Expression Profiles of Kidney Mitochondrial Proteome during the Progression of the Unilateral Ureteral Obstruction: Focus on Energy Metabolism Adaptions
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Unilateral Ureteral Obstruction (UUO) Procedure
2.3. Mitochondrial Isolation
2.4. Sample Preparation for Mass Spectrometry
2.5. Label–Free Quantification by Mass Spectrometry and Data Analysis
2.6. Proteomics Statistical and Bioinformatics Analysis
2.7. Western Blot
2.8. Statistical Analysis
3. Results
3.1. Validation of Renal Fibrotic Damage and Mitochondrial VDAC Reduction during the UUO Progression
3.2. Kidney Mitochondrial Proteome Expression Profiles during UUO Progression
3.3. OXPHOS Proteins Decrease in Renal Tissue during UUO Progression
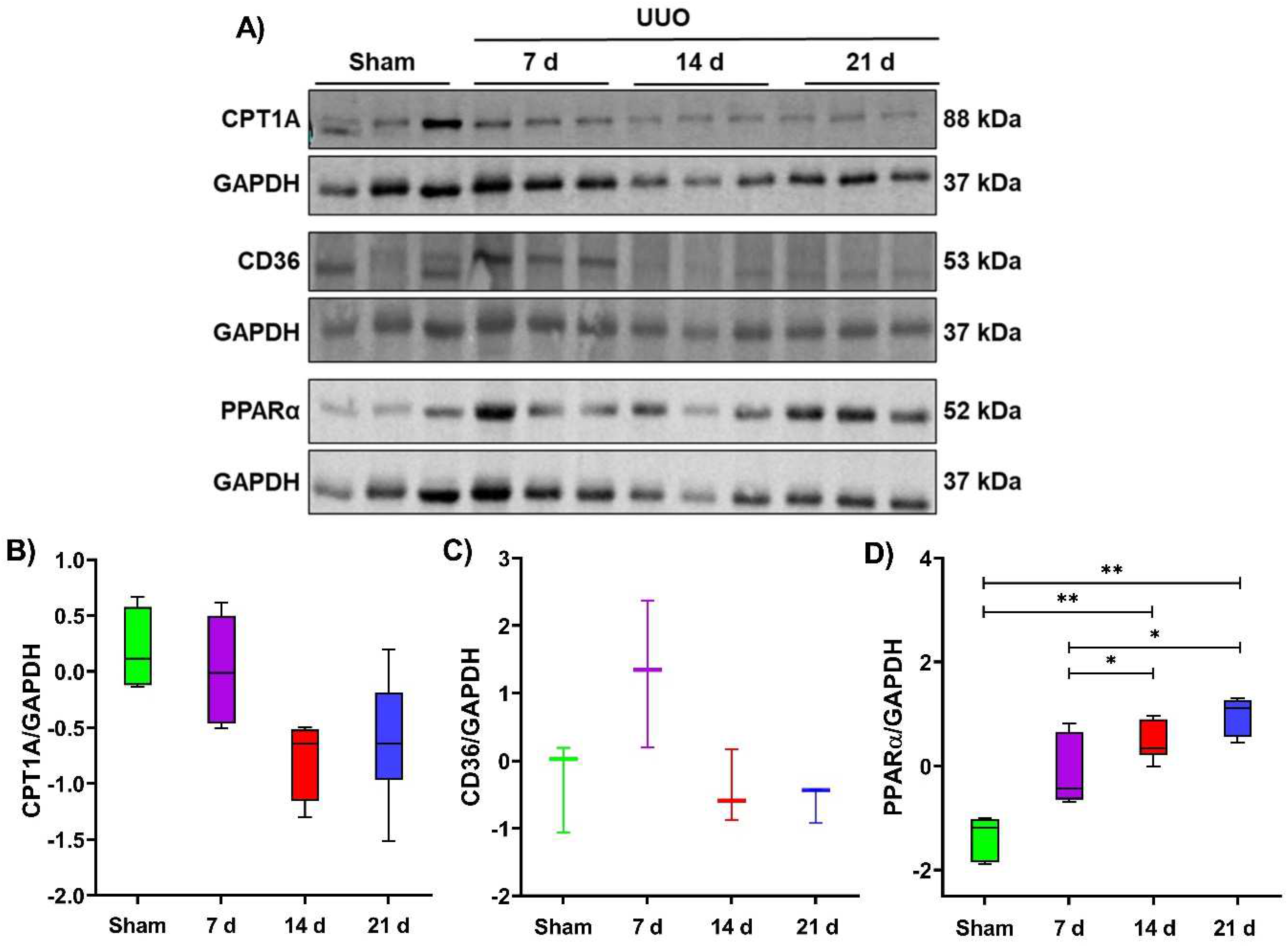
3.4. Slight Alterations in Lipid Metabolism Proteins in Renal Tissue during the UUO Progression
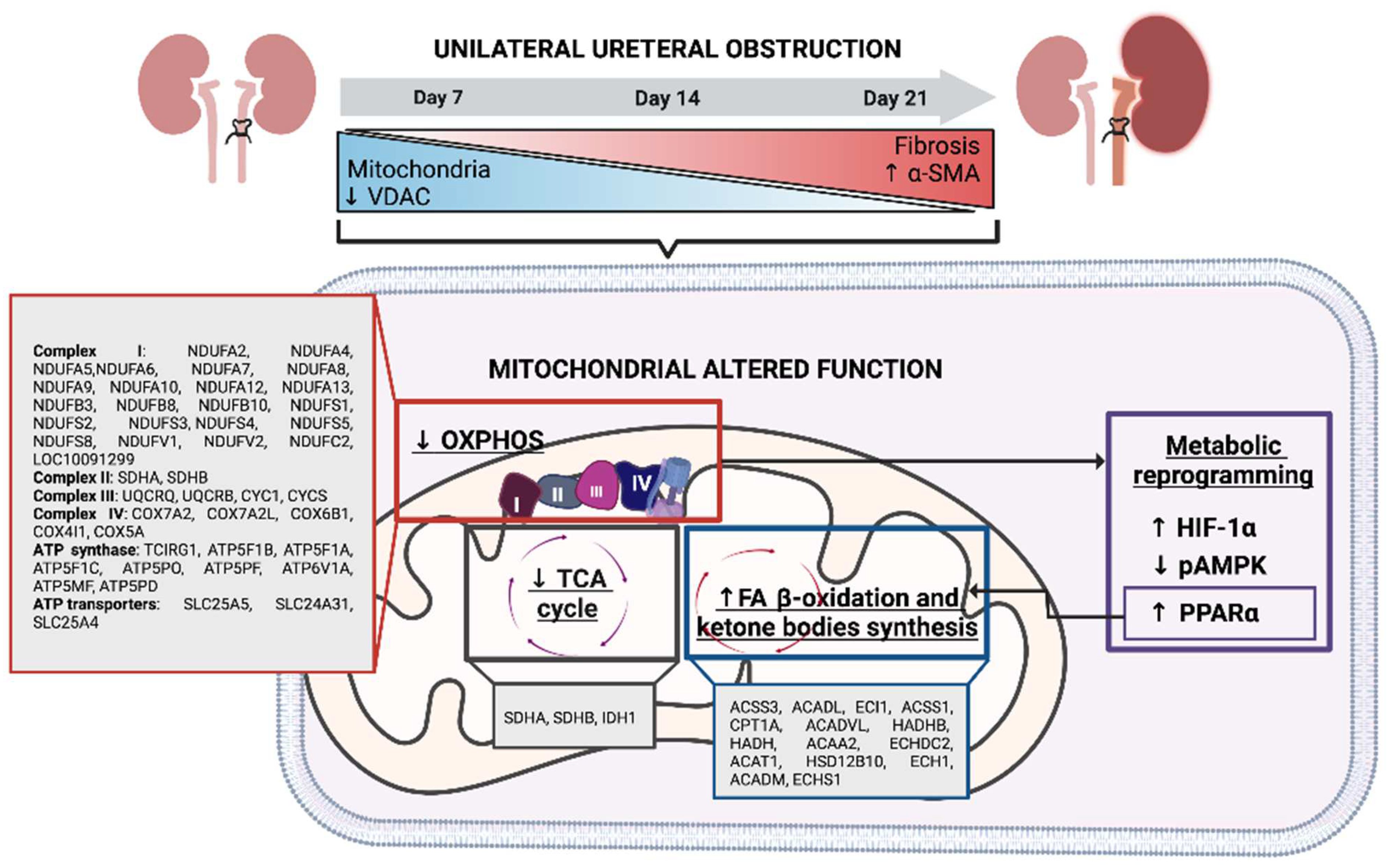
3.5. Indirect Evaluation of Metabolic Reprogramming during the UUO Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siddiqui, M.M.; McDougal, W.S. Urologic Assessment of Decreasing Renal Function. Med. Clin. N. Am. 2011, 95, 161–168. [Google Scholar] [CrossRef]
- Mourmouris, P.I.; Chiras, T.; Papatsoris, A.G. Obstructive Uropathy: From Etiopathology to Therapy. World J. Nephrol. Urol. 2014, 3, 1–6. [Google Scholar] [CrossRef]
- Krajewski, W.; Wojciechowska, J.; Dembowski, J.; Zdrojowy, R.; Szydełko, T. Hydronephrosis in the course of ureteropelvic junction obstruction—An underestimated problem? Current opinion on pathogenesis, diagnosis and treatment. Adv. Clin. Exp. Med. 2017, 26, 857–864. [Google Scholar] [CrossRef]
- Stevens, S. Obstructive Kidney Disease. Nurs. Clin. N. Am. 2018, 53, 569–578. [Google Scholar] [CrossRef]
- Chevalier, R.L.; Forbes, M.S.; Thornhill, B.A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009, 75, 1145–1152. [Google Scholar] [CrossRef]
- Zhang, J.-G.; Xing, Z.-Y.; Zha, T.-T.; Tian, X.-J.; Du, Y.-N.; Chen, J.; Xing, W. Longitudinal assessment of rabbit renal fibrosis induced by unilateral ureteral obstruction using two-dimensional susceptibility weighted imaging. J. Magn. Reson. Imaging 2018, 47, 1572–1577. [Google Scholar] [CrossRef]
- Huang, L.; Ni, J.; Duncan, T.; Song, Z.; Johnson, T.S. Development of a unilateral ureteral obstruction model in cynomolgus monkeys. Anim. Model. Exp. Med. 2021, 4, 359–368. [Google Scholar] [CrossRef]
- Atkinson, J.; Boden, T.; Mocho, J.-P.; Johnson, T. Refining the unilateral ureteral obstruction mouse model: No sham, no shame. Lab. Anim. 2021, 55, 21–29. [Google Scholar] [CrossRef]
- Chung, S.D.; Lai, T.Y.; Chien, C.T.; Yu, H.J. Activating Nrf-2 Signaling Depresses Unilateral Ureteral Obstruction-Evoked Mitochondrial Stress-Related Autophagy, Apoptosis and Pyroptosis in Kidney. PLoS ONE 2012, 7, e47299. [Google Scholar] [CrossRef]
- Xu, Y.; Ruan, S.; Wu, X.; Chen, H.; Zheng, K.; Fu, B. Autophagy and apoptosis in tubular cells following unilateral ureteral obstruction are associated with mitochondrial oxidative stress. Int. J. Mol. Med. 2013, 31, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef]
- Jiménez-Uribe, A.P.; Bellido, B.; Aparicio-Trejo, O.E.; Tapia, E.; Sánchez-Lozada, L.G.; Hernández-Santos, J.A.; Fernández-Valverde, F.; Hernández-Cruz, E.Y.; Orozco-Ibarra, M.; Pedraza-Chaverri, J. Temporal characterization of mitochondrial impairment in the unilateral ureteral obstruction model in rats. Free Radic. Biol. Med. 2021, 172, 358–371. [Google Scholar] [CrossRef]
- Hall, A.M.; Rhodes, G.J.; Sandoval, R.M.; Corridon, P.R.; Molitoris, B.A. In vivo multiphoton imaging of mitochondrial structure and function during acute kidney injury. Kidney Int. 2013, 83, 72–83. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Y.; Li, L.; Liu, S.; Wang, C.; Yuan, Y.; Yang, G.; Chen, Y.; Cheng, J.; Lu, Y.; et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics 2021, 11, 1845–1863. [Google Scholar] [CrossRef]
- Aparicio-Trejo, O.E.; Avila-Rojas, S.H.; Tapia, E.; Rojas-Morales, P.; León-Contreras, J.C.; Martínez-Klimova, E.; Hernández-Pando, R.; Sánchez- Lozada, L.G.; Pedraza-Chaverri, J. Chronic impairment of mitochondrial bioenergetics and β-oxidation promotes experimental AKI-to-CKD transition induced by folic acid. Free Radic. Biol. Med. 2020, 154, 18–32. [Google Scholar] [CrossRef]
- Aparicio-Trejo, O.E.; Reyes-Fermín, L.M.; Briones-Herrera, A.; Tapia, E.; León-Contreras, J.C.; Hernández-Pando, R.; Sánchez-Lozada, L.G.; Pedraza-Chaverri, J. Protective effects of N-acetyl-cysteine in mitochondria bioenergetics, oxidative stress, dynamics and S-glutathionylation alterations in acute kidney damage induced by folic acid. Free Radic. Biol. Med. 2019, 130, 379–396. [Google Scholar] [CrossRef]
- Liang, N.-N.; Zhao, Y.; Guo, Y.-Y.; Zhang, Z.-H.; Gao, L.; Yu, D.-X.; Xu, D.-X.; Xu, S. Mitochondria-derived reactive oxygen species are involved in renal cell ferroptosis during lipopolysaccharide-induced acute kidney injury. Int. Immunopharmacol. 2022, 107, 108687. [Google Scholar] [CrossRef]
- Aparicio-Trejo, O.E.; Rojas-Morales, P.; Avila-Rojas, S.H.; León-Contreras, J.C.; Hernández-Pando, R.; Jiménez-Uribe, A.P.; Prieto-Carrasco, R.; Sánchez-Lozada, L.G.; Pedraza-Chaverri, J.; Tapia, E. Temporal Alterations in Mitochondrial β-Oxidation and Oxidative Stress Aggravate Chronic Kidney Disease Development in 5/6 Nephrectomy Induced Renal Damage. Int. J. Mol. Sci. 2020, 21, 6512. [Google Scholar] [CrossRef]
- Wei, Q.; Su, J.; Dong, G.; Zhang, M.; Huo, Y.; Dong, Z. Glycolysis inhibitors suppress renal interstitial fibrosis via divergent effects on fibroblasts and tubular cells. Am. J. Physiol. Physiol. 2019, 316, F1162–F1172. [Google Scholar] [CrossRef]
- Ding, H.; Jiang, L.; Xu, J.; Bai, F.; Zhou, Y.; Yuan, Q.; Luo, J.; Zen, K.; Yang, J. Inhibiting aerobic glycolysis suppresses renal interstitial fibroblast activation and renal fibrosis. Am. J. Physiol. Physiol. 2017, 313, F561–F575. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Song, J.; Herrmann, J.M.; Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 2021, 22, 54–70. [Google Scholar] [CrossRef]
- Ríos-Castro, E.; Souza, G.H.M.F.; Delgadillo-Álvarez, D.M.; Ramírez-Reyes, L.; Torres-Huerta, A.L.; Velasco-Suárez, A.; Cruz-Cruz, C.; Hernández-Hernández, J.M.; Tapia-Ramírez, J. Quantitative Proteomic Analysis of MARC-145 Cells Infected with a Mexican Porcine Reproductive and Respiratory Syndrome Virus Strain Using a Label-Free Based DIA approach. J. Am. Soc. Mass Spectrom. 2020, 31, 1302–1312. [Google Scholar] [CrossRef]
- Souza, G.H.M.F.; Guest, P.C.; Martins-de-Souza, D. LC-MSE, Multiplex MS/MS, Ion Mobility, and Label-Free Quantitation in Clinical Proteomics. In Multiplex Biomarker Techniques: Methods in Molecular Biology; Humana: New York, NY, USA, 2017; pp. 57–73. [Google Scholar]
- Silva, J.C.; Gorenstein, M.V.; Li, G.-Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute Quantification of Proteins by LCMSE. Mol. Cell. Proteomics 2006, 5, 144–156. [Google Scholar] [CrossRef]
- Gómez-Caudillo, L.; Ortega-Lozano, A.; Martínez-Batallar, Á.; Rosas-Vargas, H.; Minauro-Sanmiguel, F.; Encarnación-Guevara, S. Principal component analysis on LC-MS/MS and 2DE-MALDI-TOF in glioblastoma cell lines reveals that mitochondria act as organelle sensors of the metabolic state in glioblastoma. Oncol. Rep. 2020, 44, 661–673. [Google Scholar] [CrossRef]
- Smith, A.C.; Robinson, A.J. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016, 44, D1258–D1261. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Buhlmann, P. MissForest—Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Blagotić, A.; Daróczi, G. Rapport: A Report Templating System; R Core Team: Vienna, Austria, 2012. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Knoch, B.; Müller, M.; Kersten, S. Peroxisome Proliferator-Activated Receptor Alpha Target Genes. PPAR Res. 2010, 2010, 612089. [Google Scholar] [CrossRef]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Zhu, X.; Jiang, L.; Long, M.; Wei, X.; Hou, Y.; Du, Y. Metabolic Reprogramming and Renal Fibrosis. Front. Med. 2021, 8, 746920. [Google Scholar] [CrossRef]
- Martínez-Klimova, E.; Aparicio-Trejo, O.E.; Gómez-Sierra, T.; Jiménez-Uribe, A.P.; Bellido, B.; Pedraza-Chaverri, J. Mitochondrial dysfunction and endoplasmic reticulum stress in the promotion of fibrosis in obstructive nephropathy induced by unilateral ureteral obstruction. BioFactors 2020, 46, 716–733. [Google Scholar] [CrossRef]
- Liu, H.; Li, W.; He, Q.; Xue, J.; Wang, J.; Xiong, C.; Pu, X.; Nie, Z. Mass Spectrometry Imaging of Kidney Tissue Sections of Rat Subjected to Unilateral Ureteral Obstruction. Sci. Rep. 2017, 7, 41954. [Google Scholar] [CrossRef]
- Prieto-Carrasco, R.; Silva-Palacios, A.; Rojas-Morales, P.; Aparicio-Trejo, O.E.; Medina-Reyes, E.I.; Hernández-Cruz, E.Y.; Sánchez-Garibay, C.; Salinas-Lara, C.; Pavón, N.; Roldán, F.J.; et al. Unilateral Ureteral Obstruction for 28 Days in Rats Is Not Associated with Changes in Cardiac Function or Alterations in Mitochondrial Function. Biology 2021, 10, 671. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Du, F.; Su, X.; Sun, G.; Zhou, G.; Bian, X.; Liu, N. Epigallocatechin-3-Gallate Attenuates Unilateral Ureteral Obstruction-Induced Renal Interstitial Fibrosis in Mice. J. Histochem. Cytochem. 2015, 63, 270–279. [Google Scholar] [CrossRef]
- Banerjee, S.; Wong, A.C.-Y.; Yan, X.; Wu, B.; Zhao, H.; Tibshirani, R.J.; Zare, R.N.; Brooks, J.D. Early detection of unilateral ureteral obstruction by desorption electrospray ionization mass spectrometry. Sci. Rep. 2019, 9, 11007. [Google Scholar] [CrossRef]
- Tirichen, H.; Yaigoub, H.; Xu, W.; Wu, C.; Li, R.; Li, Y. Mitochondrial Reactive Oxygen Species and Their Contribution in Chronic Kidney Disease Progression Through Oxidative Stress. Front. Physiol. 2021, 12, 398. [Google Scholar] [CrossRef]
- Granata, S.; Zaza, G.; Simone, S.; Villani, G.; Latorre, D.; Pontrelli, P.; Carella, M.; Schena, F.P.; Grandaliano, G.; Pertosa, G. Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genom. 2009, 10, 388. [Google Scholar] [CrossRef]
- Chandel, N.S.; McClintock, D.S.; Feliciano, C.E.; Wood, T.M.; Melendez, J.A.; Rodriguez, A.M.; Schumacker, P.T. Reactive Oxygen Species Generated at Mitochondrial Complex III Stabilize Hypoxia-inducible Factor-1α during Hypoxia. J. Biol. Chem. 2000, 275, 25130–25138. [Google Scholar] [CrossRef]
- Boor, P.; Celec, P.; Martin, I.V.; Villa, L.; Hodosy, J.; Klenovicsová, K.; Esposito, C.; Schäfer, S.; Albrecht-Küpper, B.; Ostendorf, T.; et al. The peroxisome proliferator-activated receptor-α agonist, BAY PP1, attenuates renal fibrosis in rats. Kidney Int. 2011, 80, 1182–1197. [Google Scholar] [CrossRef]
- Yan, Q.; Song, Y.; Zhang, L.; Chen, Z.; Yang, C.; Liu, S.; Yuan, X.; Gao, H.; Ding, G.; Wang, H. Autophagy activation contributes to lipid accumulation in tubular epithelial cells during kidney fibrosis. Cell Death Discov. 2018, 4, 39. [Google Scholar] [CrossRef]
- Ke, Q.; Yuan, Q.; Qin, N.; Shi, C.; Luo, J.; Fang, Y.; Xu, L.; Sun, Q.; Zen, K.; Jiang, L.; et al. UCP2-induced hypoxia promotes lipid accumulation and tubulointerstitial fibrosis during ischemic kidney injury. Cell Death Dis. 2020, 11, 26. [Google Scholar] [CrossRef]
- Kang, H.M.; Ahn, S.H.; Choi, P.; Ko, Y.-A.; Han, S.H.; Chinga, F.; Park, A.S.D.; Tao, J.; Sharma, K.; Pullman, J.; et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015, 21, 37–46. [Google Scholar] [CrossRef]
- Miguel, V.; Tituaña, J.; Herrero, J.I.; Herrero, L.; Serra, D.; Cuevas, P.; Barbas, C.; Puyol, D.R.; Márquez-Expósito, L.; Ruiz-Ortega, M.; et al. Renal tubule Cpt1a overexpression protects from kidney fibrosis by restoring mitochondrial homeostasis. J. Clin. Investig. 2021, 131, e14695. [Google Scholar] [CrossRef]
- Cai, T.; Ke, Q.; Fang, Y.; Wen, P.; Chen, H.; Yuan, Q.; Luo, J.; Zhang, Y.; Sun, Q.; Lv, Y.; et al. Sodium–glucose cotransporter 2 inhibition suppresses HIF-1α-mediated metabolic switch from lipid oxidation to glycolysis in kidney tubule cells of diabetic mice. Cell Death Dis. 2020, 11, 390. [Google Scholar] [CrossRef]
- Wang, R.; Kairen, C.; Li, L.; Zhang, L.; Gong, H.; Huang, X. Overexpression of NDUFV1 alleviates renal damage by improving mitochondrial function in unilateral ureteral obstruction model mice. Cell Biol. Int. 2022, 46, 381–390. [Google Scholar] [CrossRef]
- Fierro-Fernández, M.; Miguel, V.; Márquez-Expósito, L.; Nuevo-Tapioles, C.; Herrero, J.I.; Blanco-Ruiz, E.; Tituaña, J.; Castillo, C.; Cannata, P.; Monsalve, M.; et al. MiR-9-5p protects from kidney fibrosis by metabolic reprogramming. FASEB J. 2020, 34, 410–431. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Y.; Ding, G.; Huang, S.; Zhang, A.; Jia, Z. Inhibition of Mitochondrial Complex-1 Prevents the Downregulation of NKCC2 and ENaCα in Obstructive Kidney Disease. Sci. Rep. 2015, 5, 12480. [Google Scholar] [CrossRef]
- Lakhia, R.; Yheskel, M.; Flaten, A.; Quittner-Strom, E.B.; Holland, W.L.; Patel, V. PPARα agonist fenofibrate enhances fatty acid β-oxidation and attenuates polycystic kidney and liver disease in mice. Am. J. Physiol. Physiol. 2018, 314, F122–F131. [Google Scholar] [CrossRef]
- Fontecha-Barriuso, M.; Lopez-Diaz, A.M.; Guerrero-Mauvecin, J.; Miguel, V.; Ramos, A.M.; Sanchez-Niño, M.D.; Ruiz-Ortega, M.; Ortiz, A.; Sanz, A.B. Tubular Mitochondrial Dysfunction, Oxidative Stress, and Progression of Chronic Kidney Disease. Antioxidants 2022, 11, 1356. [Google Scholar] [CrossRef]
- Idrovo, J.-P.; Yang, W.-L.; Nicastro, J.; Coppa, G.F.; Wang, P. Stimulation of carnitine palmitoyltransferase 1 improves renal function and attenuates tissue damage after ischemia/reperfusion. J. Surg. Res. 2012, 177, 157–164. [Google Scholar] [CrossRef]
- Idrovo, J.-P.; Yang, W.-L.; Matsuda, A.; Nicastro, J.; Coppa, G.F.; Wang, P. Post-Treatment with the Combination of 5-Aminoimidazole-4-Carboxyamide Ribonucleoside and Carnitine Improves Renal Function After Ischemia/Reperfusion Injury. Shock 2012, 37, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega-Lozano, A.J.; Jiménez-Uribe, A.P.; Aranda-Rivera, A.K.; Gómez-Caudillo, L.; Ríos-Castro, E.; Tapia, E.; Bellido, B.; Aparicio-Trejo, O.E.; Sánchez-Lozada, L.G.; Pedraza-Chaverri, J. Expression Profiles of Kidney Mitochondrial Proteome during the Progression of the Unilateral Ureteral Obstruction: Focus on Energy Metabolism Adaptions. Metabolites 2022, 12, 936. https://doi.org/10.3390/metabo12100936
Ortega-Lozano AJ, Jiménez-Uribe AP, Aranda-Rivera AK, Gómez-Caudillo L, Ríos-Castro E, Tapia E, Bellido B, Aparicio-Trejo OE, Sánchez-Lozada LG, Pedraza-Chaverri J. Expression Profiles of Kidney Mitochondrial Proteome during the Progression of the Unilateral Ureteral Obstruction: Focus on Energy Metabolism Adaptions. Metabolites. 2022; 12(10):936. https://doi.org/10.3390/metabo12100936
Chicago/Turabian StyleOrtega-Lozano, Ariadna Jazmín, Alexis Paulina Jiménez-Uribe, Ana Karina Aranda-Rivera, Leopoldo Gómez-Caudillo, Emmanuel Ríos-Castro, Edilia Tapia, Belen Bellido, Omar Emiliano Aparicio-Trejo, Laura Gabriela Sánchez-Lozada, and José Pedraza-Chaverri. 2022. "Expression Profiles of Kidney Mitochondrial Proteome during the Progression of the Unilateral Ureteral Obstruction: Focus on Energy Metabolism Adaptions" Metabolites 12, no. 10: 936. https://doi.org/10.3390/metabo12100936
APA StyleOrtega-Lozano, A. J., Jiménez-Uribe, A. P., Aranda-Rivera, A. K., Gómez-Caudillo, L., Ríos-Castro, E., Tapia, E., Bellido, B., Aparicio-Trejo, O. E., Sánchez-Lozada, L. G., & Pedraza-Chaverri, J. (2022). Expression Profiles of Kidney Mitochondrial Proteome during the Progression of the Unilateral Ureteral Obstruction: Focus on Energy Metabolism Adaptions. Metabolites, 12(10), 936. https://doi.org/10.3390/metabo12100936








