Proximate Composition
2.4.3. Enzyme Activity of Muscle and Hepatopancreas
2.4.4. Apparent Digestibility
2.4.5. Histological Observation
2.4.6. Muscle Textural Properties
2.4.7. Gene Expression Quantification
2.4.8. Intestine Microbiome Analysis
2.5. Statistical Analysis
3. Results
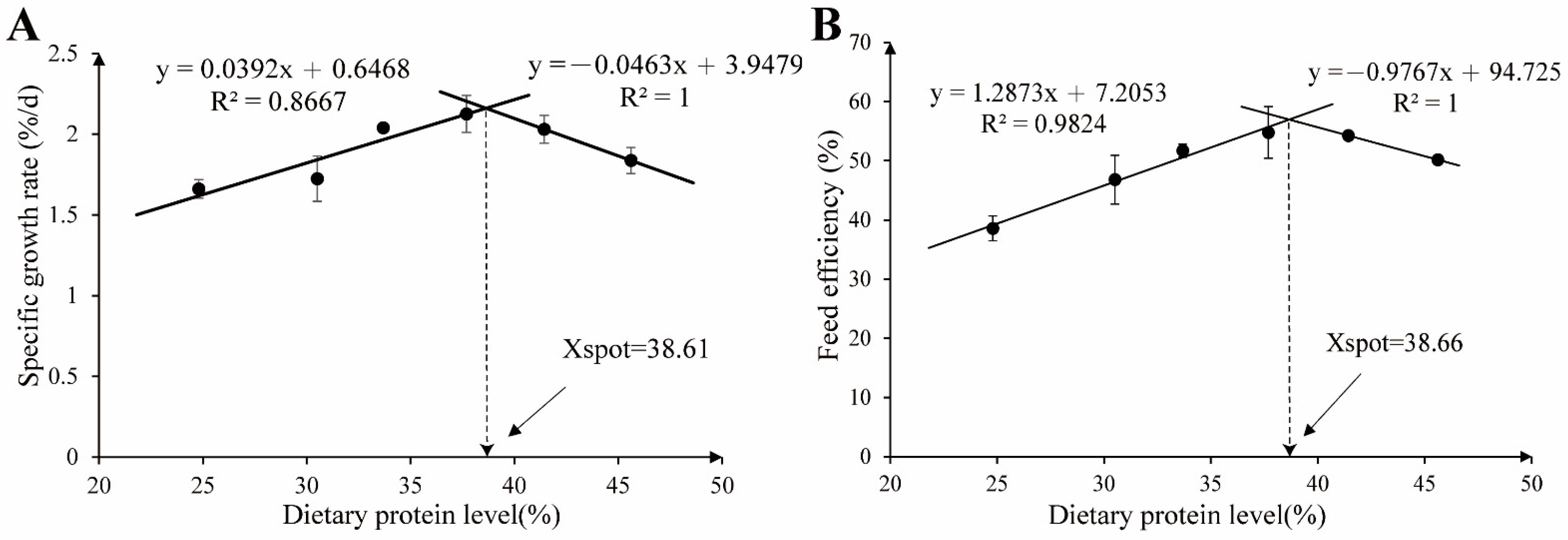
3.1. Growth Performance, Apparent Digestibility, and Morphology Parameters
3.2. Body and Dorsal Muscle Proximate Composition
3.3. Serum Biochemical Indices and Immune Enzyme Activity
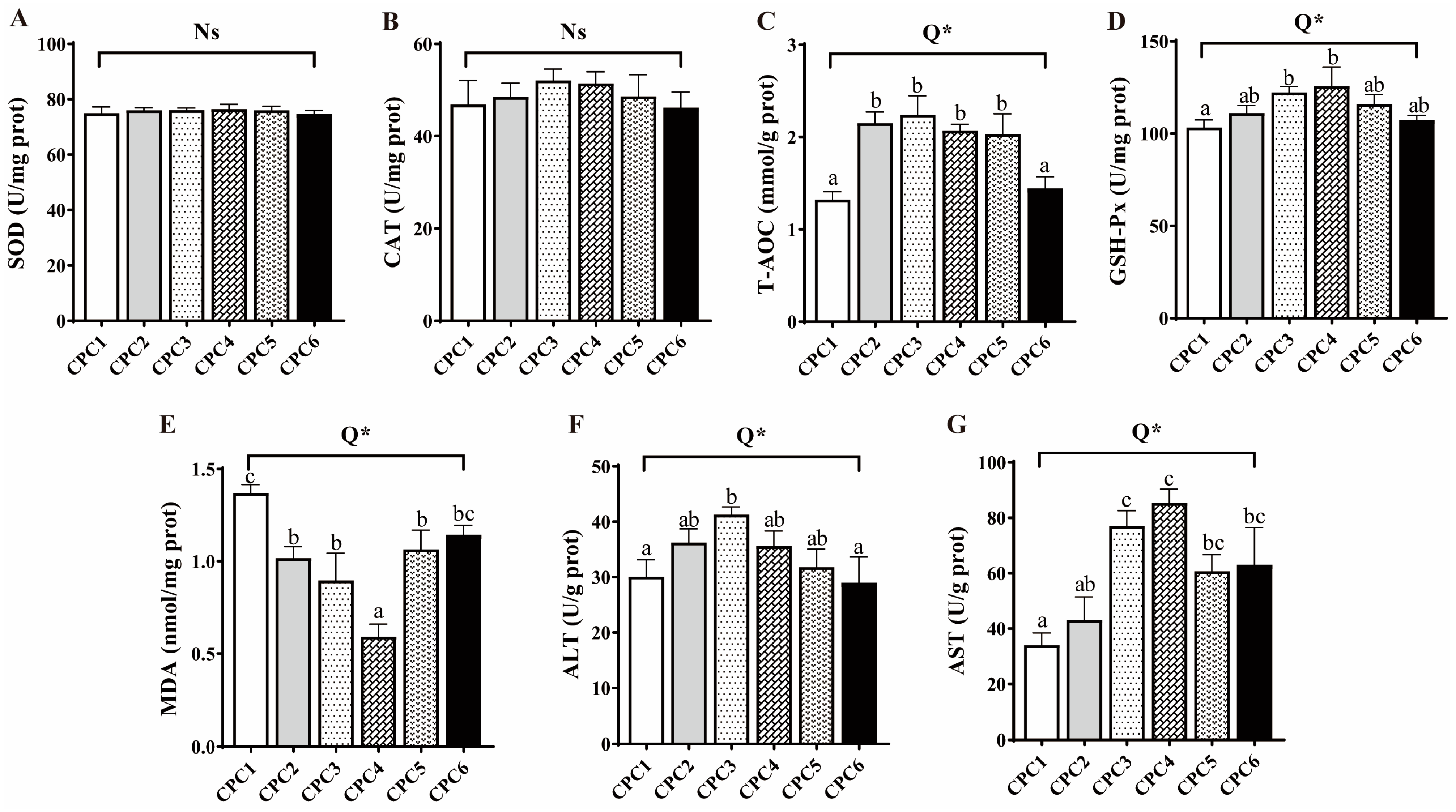
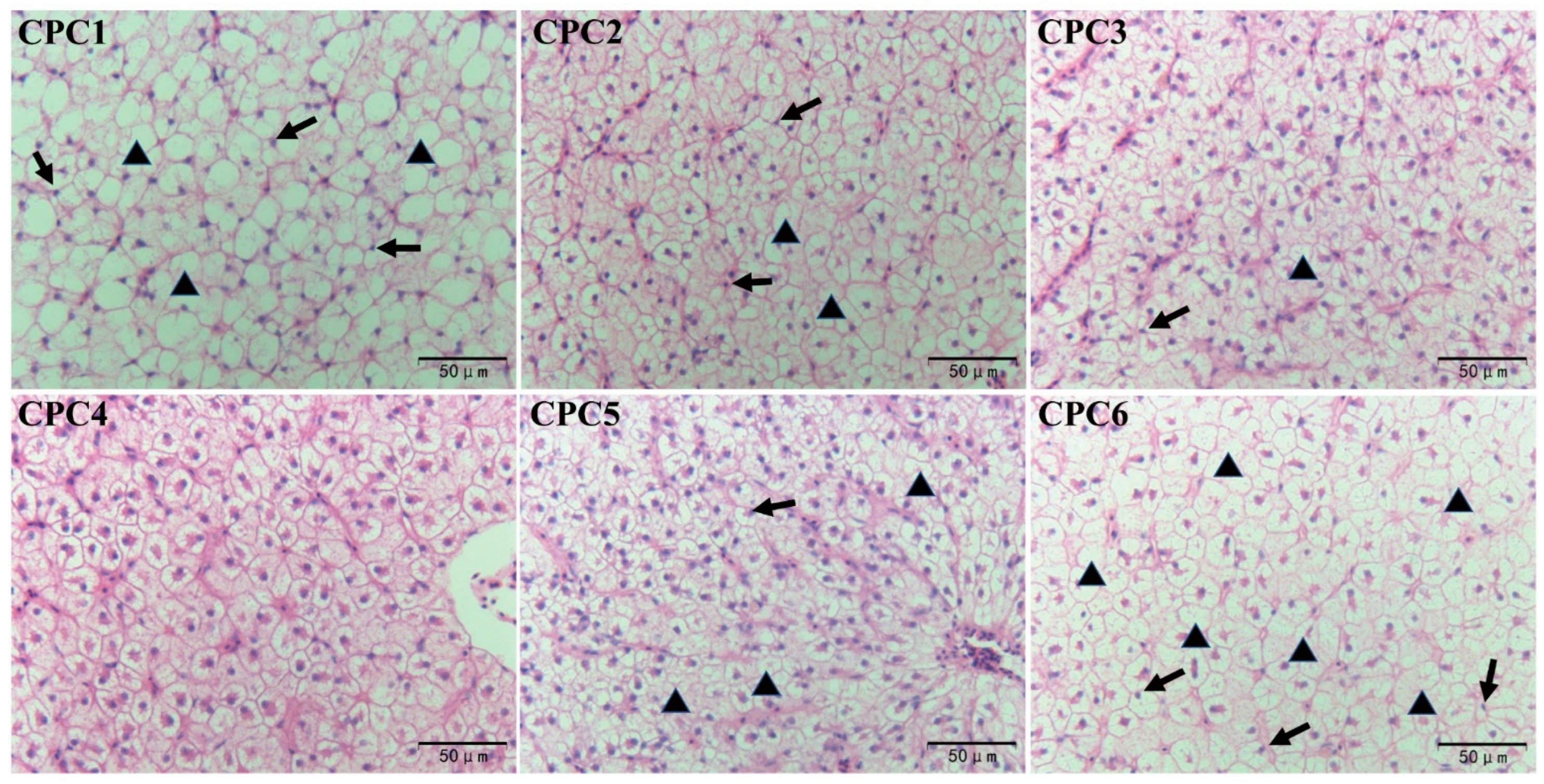
3.4. Antioxidative Capacity, Metabolic Enzymes, and Histological Observation of Hepatopancreas
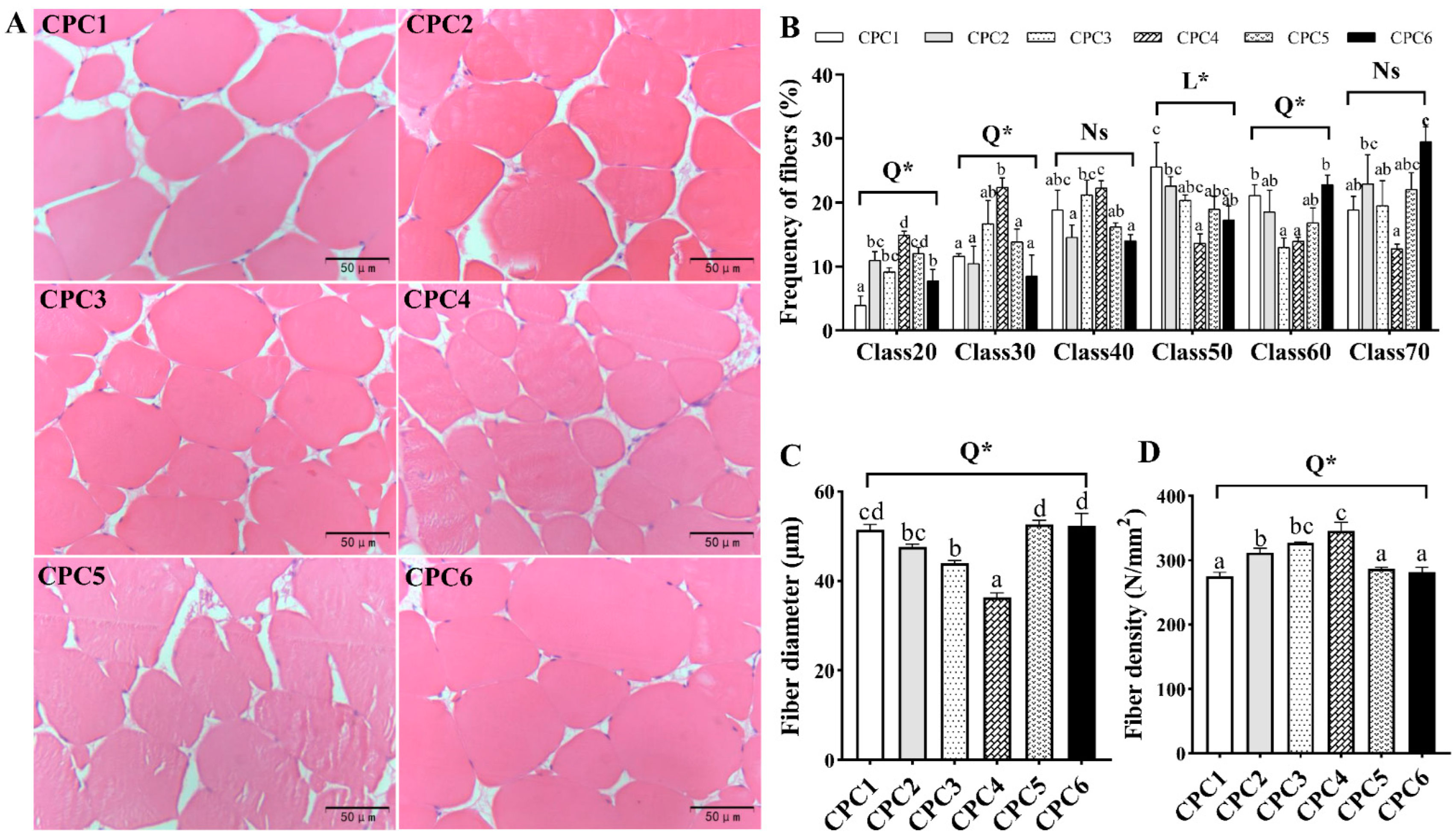
3.5. Muscle Texture Analysis
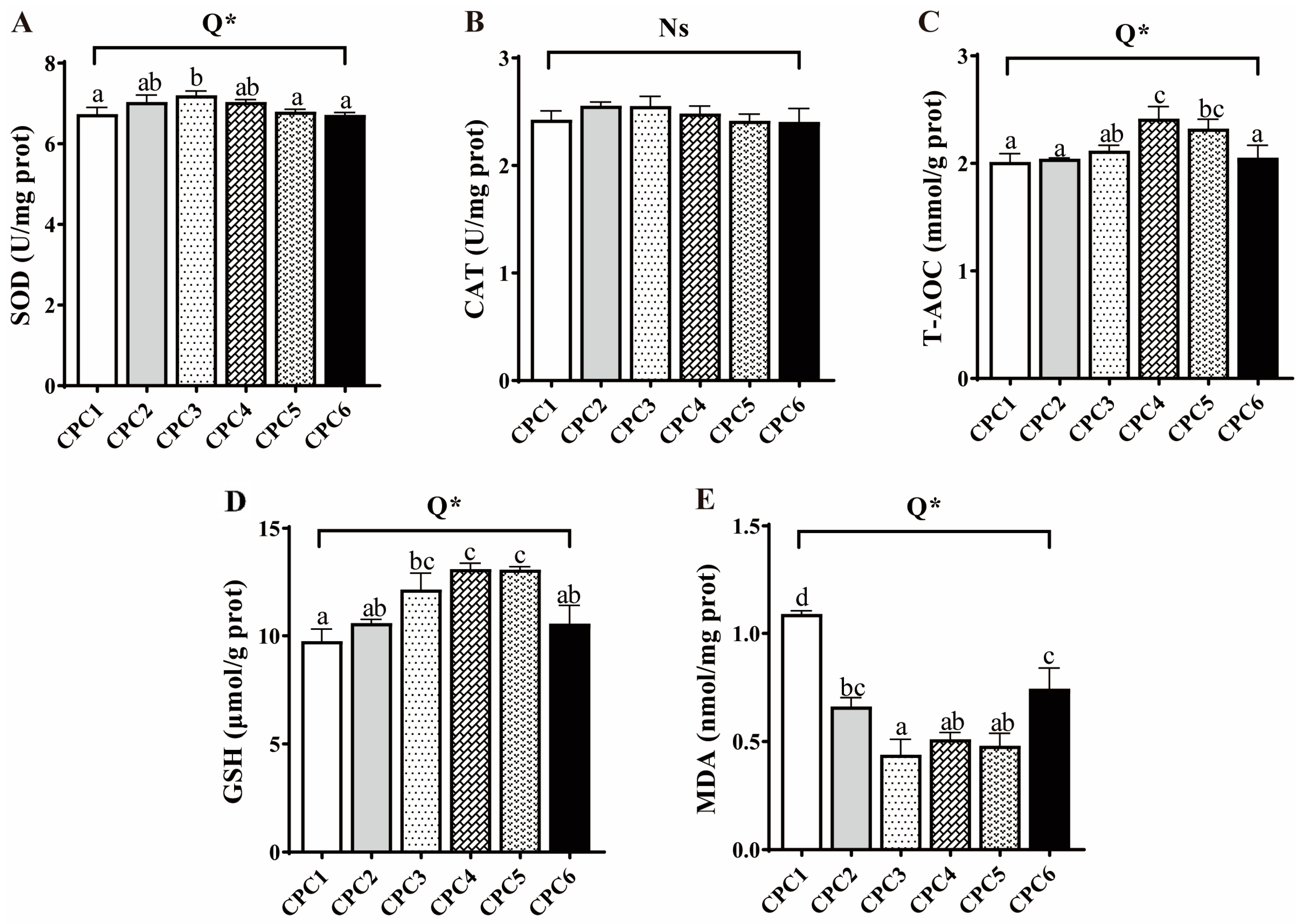
3.6. Antioxidative Capacity and Histological Observation of Muscle
3.7. Muscle-Related Genes Expression
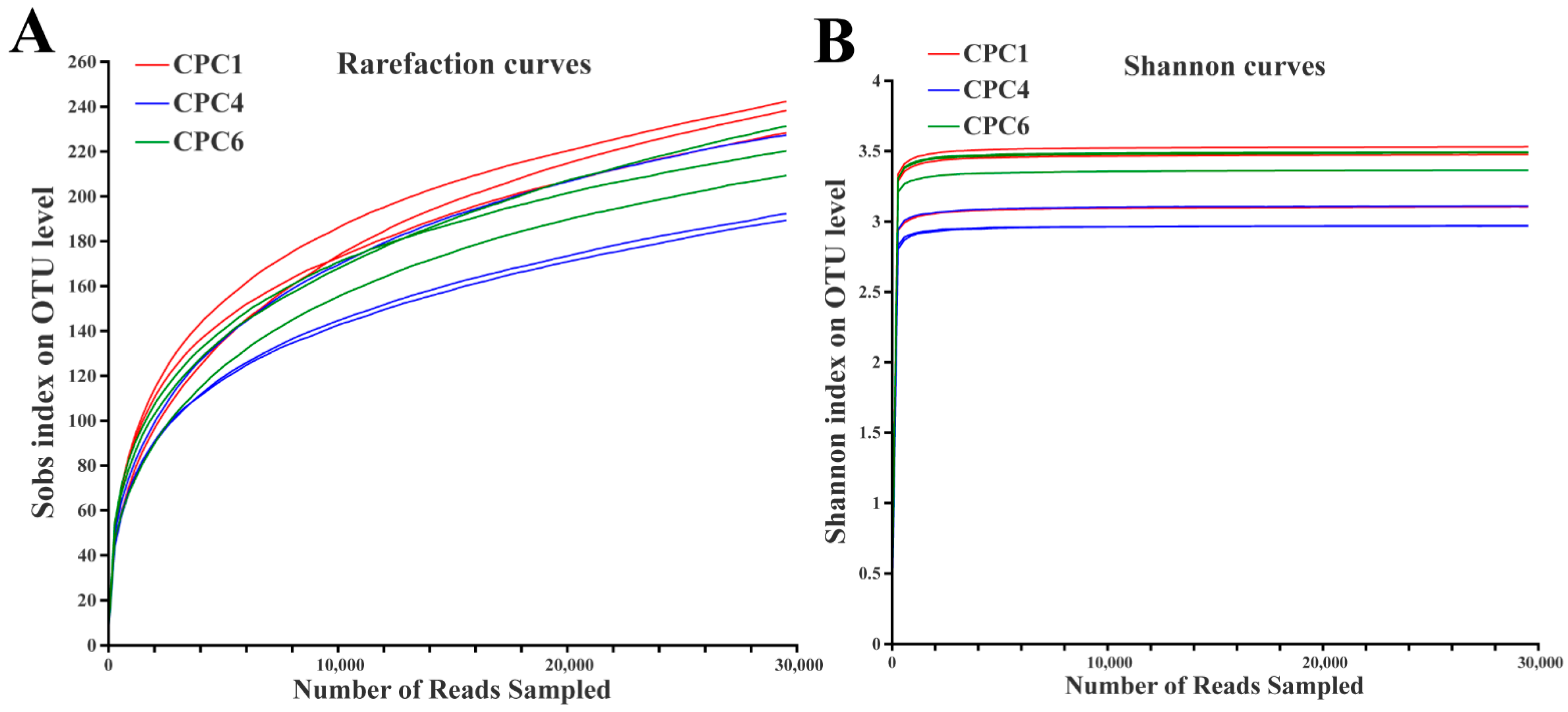
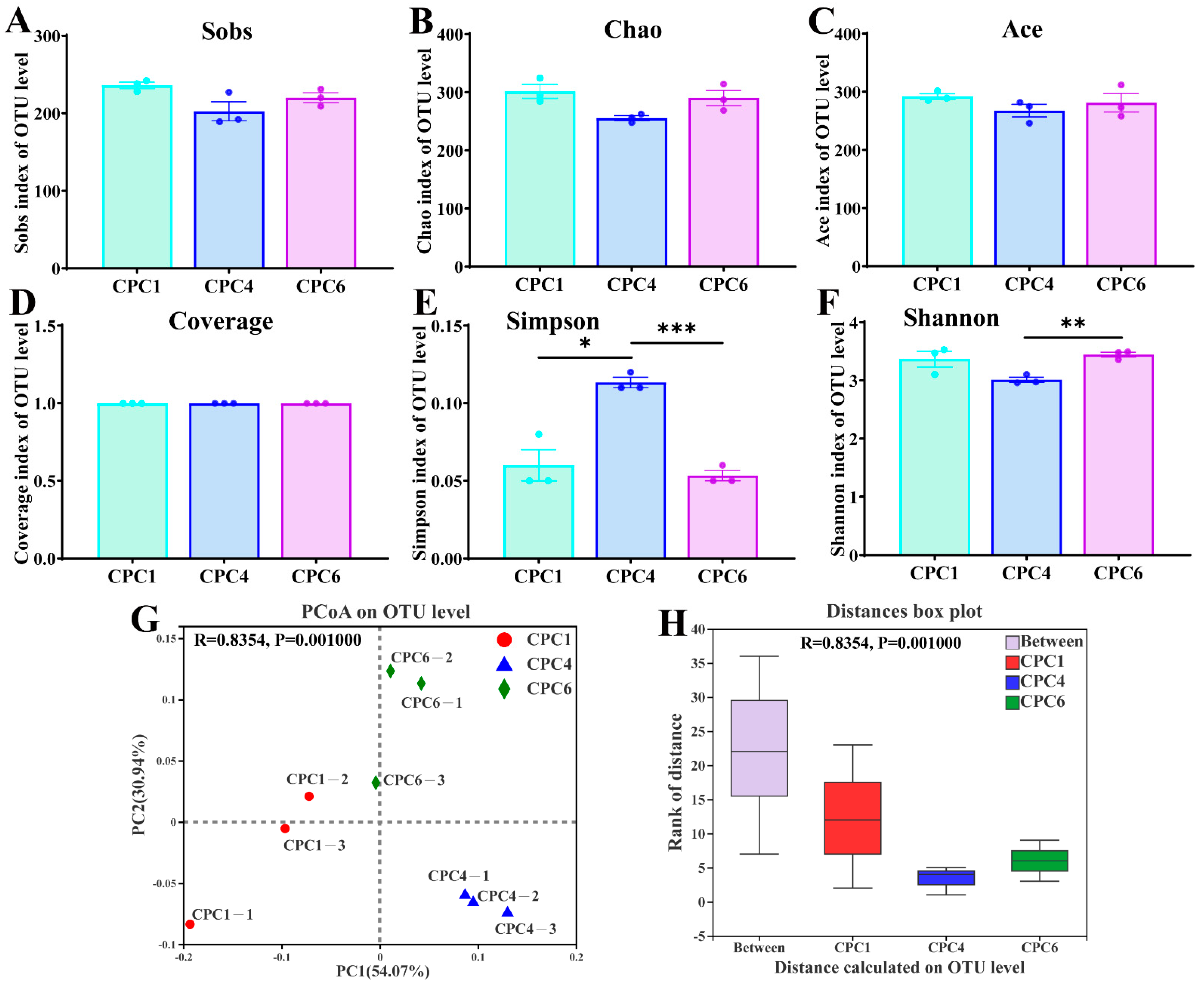
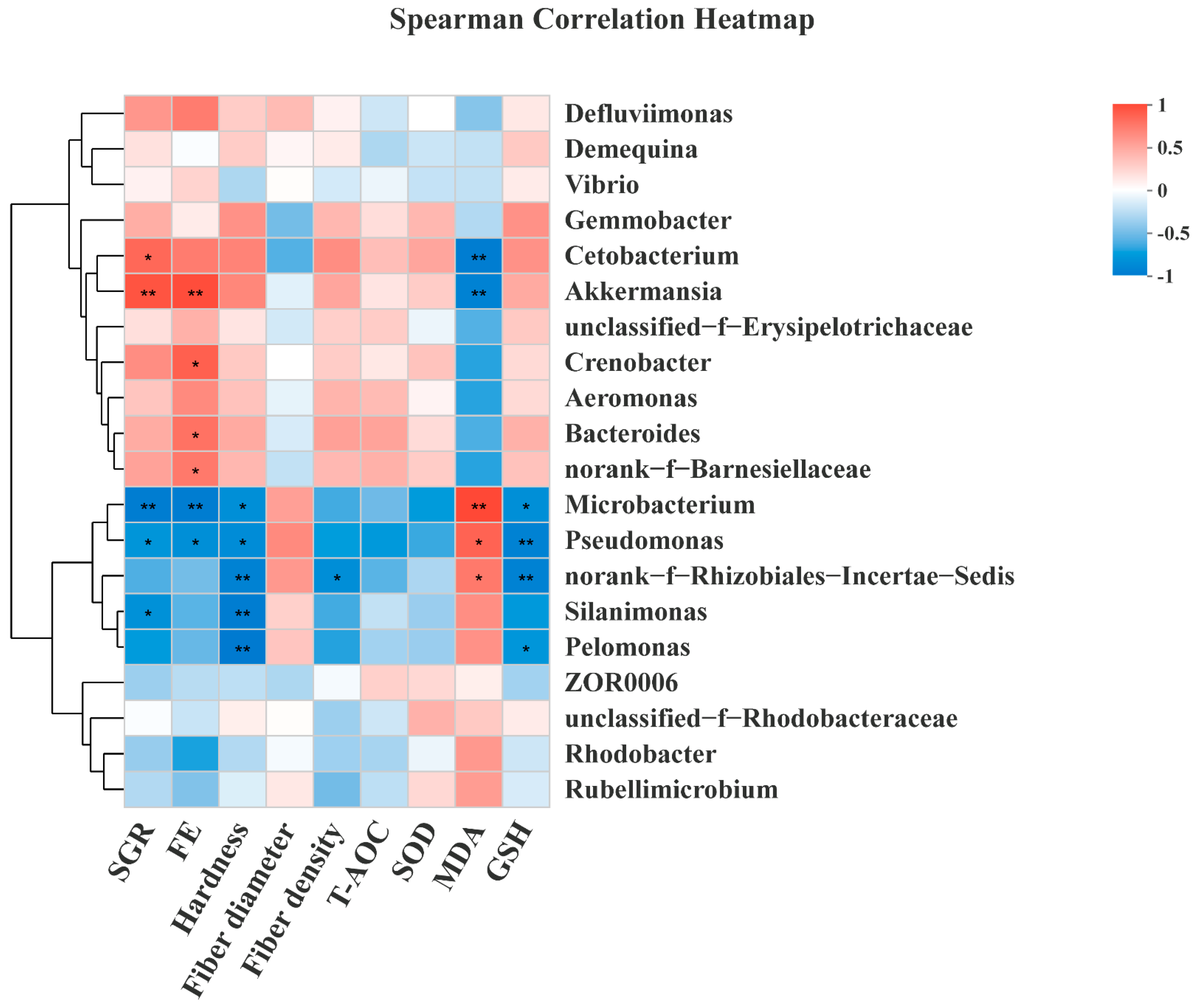
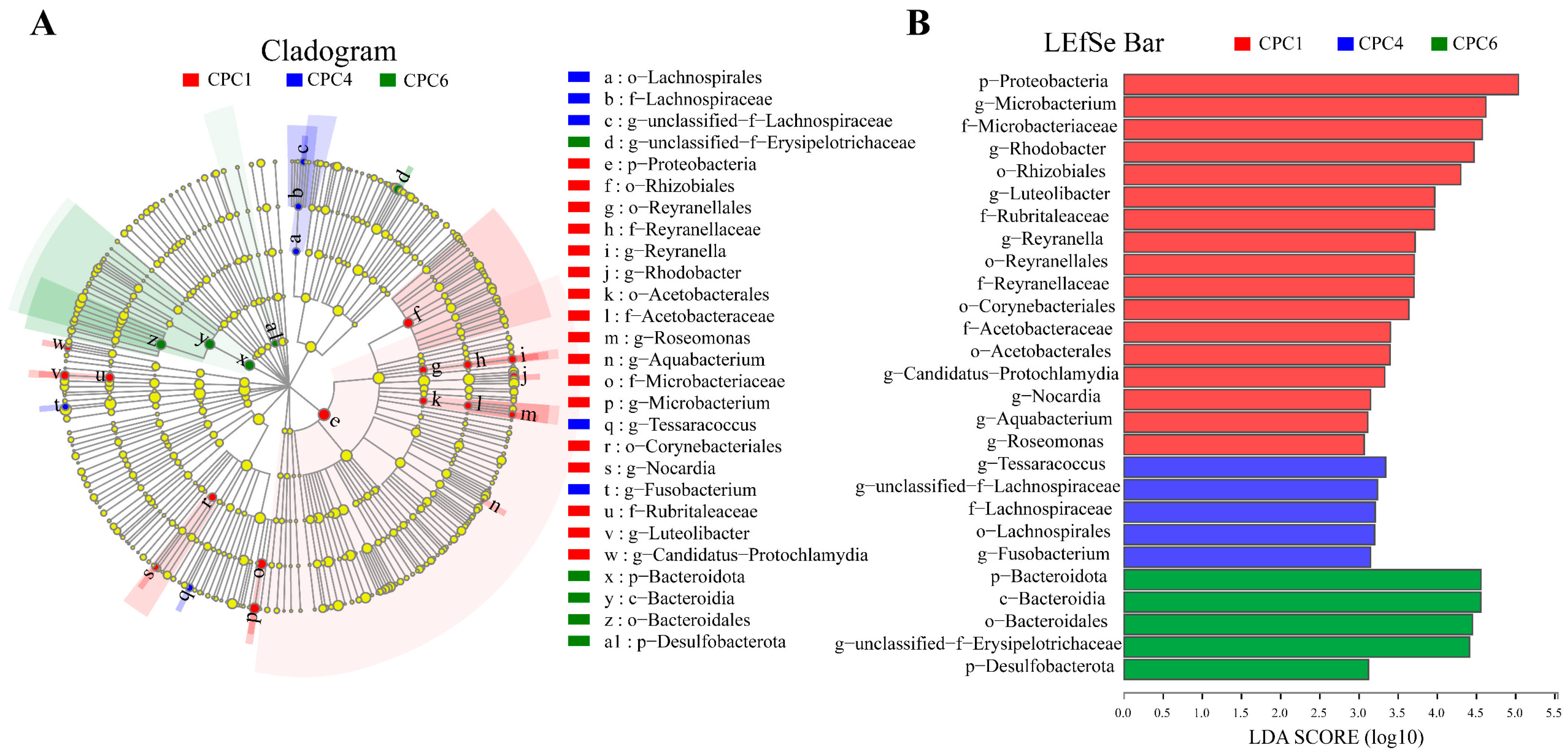
3.8. Intestinal Microbiota of Grass Carp
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olsen, R.L.; Hasan, M.R. A limited supply of fishmeal: Impact on future increases in global aquaculture production. Trends Food Sci. Technol. 2012, 27, 120–128. [Google Scholar] [CrossRef]
- Montoya-Camacho, N.; Marquez-Ríos, E.; Castillo-Yáñez, F.J.; Cárdenas López, J.L.; López-Elías, J.A.; Ruíz-Cruz, S.; Jiménez-Ruíz, E.I.; Rivas-Vega, M.E.; Ocaño-Higuera, V.M. Advances in the use of alternative protein sources for tilapia feeding. Rev. Aquac. 2019, 11, 515–526. [Google Scholar] [CrossRef]
- Xie, X.; Wang, J.; Guan, Y.; Xing, S.; Liang, X.; Xue, M.; Wang, J.; Chang, Y.; Leclercq, E. Cottonseed protein concentrate as fishmeal alternative for largemouth bass (Micropterus salmoides) supplemented a yeast-based paraprobiotic: Effects on growth performance, gut health and microbiome. Aquaculture 2022, 551, 737898. [Google Scholar] [CrossRef]
- Xu, X.; Yang, H.; Zhang, C.; Bian, Y.; Yao, W.; Xu, Z.; Wang, Y.; Li, X.; Leng, X. Effects of replacing fishmeal with cottonseed protein concentrate on growth performance, flesh quality and gossypol deposition of largemouth bass (Micropterus salmoides). Aquaculture 2022, 548, 737551. [Google Scholar] [CrossRef]
- Chen, G.; Yin, B.; Liu, H.; Tan, B.; Dong, X.; Yang, Q.; Chi, S.; Zhang, S. Effects of fishmeal replacement with cottonseed protein concentrate on growth, digestive proteinase, intestinal morphology and microflora in pearl gentian grouper (♀ Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatu). Aquac. Res. 2020, 51, 2870–2884. [Google Scholar] [CrossRef]
- Wu, S.; Wang, G.; Angert, E.R.; Wang, W.; Li, W.; Zou, H. Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS ONE 2012, 7, e30440. [Google Scholar] [CrossRef]
- Lin, S.; Milardi, M.; Gao, Y.; Wong, M.H. Sustainable management of non-native grass carp as a protein source, weed-control agent and sport fish. Aquac. Res. 2022. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United States (FAO). Global Aquaculture Production Quantity. 1950–2020. Available online: https://www.fao.org/fishery/statistics-query/en/aquaculture/aquaculture_quantity (accessed on 24 October 2022).
- Dabrowski, K. Protein requirements of grass carp fry (Ctenopharyngodon idella Val.). Aquaculture 1977, 12, 63–73. [Google Scholar] [CrossRef]
- Jin, Y.; Tian, L.X.; Xie, S.W.; Guo, D.Q.; Yang, H.J.; Liang, G.Y.; Liu, Y.J. Interactions between dietary protein levels, growth performance, feed utilization, gene expression and metabolic products in juvenile grass carp (Ctenopharyngodon idella). Aquaculture 2015, 437, 75–83. [Google Scholar] [CrossRef]
- Xu, J.; Wu, P.; Jiang, W.D.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Tang, W.N.; Zhang, Y.A.; Zhou, X.Q.; et al. Optimal dietary protein level improved growth, disease resistance, intestinal immune and physical barrier function of young grass carp (Ctenopharyngodon idella). Fish Shellfish. Immunol. 2016, 55, 64–87. [Google Scholar] [CrossRef]
- Xu, J.; Feng, L.; Jiang, W.D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Zhou, X.Q. Different dietary protein levels affect flesh quality, fatty acids and alter gene expression of Nrf2-mediated antioxidant enzymes in the muscle of grass carp (Ctenopharyngodon idella). Aquaculture 2018, 493, 272–282. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Y.; Feng, L.; Jiang, W.D.; Kuang, S.Y.; Jiang, J.; Li, S.H.; Tang, L.; Zhou, X.Q. Effects of dietary arginine supplementation on growth performance, flesh quality, muscle antioxidant capacity and antioxidant-related signalling molecule expression in young grass carp (Ctenopharyngodon idella). Food Chem. 2015, 167, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.A. Muscle development and growth: Potential implications for flesh quality in fish. Aquaculture 1999, 177, 99–115. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.Y.; Jiang, Q.; Zhou, X.Q.; Feng, L.; Liu, Y.; Jiang, W.D.; Wu, P.; Zhou, J.; Zhao, J. Leucine improved growth performance, muscle growth, and muscle protein deposition through AKT/TOR and AKT/FOXO3 a signaling pathways in hybrid catfish Pelteobagrus vachelli × Leiocassis longirostris. Cells 2020, 9, 327. [Google Scholar] [CrossRef]
- Abouel Azm, F.R.; Kong, F.; Tan, Q.; Zhu, Y.; Yu, H.; Yao, J.; Luo, Z. Effects of replacement of dietary rapeseed meal by distiller’s dried grains with solubles (DDGS) on growth performance, muscle texture, health and expression of muscle-related genes in grass carp (Ctenopharyngodon idellus). Aquaculture 2021, 533, 736169. [Google Scholar] [CrossRef]
- Xu, W.H.; Guo, H.H.; Chen, S.J.; Wang, Y.Z.; Lin, Z.H.; Huang, X.D.; Tang, H.J.; He, Y.H.; Sun, J.J.; Gan, L. Transcriptome analysis revealed changes of multiple genes involved in muscle hardness in grass carp (Ctenopharyngodon idellus) fed with faba bean meal. Food Chem. 2020, 314, 126205. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.L.; Zeng, Q.X.; Zhu, Z.W. Different changes in mastication between crisp grass carp (Ctenopharyngodon idellus C. et V) and grass carp (Ctenopharyngodon idellus) after heating: The relationship between texture and ultrastructure in muscle tissue. Food Res. Int. 2009, 42, 271–278. [Google Scholar] [CrossRef]
- Kong, F.; Abouel Azm, F.R.; Wang, X.; Zhu, Y.; Yu, H.; Yao, J.; Luo, Z.; Tan, Q. Effects of replacement of dietary cottonseed meal by distiller’s dried grains with solubles on growth performance, muscle texture, health and expression of muscle-related genes in grass carp (Ctenopharyngodon idellus). Aquac. Nutr. 2021, 27, 1255–1266. [Google Scholar] [CrossRef]
- Wang, X.Y.; Liu, G.Q.; Xie, S.Q.; Pan, L.; Tan, Q.S. Growth and meat quality of grass carp (Ctenopharyngodon idellus) responded to dietary protein (soybean meal) level through the muscle metabolism and gene expression of myosin heavy chains. Front. Nutr. 2022, 9, 833924. [Google Scholar] [CrossRef]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef]
- Eroglu, A.; Dogan, Z.; Kanak, E.; Atli, G.; Canli, M. Effects of heavy metals (Cd, Cu, Cr, Pb, Zn) on fish glutathione metabolism. Environ. Sci. Pollut. Res. 2015, 22, 3229–3237. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Wu, D.; Zhang, Y.; Li, J.; Xu, Q.; Wang, L. Carbonate alkalinity and dietary protein levels affected growth performance, intestinal immune responses and intestinal microflora in Songpu mirror carp (Cyprinus carpio Songpu). Aquaculture 2021, 545, 737135. [Google Scholar] [CrossRef]
- Przewłócka, K.; Folwarski, M.; Kaźmierczak-Siedlecka, K.; Skonieczna-Żydecka, K.; Kaczor, J.J. Gut-muscle axis exists and may affect skeletal muscle adaptation to training. Nutrients 2020, 12, 1451. [Google Scholar] [CrossRef] [PubMed]
- AOAC. Official Methods of Analysis of the Association of Official Analytical Chemists International, 16th ed.; AOAC: Arlington, VA, USA, 1995. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M.; Ahmad, M.H.; Khattab, Y.A.; Shalaby, A.M. Effect of dietary protein level, initial body weight, and their interaction on the growth, feed utilization, and physiological alterations of Nile tilapia, Oreochromis niloticus (L.). Aquaculture 2010, 298, 267–274. [Google Scholar] [CrossRef]
- Mcgoogan, B.B.; Gatlin Iii, D.M. Dietary manipulations affecting growth and nitrogenous waste production of red drum, Sciaenops ocellatus I. Effects of dietary protein and energy levels. Aquaculture 1999, 178, 333–348. [Google Scholar] [CrossRef]
- Bassily, N.; Michael, K.; Said, A. Blood urea content for evaluating dietary protein quality. Mol. Nutr. Food Res. 1982, 26, 759–764. [Google Scholar] [CrossRef]
- Lone, K.; Inch, B.; Matty, A. Changes in the blood chemistry of rainbow trout, Salmo gairdneri Rich, in relation to dietary protein level, and an anabolic steroid hormone, ethylestrenol. J. Fish Biol. 1982, 20, 597–606. [Google Scholar] [CrossRef]
- Huang, J.F.; Xu, Q.Y.; Chang, Y.M. Effects of temperature and dietary protein on gene expression of Hsp70 and Wap65 and immunity of juvenile mirror carp (Cyprinus carpio). Aquac. Res. 2015, 46, 2776–2788. [Google Scholar] [CrossRef]
- Martínez-Álvarez, R.M.; Morales, A.E.; Sanz, A. Antioxidant defenses in fish: Biotic and abiotic factors. Rev. Fish Biol. Fish. 2005, 15, 75–88. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Meng, H.; Kong, X.; Cheng, X.; Ma, T.; He, H.; Du, W.; Yang, S.; Li, S.; Zhang, L. Combined effects of polyethylene and organic contaminant on zebrafish (Danio rerio): Accumulation of 9-Nitroanthracene, biomarkers and intestinal microbiota. Environ. Pollut. 2021, 277, 116767. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, C.; Sakata, T.; Sugita, H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 2008, 46, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Anhê, F.F.; Pilon, G.; Roy, D.; Desjardins, Y.; Levy, E.; Marette, A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes 2016, 7, 146–153. [Google Scholar] [CrossRef]
- Kan, H.; Zhao, F.; Zhang, X.X.; Ren, H.; Gao, S. Correlations of gut microbial community shift with hepatic damage and growth inhibition of Carassius auratus induced by pentachlorophenol exposure. Environ. Sci. Technol. 2015, 49, 11894–11902. [Google Scholar] [CrossRef]
- Rimoldi, S.; Gliozheni, E.; Ascione, C.; Gini, E.; Terova, G. Effect of a specific composition of short-and medium-chain fatty acid 1-Monoglycerides on growth performances and gut microbiota of gilthead sea bream (Sparus aurata). PeerJ 2018, 6, e5355. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Abdel-Wahab, A.F.; Mahmoud, W.; Al-Harizy, R.M. Targeting glucose metabolism to suppress cancer progression: Prospective of anti-glycolytic cancer therapy. Pharmacol. Res. 2019, 150, 104511. [Google Scholar] [CrossRef]
- Cai, W.; Liang, X.F.; Yuan, X.; Liu, L.; He, S.; Li, J.; Li, B.; Xue, M. Different strategies of grass carp (Ctenopharyngodon idella) responding to insufficient or excessive dietary carbohydrate. Aquaculture 2018, 497, 292–298. [Google Scholar] [CrossRef]
- Serrano, J.A.; Nematipour, G.R.; Gatlin Iii, D.M. Dietary protein requirement of the red drum (Sciaenops ocellatus) and relative use of dietary carbohydrate and lipid. Aquaculture 1992, 101, 283–291. [Google Scholar] [CrossRef]
- Garling, D.L., Jr.; Wilson, R.P. Effects of dietary carbohydrate-to-lipid ratios on growth and body composition of fingerling channel catfish. Prog. Fish Cult. 1977, 39, 43–47. [Google Scholar] [CrossRef]



| CPC1 | CPC2 | CPC3 | CPC4 | CPC5 | CPC6 | |
|---|---|---|---|---|---|---|
| Cottonseed protein concentrate (CPC) 1 | 38.40 | 44.50 | 50.70 | 56.90 | 63.10 | 69.2 |
| Soy oil:Fish oil (1:1) | 5.90 | 5.76 | 5.62 | 5.47 | 5.32 | 5.14 |
| Corn starch | 18.00 | 16.80 | 15.60 | 14.40 | 13.20 | 11.98 |
| Microcrystalline cellulose | 34.30 | 29.54 | 24.68 | 19.83 | 14.98 | 10.29 |
| Vitamin and mineral premix 2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Calcium biphosphate | 1.80 | 1.80 | 1.80 | 1.80 | 1.80 | 1.80 |
| Choline chloride (50%) | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| Yttrium oxide | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 |
| Proximate composition (%) | ||||||
| Moisture | 6.55 | 7.14 | 6.67 | 7.42 | 7.42 | 7.88 |
| Crude protein (dry matter) | 24.80 | 30.51 | 33.68 | 37.69 | 41.43 | 45.61 |
| Crude lipid (dry matter) | 7.92 | 8.04 | 7.91 | 7.95 | 7.97 | 8.02 |
| Ash (dry matter) | 5.70 | 6.52 | 6.67 | 7.15 | 7.63 | 8.29 |
| Genes 1 | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplification Efficiency (%) | Accession Number |
|---|---|---|---|---|
| myod | ATGGAGTTGTCGGATATTCCCTTC | GCGGTCAGCGTTGGTTGTT | 104.45 | MG544985 |
| myog | TTACGAAGGCGGCGATAACTT | TGGTGAGGAGACATGGACAGA | 101.18 | JQ793897 |
| myf5 | GTGCCTGTGCCTCATCTCCT | AATGCGTGGTTCACCTTCTTCA | 92.41 | GU290227 |
| mrf4 | TCGCTCCTGTATTGATGTTGATGA | GCTCCTGTCTCGCATTCGTT | 107.98 | KT899334 |
| fgf6a | CGCATACGAGTCTTCCAT | CCTACGAGAACATCCAACA | 102.95 | MK050993 |
| fgf6b | TCCAGTCCGCTTCCGAGTA | AGATGAAACCCGATGCCTACA | 91.14 | MK050992 |
| mstn | CTGACGCCAAGTTCCACATACA | CGACTCTGCTTCAAGTTCTTCTCT | 99.15 | KP719016 |
| myhc-7 | AACTGCGCTGTAACGGTGTA | AGTGTGCCCAAACCTGTACT | 101.85 | MW113233 |
| myhc-2 | ACAGTGGCCAGCATTGATGA | TCCGCAGAGTTCAAACCCAA | 101.15 | MW113235 |
| myhc-4 | ACTCCGCTGACATGCTGAAA | TGTCCAGCACACCAATGAAGA | 103.78 | MW113236 |
| myhc-1 | TTCCGTTGTTGTGTCAGGCT | TACTGGATGACGCGTTTGGT | 99.12 | MW113234 |
| igf-II | TCTGTGGCAGTCCTCAACAAC | TTCCGCAACTTCTTCGCTCTT | 97.78 | EF062860 |
| tor | TCCCACTTTCCACCAACT | ACACCTCCACCTTCTCCA | 105.68 | JX854449 |
| s6k1 | ACATAAAGCAGCCTGACG | TGGAGGAGGTAATGGACG | 101.51 | EF373673 |
| 4e-bp1 | GCTGGCTGAGTTTGTGGTTG | CGAGTCGTGCTAAAAAGGGTC | 99.59 | KT757305 |
| β-actin | TATGTTGGTGACGAGGCTCA | GCAGCTCGTTGTAGAAGGTG | 98.82 | M25013 |
| ef1α | TGACTGTGCCGTGCTGAT | CGCTGACTTCCTTGGTGATT | 99.51 | GQ266394 |
| Items 2 | Diet Treatments | PSE 3 | Orthogonal Contrast 4 | Regression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPC1 | CPC2 | CPC3 | CPC4 | CPC5 | CPC6 | Linear | Quadratic | Cubic | Model 5 | (Pr > F) 6 | R2 | ||
| IBW (g) | 4.70 | 4.69 | 4.70 | 4.67 | 4.67 | 4.67 | 0.03 | 0.903 | 0.447 | 0.281 | Ns | - | - |
| FBW (g) | 11.83 a | 12.39 ab | 14.72 bc | 15.42 c | 14.61 bc | 13.11 abc | 1.23 | 0.040 | 0.004 | 0.234 | Qd | 0.003 | 0.541 |
| SR (%) | 96.67 | 97.78 | 97.78 | 95.56 | 97.78 | 97.78 | 3.77 | 0.858 | 0.870 | 0.630 | Ns | - | - |
| SGR (%/d) | 1.66 a | 1.72 a | 2.04 b | 2.13 b | 2.03 b | 1.84 ab | 0.16 | 0.030 | 0.005 | 0.216 | 2 SBL-LL | 0.003 | 0.887 |
| FR (%/d) | 4.09 b | 3.50 a | 3.63 a | 3.55 a | 3.44 a | 3.44 a | 0.18 | 0.001 | 0.060 | 0.087 | Ln | 0.004 | 0.410 |
| FE (%) | 38.58 a | 46.81 ab | 51.72 b | 54.78 b | 54.26 b | 50.18 b | 5.03 | 0.005 | 0.009 | 0.872 | 2 SBL-LL | 0.000 | 0.982 |
| PER | 1.56 b | 1.53 b | 1.54 b | 1.45 b | 1.31 ab | 1.10 a | 0.15 | 0.001 | 0.078 | 0.750 | Ln | 0.000 | 0.528 |
| ADCd (%) | 66.89 c | 72.02 d | 71.18 d | 71.60 d | 61.51 b | 56.27 a | 1.85 | 0.000 | 0.000 | 0.214 | Qd | 0.000 | 0.881 |
| ADCp (%) | 64.43 a | 67.03 a | 69.86 ab | 70.62 ab | 75.99 bc | 79.77 c | 3.54 | 0.000 | 0.409 | 0.698 | Ln | 0.000 | 0.734 |
| CF (%) | 1.70 a | 1.77 ab | 1.84 bc | 1.89 c | 1.84 bc | 1.79 abc | 0.05 | 0.032 | 0.003 | 0.680 | Qd | 0.001 | 0.603 |
| HSI (%) | 1.86 a | 2.14 cd | 2.22 d | 2.28 d | 2.02 bc | 1.91 ab | 0.08 | 0.917 | 0.000 | 0.187 | Qd | 0.000 | 0.777 |
| MFI (%) | 1.29 a | 1.52 b | 1.85 c | 2.25 d | 2.40 e | 1.58 b | 0.04 | 0.000 | 0.000 | 0.000 | Qd | 0.000 | 0.741 |
| Diet Treatments | PSE 2 | Orthogonal Contrast 3 | Regression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPC1 | CPC2 | CPC3 | CPC4 | CPC5 | CPC6 | Linear | Quadratic | Cubic | Model 4 | (Pr > F) 5 | R2 | ||
| Whole body | |||||||||||||
| Moisture | 72.03 | 72.54 | 72.38 | 71.55 | 72.38 | 71.32 | 0.70 | 0.180 | 0.317 | 0.869 | Ns | - | - |
| Crude Protein | 13.65 | 13.68 | 13.77 | 13.77 | 13.72 | 13.64 | 0.24 | 0.951 | 0.415 | 0.863 | Ns | - | - |
| Crude Lipid | 9.90 | 9.95 | 10.84 | 11.12 | 10.85 | 10.80 | 0.45 | 0.005 | 0.051 | 0.416 | Ln | 0.007 | 0.375 |
| Ash | 3.14 | 3.33 | 2.66 | 3.13 | 2.59 | 3.09 | 0.41 | 0.344 | 0.360 | 0.357 | Ns | - | - |
| Dorsal muscle | |||||||||||||
| Moisture | 77.20 | 76.68 | 76.19 | 77.03 | 77.45 | 77.13 | 0.72 | 0.434 | 0.249 | 0.130 | Ns | - | - |
| Crude Protein | 18.18 | 18.69 | 18.68 | 18.72 | 18.35 | 18.23 | 0.34 | 0.649 | 0.026 | 0.368 | Qd | 0.049 | 0.331 |
| Crude Lipid | 3.72 | 3.89 | 3.88 | 3.88 | 3.77 | 3.63 | 0.31 | 0.610 | 0.252 | 0.876 | Ns | - | - |
| Ash | 0.44 | 0.45 | 0.46 | 0.39 | 0.44 | 0.50 | 0.05 | 0.514 | 0.197 | 0.181 | Ns | - | - |
| Diet Treatments | PSE 3 | Orthogonal Contrast 4 | Regression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indices 2 | CPC1 | CPC2 | CPC3 | CPC4 | CPC5 | CPC6 | Linear | Quadratic | Cubic | Model 5 | (Pr > F) 6 | R2 | |
| TP (g/L) | 38.00 | 38.57 | 38.17 | 38.63 | 40.97 | 42.00 | 2.43 | 0.036 | 0.322 | 0.945 | Ln | 0.021 | 0.291 |
| GLU (mmol/L) | 5.60 | 5.00 | 4.70 | 4.97 | 5.43 | 5.67 | 1.10 | 0.726 | 0.236 | 0.665 | Ns | - | - |
| UN (mmol/L) | 0.50 a | 0.53 a | 0.63 a | 0.77 ab | 1.07 bc | 1.10 c | 0.17 | 0.000 | 0.391 | 0.355 | Ln | 0.000 | 0.696 |
| TG (mmol/L) | 2.50 a | 2.85 a | 3.57 b | 3.99 b | 3.68 b | 3.57 b | 0.35 | 0.000 | 0.004 | 0.448 | Qd | 0.000 | 0.710 |
| CHOL (mmol/L) | 6.70 | 6.95 | 7.17 | 6.75 | 6.67 | 6.60 | 0.63 | 0.567 | 0.424 | 0.530 | Ns | - | - |
| HDL (mmol/L) | 2.69 | 2.70 | 2.73 | 2.75 | 2.78 | 2.77 | 0.32 | 0.202 | 0.778 | 0.776 | Ns | - | - |
| LDL (mmol/L) | 4.22 a | 4.37 ab | 4.61 b | 4.44 ab | 4.33 ab | 4.30 ab | 0.18 | 0.884 | 0.027 | 0.334 | Qd | 0.069 | 0.300 |
| C3 (μg/mL) | 122.62 a | 125.61 ab | 127.58 ab | 130.02 bc | 133.96 c | 124.87 ab | 2.87 | 0.016 | 0.005 | 0.025 | Qd | 0.008 | 0.471 |
| IgM (μg/mL) | 32.20 a | 32.29 a | 38.00 b | 38.83 b | 38.42 b | 40.42 b | 2.08 | 0.000 | 0.199 | 0.751 | Ln | 0.000 | 0.650 |
| LYS (ng/mL) | 181.00 | 187.22 | 178.87 | 179.95 | 180.99 | 176.60 | 6.50 | 0.230 | 0.660 | 0.737 | Ns | - | - |
| Diet Treatments | PSE 2 | Orthogonal Contrast 3 | Regression | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPC1 | CPC2 | CPC3 | CPC4 | CPC5 | CPC6 | Linear | Quadratic | Cubic | Model 4 | (Pr > F) 5 | R2 | ||
| Cooking loss (%) | 29.38 a | 31.17 ab | 33.78 abc | 35.54 bc | 35.58 bc | 38.16 c | 2.71 | 0.001 | 0.677 | 0.781 | Ln | 0.000 | 0.611 |
| Hardness (g) | 1037.24 a | 1188.03 b | 1353.18 c | 1476.93 d | 1151.90 ab | 1104.84 ab | 65.95 | 0.289 | 0.000 | 0.854 | Qd | 0.000 | 0.687 |
| Springiness | 0.53 | 0.51 | 0.48 | 0.48 | 0.52 | 0.50 | 0.10 | 0.448 | 0.145 | 0.442 | Ns | - | - |
| Cohesiveness | 0.46 a | 0.47 ab | 0.48 abc | 0.49 bc | 0.50 c | 0.47 ab | 0.00 | 0.020 | 0.013 | 0.106 | Qd | 0.007 | 0.480 |
| Gumminess | 504.58 a | 557.33 ab | 660.12 bc | 741.68 c | 683.29 c | 557.53 ab | 59.94 | 0.028 | 0.000 | 0.065 | Qd | 0.000 | 0.637 |
| Chewiness (g) | 279.51 a | 325.41 b | 367.64 c | 433.60 d | 313.62 b | 305.25 ab | 15.19 | 0.051 | 0.000 | 0.663 | Qd | 0.000 | 0.645 |
| Resilience (g/s) | 0.26 c | 0.24 bc | 0.22 ab | 0.21 a | 0.22 ab | 0.23 ab | 0.00 | 0.004 | 0.001 | 0.794 | Qd | 0.000 | 0.717 |
| pH | 5.40 b | 5.34 ab | 5.31 ab | 5.30 ab | 5.31 ab | 5.22 a | 0.09 | 0.042 | 0.918 | 0.361 | Ln | 0.023 | 0.282 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Zhou, M.; Wang, X.; Mao, X.; Long, X.; Xie, S.; Han, D.; Tan, Q. Effects of Dietary Cottonseed Protein Concentrate Levels on Growth Performance, Health Status, Flesh Quality and Intestinal Microbiota of Grass Carp (Ctenopharyngodon idellus). Metabolites 2022, 12, 1046. https://doi.org/10.3390/metabo12111046
Liu G, Zhou M, Wang X, Mao X, Long X, Xie S, Han D, Tan Q. Effects of Dietary Cottonseed Protein Concentrate Levels on Growth Performance, Health Status, Flesh Quality and Intestinal Microbiota of Grass Carp (Ctenopharyngodon idellus). Metabolites. 2022; 12(11):1046. https://doi.org/10.3390/metabo12111046
Chicago/Turabian StyleLiu, Guoqing, Meng Zhou, Xiaoyu Wang, Xiangjie Mao, Xianmei Long, Shouqi Xie, Dong Han, and Qingsong Tan. 2022. "Effects of Dietary Cottonseed Protein Concentrate Levels on Growth Performance, Health Status, Flesh Quality and Intestinal Microbiota of Grass Carp (Ctenopharyngodon idellus)" Metabolites 12, no. 11: 1046. https://doi.org/10.3390/metabo12111046
APA StyleLiu, G., Zhou, M., Wang, X., Mao, X., Long, X., Xie, S., Han, D., & Tan, Q. (2022). Effects of Dietary Cottonseed Protein Concentrate Levels on Growth Performance, Health Status, Flesh Quality and Intestinal Microbiota of Grass Carp (Ctenopharyngodon idellus). Metabolites, 12(11), 1046. https://doi.org/10.3390/metabo12111046








