Taxonomic and Metabolite Diversities of Moss-Associated Actinobacteria from Thailand
Abstract
1. Introduction
2. Results
2.1. Isolation of Actinobacteria from Mosses
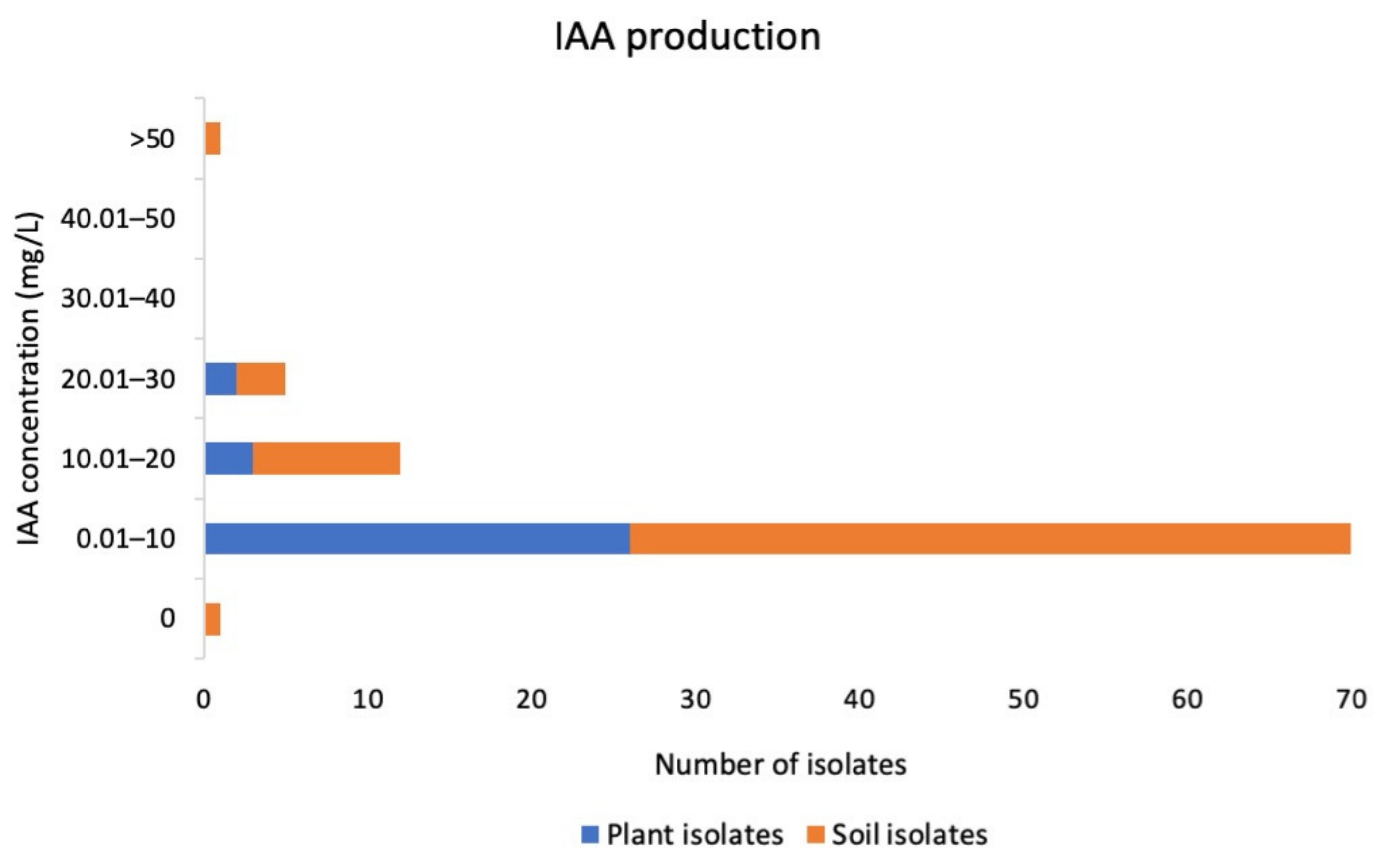
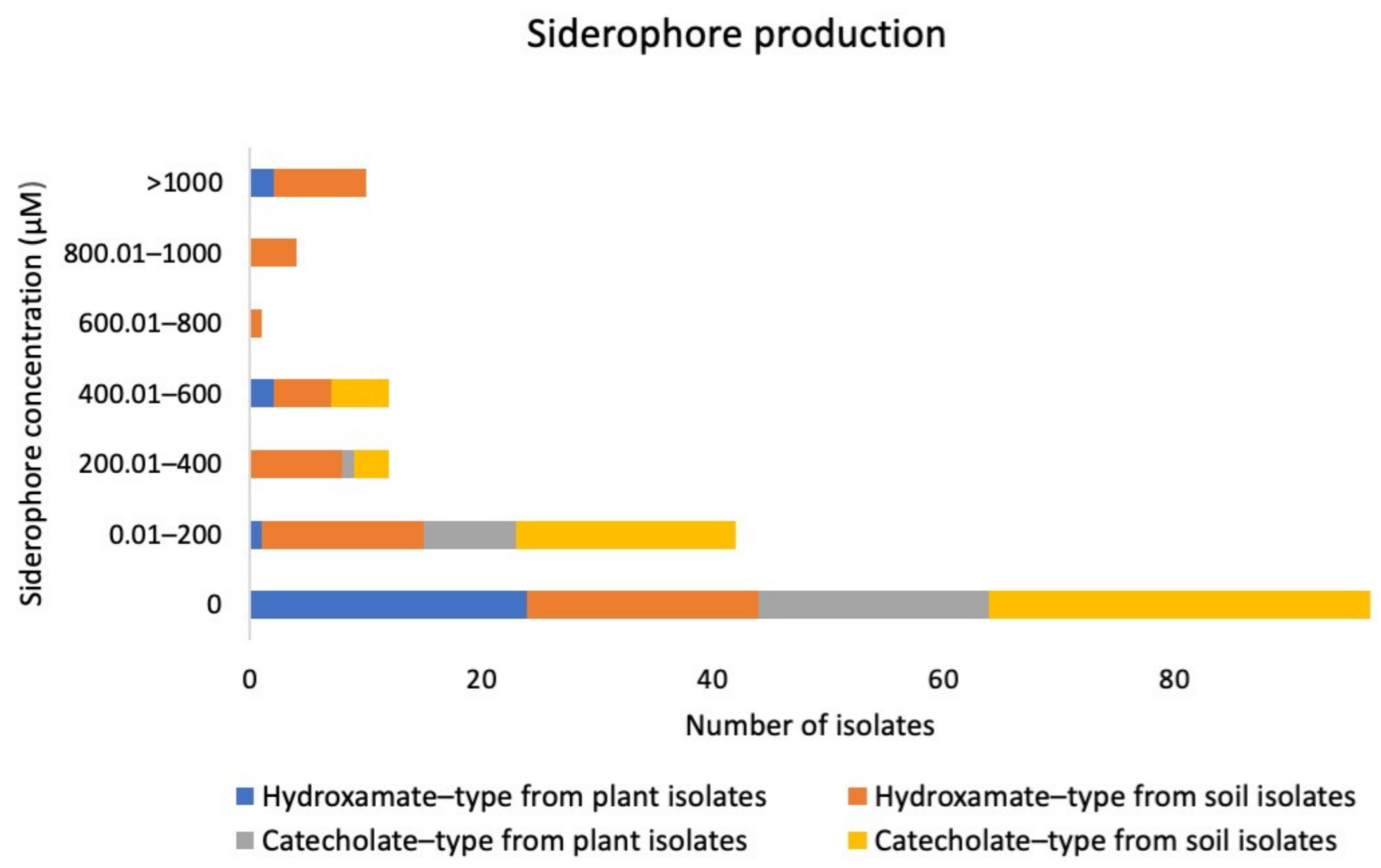
2.2. Testing for Plant Growth Promoting Potential
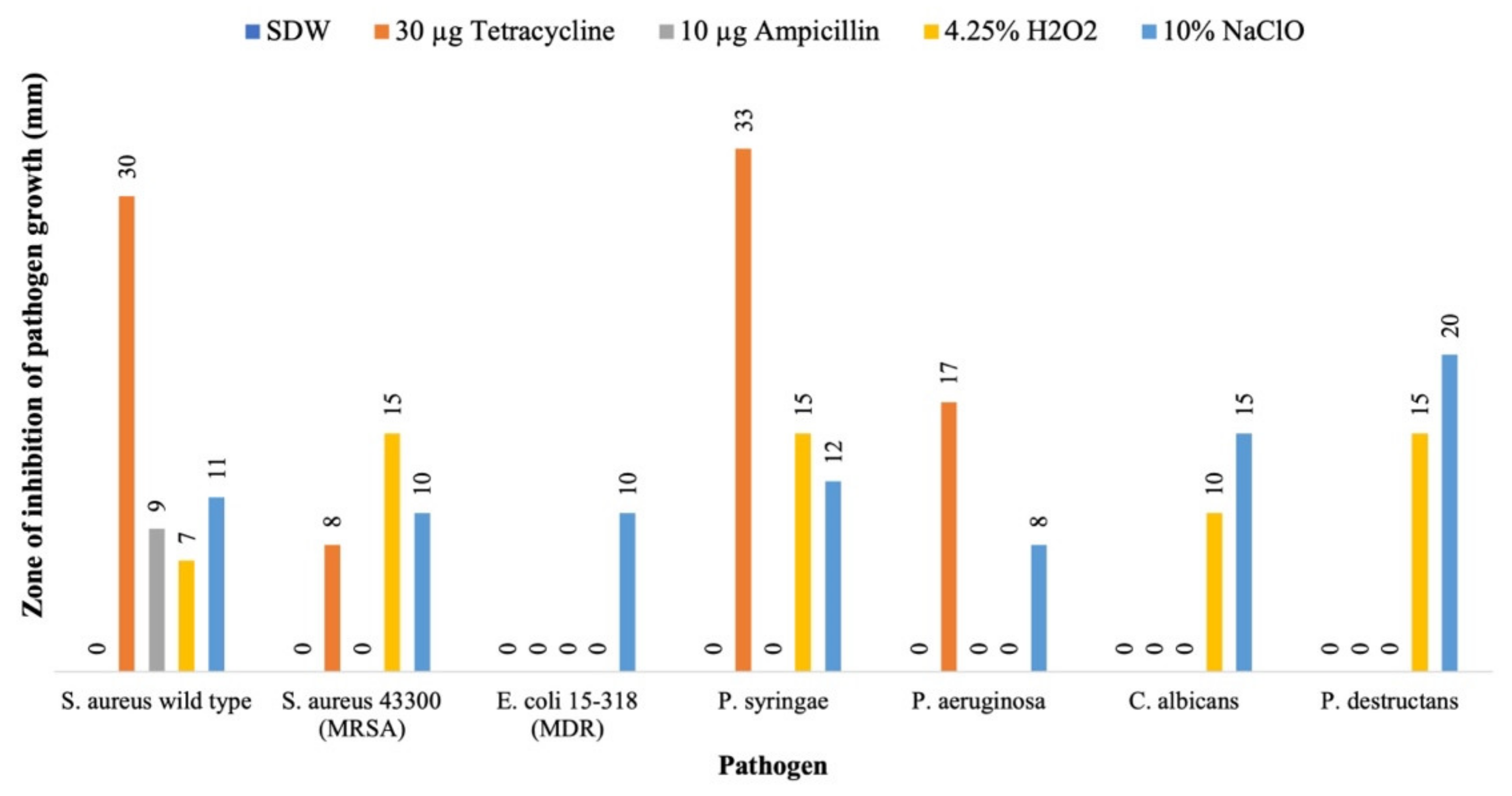
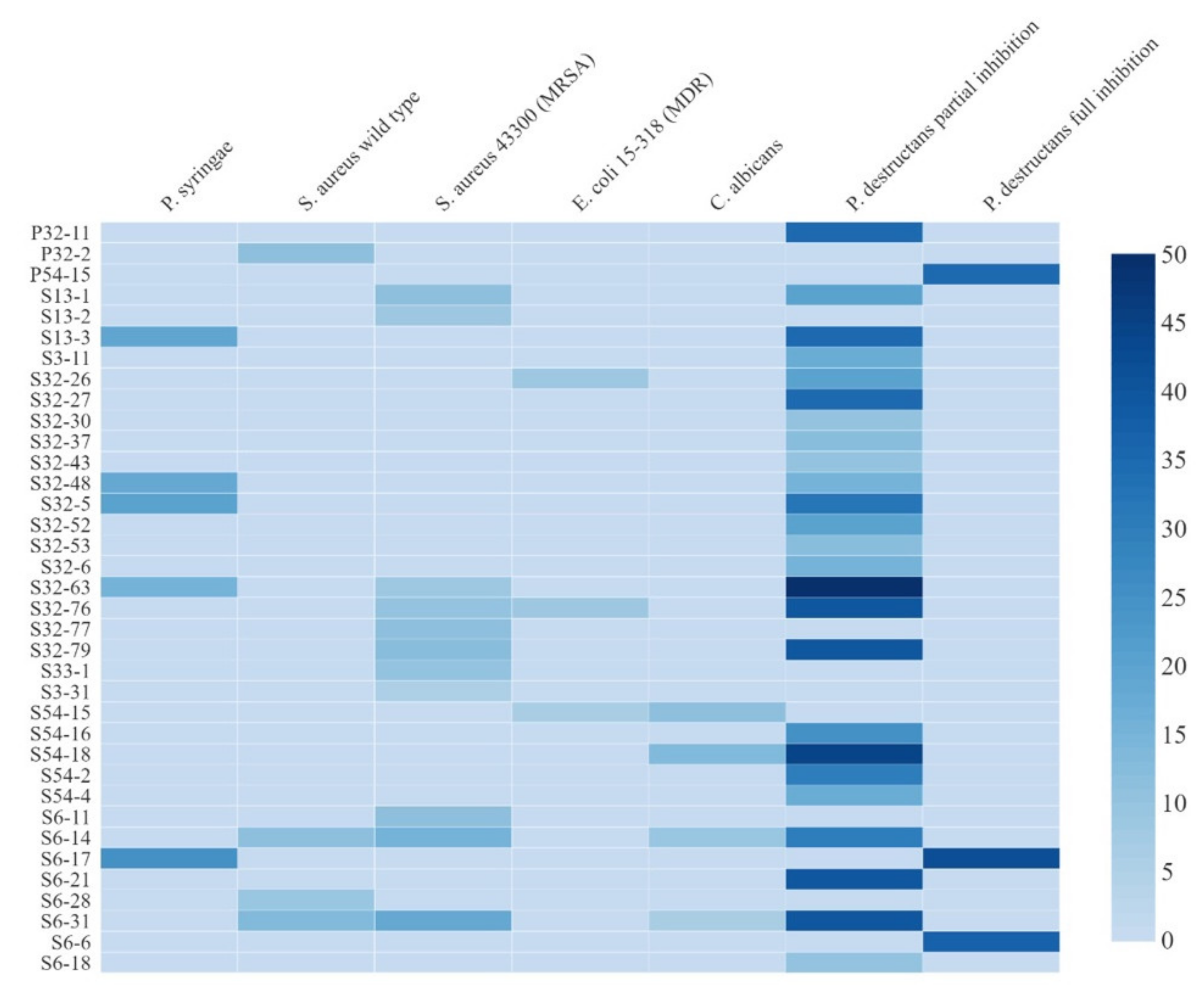
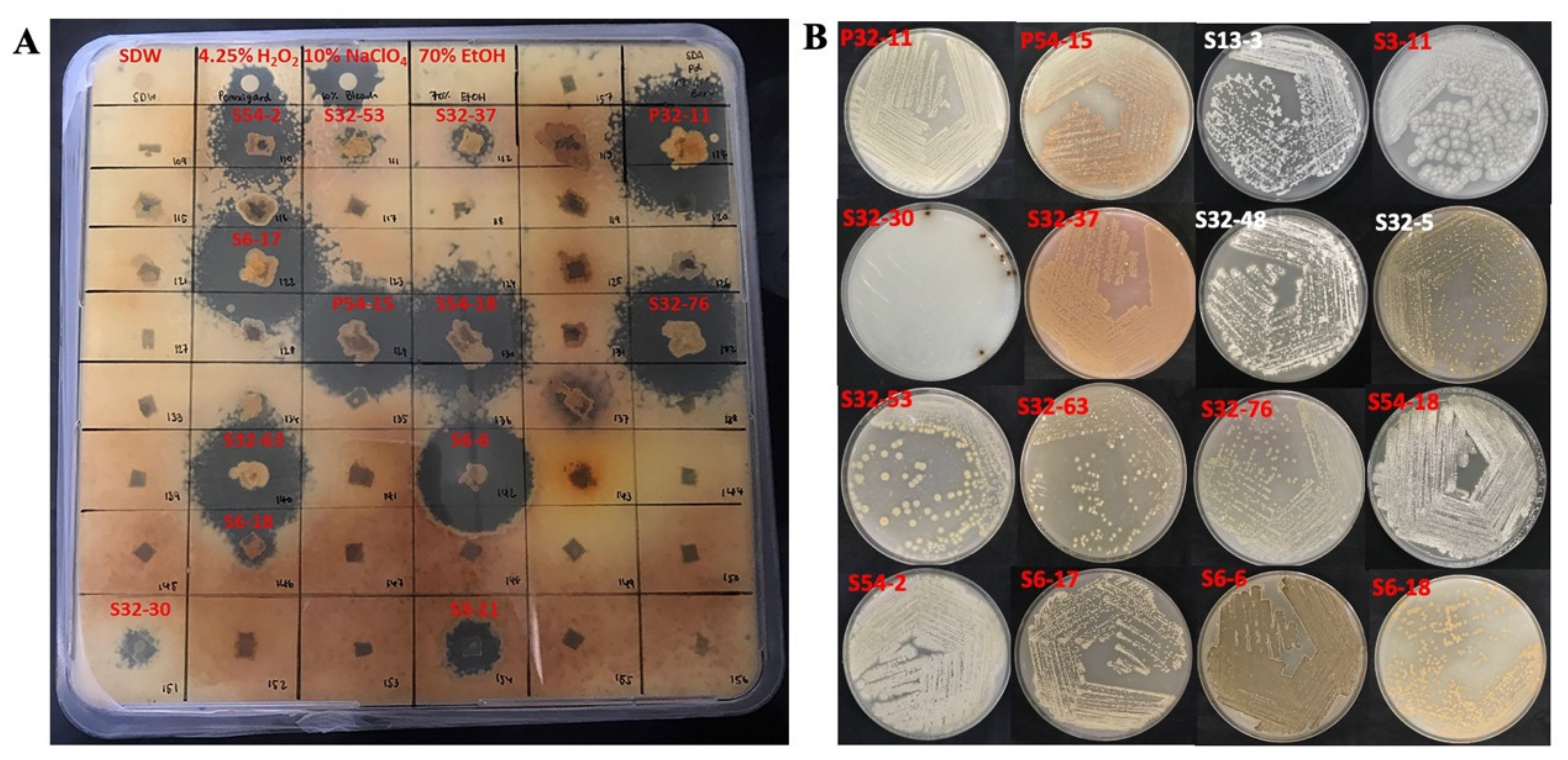
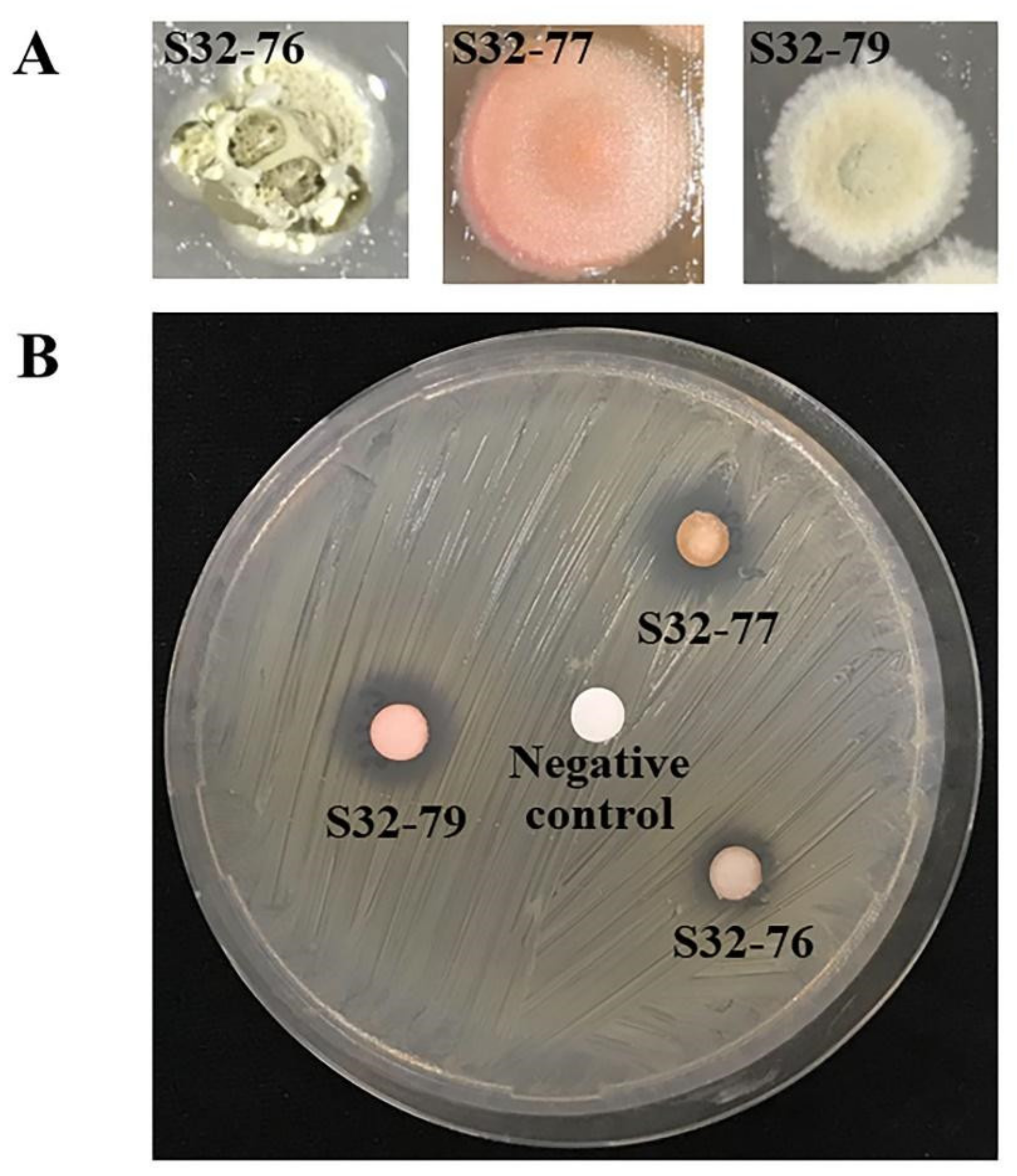
2.3. Evaluation of Antimicrobial Activity
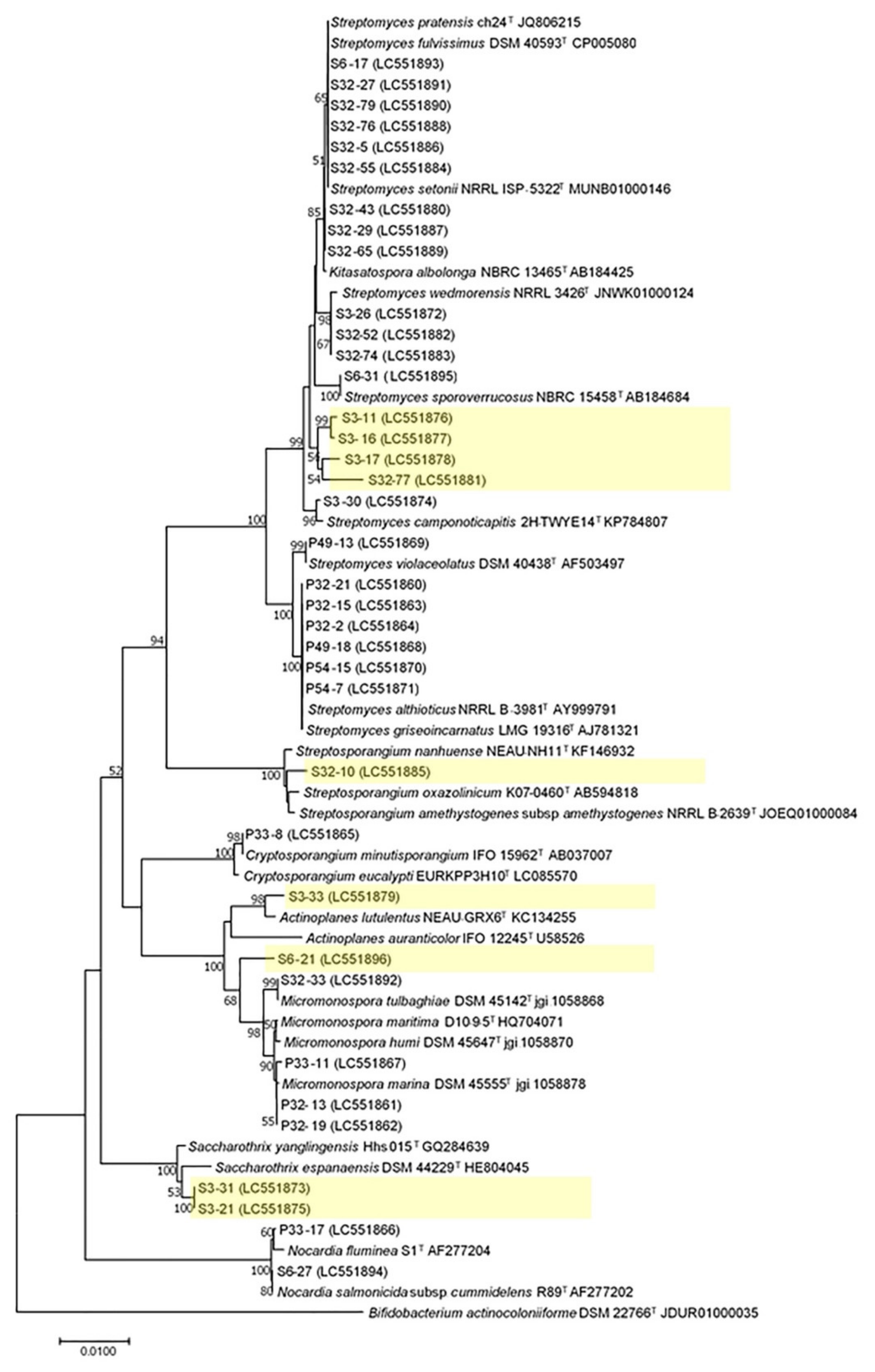
2.4. 16S rRNA Analysis of Actinobacteria from Mosses
2.5. GC-MS Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Isolation of Actinobacteria from Mosses and Moss-Associated Soils
4.3. In Vitro Quantifications of Metabolites with Plant Growth Promoting Potentials
4.4. Evaluation of Antimicrobial Activities
4.5. Identification of Actinobacteria from Mosses
4.6. GC-MS Analysis of Metabolites
4.7. Bioassay of Secreted Extracellular Compounds against MRSA
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, R.; Dubey, A.K. Diversity and Applications of Endophytic Actinobacteria of Plants in Special and Other Ecological Niches. Front. Microbiol. 2018, 9, 1767. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xing, K.; Jiang, J.-H.; Xu, L.-H.; Li, W.-J. Biodiversity, Bioactive Natural Products and Biotechnological Potential of Plant-Associated Endophytic Actinobacteria. Appl. Microbiol. Biotechnol. 2011, 89, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Golinska, P.; Wypij, M.; Agarkar, G.; Rathod, D.; Dahm, H.; Rai, M. Endophytic Actinobacteria of Medicinal Plants: Diversity and Bioactivity. Antonie van Leeuwenhoek 2015, 267–289. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shi, L.; Li, W.; Wang, J.; Wang, H.; Tian, Y.; Xiang, W.; Wang, X. Streptomyces tritici sp. nov., a Novel Actinomycete Isolated from Rhizosphere Soil of Wheat (Triticum aestivum L.). Int. J. Syst. Evol. Microbiol. 2018, 68, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam Palaniyandi, S.; Yang, S.H.; Damodharan, K.; Suh, J.-W. Genetic and Functional Characterization of Culturable Plant-Beneficial Actinobacteria Associated with Yam Rhizosphere. J. Basic Microbiol. 2013, 53, 985–995. [Google Scholar] [CrossRef]
- Carro, L.; Pukall, R.; Spröer, C.; Kroppenstedt, R.M.; Trujillo, M.E. Micromonospora cremea sp. nov. and Micromonospora zamorensis sp. nov., Isolated from the Rhizosphere of Pisum sativum. Int. J. Syst. Evol. Microbiol. 2012, 62((Pt. 12)), 2971–2977. [Google Scholar] [CrossRef]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant Growth-Promoting Actinobacteria: A New Strategy for Enhancing Sustainable Production and Protection of Grain Legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef]
- Meschke, H.; Walter, S.; Schrempf, H. Characterization and Localization of Prodiginines from Streptomyces lividans Suppressing Verticillium dahliae in the Absence or Presence of Arabidopsis thaliana. Environ. Microbiol. 2012, 14, 940–952. [Google Scholar] [CrossRef]
- Yandigeri, M.S.; Meena, K.K.; Singh, D.; Malviya, N.; Singh, D.P.; Solanki, M.K.; Yadav, A.K.; Arora, D.K. Drought-Tolerant Endophytic Actinobacteria Promote Growth of Wheat (Triticum aestivum) under Water Stress Conditions. Plant Growth Regul. 2012, 68, 411–420. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant Growth Promoting Rhizobacteria Isolated from Halophytes and Drought-Tolerant Plants: Genomic Characterisation and Exploration of Phyto-Beneficial Traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Jog, R.; Nareshkumar, G.; Rajkumar, S. Enhancing Soil Health and Plant Growth Promotion by Actinomycetes. In Plant Growth Promoting Actinobacteria: A New Avenue for Enhancing the Productivity and Soil Fertility of Grain Legumes; Subramaniam, G., Arumugam, S., Rajendran, V., Eds.; Springer: Singapore, 2016; pp. 33–45. [Google Scholar]
- Insuk, C.; Kuncharoen, N.; Cheeptham, N.; Tanasupawat, S.; Pathom-Aree, W. Bryophytes Harbor Cultivable Actinobacteria With Plant Growth Promoting Potential. Front. Microbiol. 2020, 11, 563047. [Google Scholar] [CrossRef]
- Liu, X.L.; Liu, S.L.; Liu, M.; Kong, B.H.; Liu, L.; Li, Y.H. A Primary Assessment of the Endophytic Bacterial Community in a Xerophilous Moss (Grimmia montana) Using Molecular Method and Cultivated Isolates. Braz. J. Microbiol. 2014, 45, 163–173. [Google Scholar] [CrossRef]
- Turetsky, M.R.; Bond-Lamberty, B.; Euskirchen, E.; Talbot, J.; Frolking, S.; McGuire, A.D.; Tuittila, E.-S. The Resilience and Functional Role of Moss in Boreal and Arctic Ecosystems. New Phytol. 2012, 196, 49–67. [Google Scholar] [CrossRef]
- Holland-Moritz, H.; Stuart, J.E.M.; Lewis, L.R.; Miller, S.N.; Mack, M.C.; Ponciano, J.M.; McDaniel, S.F.; Fierer, N. The Bacterial Communities of Alaskan Mosses and Their Contributions to N2-Fixation. Microbiome 2021, 9, 53. [Google Scholar] [CrossRef]
- Hsing, W.; Canale-Parola, E. Cellobiose Chemotaxis by the Cellulolytic Bacterium Cellulomonas gelida. J. Bacteriol. 1992, 174, 7996–8002. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Varvaro, L.; Parisi, L.; Audergon, J.-M.; Morris, C.E. Disease and Frost Damage of Woody Plants Caused by Pseudomonas syringae. Adv. Agron. 2014, 126, 235–295. [Google Scholar] [CrossRef]
- Balestra, G.M.; Perestrelo, L.; Mazzaglia, A.; Rossetti, A. First Report of Blossom Blight Caused by Pseudomonas syringae on Kiwifruit Plants in Portugal. J. Plant Pathol. 2009, 91, 231. [Google Scholar]
- Hall, B.H.; Cother, E.J.; Noble, D.; McMahon, R.; Wicks, T.J. First Report of Pseudomonas syringae on Olives (Olea europaea) in South Australia. Australas. Plant Pathol. 2003, 32, 119–120. [Google Scholar] [CrossRef]
- Kotan, R.; Şahin, F. First Record of Bacterial Canker Caused by Pseudomonas syringae pv. syringae, on Apricot Trees in Turkey. Plant Pathol. 2002, 51, 798. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; Cazorla, F.M.; de Vicente, A. Pseudomonas syringae pv. syringae Associated with Mango Trees, a Particular Pathogen within the “Hodgepodge” of the Pseudomonas syringae Complex. Front. Plant Sci. 2019, 10, 570. [Google Scholar] [CrossRef]
- Sitaraman, R. Pseudomonas spp. as Models for Plant-Microbe Interactions. Front. Plant Sci. 2015, 6, 787. [Google Scholar] [CrossRef]
- Gao, J.; Wang, Y.; Wang, C.W.; Lu, B.H. First Report of Bacterial Root Rot of Ginseng Caused by Pseudomonas aeruginosa in China. Plant Dis. 2014, 98, 1577. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Déziel, E.; Schweizer, H.P.; Rahme, L.G.; Fall, R.; Vivanco, J.M. Pseudomonas aeruginosa-Plant Root Interactions. Pathogenicity, Biofilm Formation, and Root Exudation. Plant Physiol. 2004, 134, 320–331. [Google Scholar] [CrossRef]
- Huang, C.J.; Lin, C.H. First Report of Pseudomonas aeruginosa Causing Internal Brown Rot of Stored Onion Bulbs in Taiwan. J. Plant Pathol. 2017, 99, 817. [Google Scholar]
- Frank, C.L.; Davis, A.D.; Herzog, C. The Evolution of a Bat Population with White-Nose Syndrome (WNS) Reveals a Shift from an Epizootic to an Enzootic Phase. Front. Zool. 2019, 16, 40. [Google Scholar] [CrossRef]
- Boyles, J.G.; Cryan, P.M.; McCracken, G.F.; Kunz, T.H. Conservation. Economic Importance of Bats in Agriculture. Science 2011, 332, 41–42. [Google Scholar] [CrossRef]
- Medina, E.; Pieper, D.H. Tackling Threats and Future Problems of Multidrug-Resistant Bacteria. Curr. Top. Microbiol. Immunol. 2016, 398, 3–33. [Google Scholar]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Asghari, G.; Jalali, M.; Sadoughi, E. Antimicrobial Activity and Chemical Composition of Essential Oil from the Seeds of Artemisia aucheri Boiss. Jundishapur J. Nat. Pharm. Prod. 2012, 7, 11–15. [Google Scholar] [CrossRef]
- Lilley, B.D.; Brewer, J.H. The Selective Antibacterial Action of Phenylethyl Alcohol. J. Am. Pharm. Assoc. 1953, 42, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Narendhran, S.; Rajiv, P.; Vanathi, P.; Sivaraj, R. Spectroscopic Analysis of Bioactive Compounds from Streptomyces cavouresis KUV39: Evaluation of Antioxidant and Cytotoxicity Activity. Int. J. Pharm. Pharm. Sci. 2014, 6, 319–322. [Google Scholar]
- Naeim, H.; El-Hawiet, A.; Abdel Rahman, R.A.; Hussein, A.; El Demellawy, M.A.; Embaby, A.M. Antibacterial Activity of Centaurea pumilio L. Root and Aerial Part Extracts against Some Multidrug Resistant Bacteria. BMC Complement. Med. Ther. 2020, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Falade, O.E.; Oyetayo, V.O.; Awala, S.I. Evaluation of the Mycochemical Composition and Antimicrobial Potency of Wild Macrofungus, Rigidoporus microporus (Sw). J. Phytopharm. 2017, 6, 115–125. [Google Scholar] [CrossRef]
- Kumari, N.; Menghani, E.; Mithal, R. GCMS Analysis of Compounds Extracted from Actinomycetes AIA6 Isolates and Study of Its Antimicrobial Efficacy. Indian J. Chem. Technol. 2019, 26, 362–370. [Google Scholar]
- Judzentiene, A.; Budiene, J.; Svediene, J.; Garjonyte, R. Toxic, Radical Scavenging, and Antifungal Activity of Rhododendron tomentosum H. Essential Oils. Molecules 2020, 25, 1676. [Google Scholar] [CrossRef]
- Dinesh, M.; Deepika, S.; HarishKumar, R.; Selvaraj, C.I.; Roopan, S.M. Evaluation of Octyl-β-D-Glucopyranoside (OGP) for Cytotoxic, Hemolytic, Thrombolytic, and Antibacterial Activity. Appl. Biochem. Biotechnol. 2018, 185, 450–463. [Google Scholar] [CrossRef]
- Bratty, M.A.; Al Bratty, M.; Makeen, H.A.; Alhazmi, H.A.; Syame, S.M.; Abdalla, A.N.; Homeida, H.E.; Sultana, S.; Ahsan, W.; Khalid, A. Phytochemical, Cytotoxic, and Antimicrobial Evaluation of the Fruits of Miswak Plant, Salvadora persica L. J. Chem. 2020, 2020, 4521951. [Google Scholar] [CrossRef]
- Liu, H.; Huang, J.; Yang, S.; Li, J.; Zhou, L. Chemical Composition, Algicidal, Antimicrobial, and Antioxidant Activities of the Essential Oils of Taiwania flousiana Gaussen. Molecules 2020, 25, 967. [Google Scholar] [CrossRef]
- Dunkić, V.; Kosalec, I.; Kosir, I.J.; Potocnik, T.; Cerenak, A.; Koncic, M.Z.; Vitali, D.; Muller, I.D.; Kopricanec, M.; Bezic, N.; et al. Antioxidant and Antimicrobial Properties of Veronica spicata L. (Plantaginaceae). Curr. Drug Targets 2015, 16, 1660–1670. [Google Scholar] [CrossRef]
- Nafis, A.; Kasrati, A.; Jamali, C.A.; Custódio, L.; Vitalini, S.; Iriti, M.; Hassani, L. A Comparative Study of the in vitro Antimicrobial and Synergistic Effect of Essential Oils from Laurus nobilis L. and Prunus armeniaca L. from Morocco with Antimicrobial Drugs: New Approach for Health Promoting Products. Antibiotics 2020, 9, 140. [Google Scholar] [CrossRef]
- Shanthakumar, S.P.; Duraisamy, P.; Vishwanath, G.; Selvanesan, B.C.; Ramaraj, V.; Vasantharaj David, B. Broad Spectrum Antimicrobial Compounds from the Bacterium Exiguobacterium mexicanum MSSRFS9. Microbiol. Res. 2015, 178, 59–65. [Google Scholar] [CrossRef]
- Tasco, A.J.H.; Rueda, R.Y.R.; Alvarez, C.J.; Sartori, F.T.; Sacilotto, A.C.B.; Ito, I.Y.; Vichnewski, W.; Salvador, M.J. Antibacterial and Antifungal Properties of Crude Extracts and Isolated Compounds from Lychnophora markgravii. Nat. Prod. Res. 2020, 34, 863–867. [Google Scholar] [CrossRef]
- Alhazmi, H.A.; Sultana, S.; Khan, A.; Al-Bratty, M. GC-MS Analysis and Antimicrobial Activity of Ethanolic Extract of Calotropis procera (Ait.) R. Br. Leaves. J. Chem. Pharm. Res. 2018, 10, 45–49. [Google Scholar]
- Ravi, R.; Zulkrnin, N.S.H.; Rozhan, N.N.; Yusoff, N.R.N.; Rasat, M.S.M.; Ahmad, M.I.; Hamzah, Z.; Ishak, I.H.; Amin, M.F.M. Evaluation of Two Different Solvents for Azolla pinnata Extracts on Chemical Compositions and Larvicidal Activity against Aedes albopictus (Diptera: Culicidae). J. Chem. 2018, 2018, 7453816. [Google Scholar] [CrossRef]
- Osuntokun, O.T.; Cristina, G.M. Bio Isolation, Chemical Purification, Identification, Antimicrobial and Synergistic Efficacy of Extracted Essential Oils from Stem Bark Extract of Spondias mombin (Linn). Int. J. Mol. Biol. 2019, 4, 135–143. [Google Scholar] [CrossRef]
- Sengupta, S.; Nandi, I.; Bhattacharyya, D.K.; Ghosh, M. Anti-Oxidant and Anti-Bacterial Properties of 1-Octacosanol Isolated from Rice Bran Wax. J. Plant Biochem. Physiol. 2018, 6, 2. [Google Scholar] [CrossRef]
- Swamy, M.K.; Arumugam, G.; Kaur, R.; Ghasemzadeh, A.; Yusoff, M.M.; Sinniah, U.R. GC-MS Based Metabolite Profiling, Antioxidant and Antimicrobial Properties of Different Solvent Extracts of Malaysian Leaves. Evid.-Based. Complement. Alternat. Med. 2017, 2017, 1517683. [Google Scholar] [CrossRef]
- Değirmenci, H.; Erkurt, H. Relationship between Volatile Components, Antimicrobial and Antioxidant Properties of the Essential Oil, Hydrosol and Extracts of Citrus aurantium L. Flowers. J. Infect. Public Health 2020, 13, 58–67. [Google Scholar] [CrossRef]
- Vinuchakkaravarthy, T.; Kumaravel, K.P.; Ravichandran, S.; Velmurugan, D. Active Compound from the Leaves of Vitex Negundo L. Shows Anti-Inflammatory Activity with Evidence of Inhibition for Secretory Phospholipase A(2) through Molecular Docking. Bioinformation 2011, 7, 199–206. [Google Scholar] [CrossRef]
- De Guzman Alvindia, D.; Mangoba, M.A.A. Bioactivities of Allium longicuspis Regel against Anthracnose of Mango Caused by Colletotrichum gloeosporioides (Penz.). Sci. Rep. 2020, 10, 11367. [Google Scholar] [CrossRef]
- Yassin, M.T.; Mostafa, A.A.; Al-Askar, A.A. Anticandidal and Anti-Carcinogenic Activities of Mentha longifolia (Wild Mint) Extracts in vitro. J. King Saud Univ.—Sci. 2020, 32, 2046–2052. [Google Scholar] [CrossRef]
- Segueni, N.; Zellagui, A.; Boulechfar, S.; Derouiche, K.; Rhouati, S. Essential Oil of Hertia cheirifolia Leaves: Chemical Composition, Antibacterial and Antioxidant Activities. J. Mater. Environ. Sci. 2017, 8, 551–556. [Google Scholar]
- Mar, A.; Pripdeevech, P. Chemical Composition and Antibacterial Activity of Essential Oil and Extracts of Citharexylum spinosum Flowers from Thailand. Nat. Prod. Commun. 2014, 9, 707–710. [Google Scholar] [CrossRef]
- Guzmán-López, O.; Trigos, Á.; Fernández, F.J.; de Jesús Yañez-Morales, M.; Saucedo-Castañeda, G. Tyrosol and Tryptophol Produced by Ceratocystis adiposa. World J. Microbiol. Biotechnol. 2007, 23, 1473–1477. [Google Scholar] [CrossRef]
- Mujeeb, F.; Bajpai, P.; Pathak, N. Phytochemical Evaluation, Antimicrobial Activity, and Determination of Bioactive Components from Leaves of Aegle marmelos. Biomed Res. Int. 2014, 2014, 497606. [Google Scholar] [CrossRef]
- Faridha Begum, I.; Mohankumar, R.; Jeevan, M.; Ramani, K. GC-MS Analysis of Bio-Active Molecules Derived from FMR19 and the Antimicrobial Activity Against Bacterial Pathogens and MDROs. Indian J. Microbiol. 2016, 56, 426–432. [Google Scholar] [CrossRef]
- Weinstein, L. The Action of Urea and Some of Its Derivatives on Bacteria; the Effect of Combining Urea or Urethane with Penicillin on Mixed Cultures of Gram-Positive and Gram-Negative Bacteria. J. Immunol. 1946, 54, 145–149. [Google Scholar]
- Zhang, J.-L.; Yao, J.; Zhuge, J.-N.; Zhang, Y.-J. Antibacterial Activity of Erythritol on Periodontal Pathogen. Shanghai Kou Qiang Yi Xue 2019, 28, 362–367. [Google Scholar] [PubMed]
- Loimaranta, V.; Mazurel, D.; Deng, D.; Söderling, E. Xylitol and Erythritol Inhibit Real-Time Biofilm Formation of Streptococcus mutans. BMC Microbiol. 2020, 20, 184. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Kim, S.-W.; Lee, H.-K.; Kim, I.-D.; Lee, H.; Lee, J.-K. Anti-Oxidative Effects of 4-Hydroxybenzyl Alcohol in Astrocytes Confer Protective Effects in Autocrine and Paracrine Manners. PLoS ONE 2017, 12, e0177322. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.-F.; Shang, X.; Zhang, Z.-J.; Zhang, Y.-N. Phytochemical Composition and Antibacterial Activity of the Essential Oils from Different Parts of Sea Buckthorn (Hippophae rhamnoides L.). J. Food Drug Anal. 2017, 25, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Radmerikhi, S.; Formantes, B.; Fajardo, K.R.; Azul, E. Antimicrobial Effect of Different Xylitol Concentrations on Streptococcus mutans and Lactobacillus acidophilus Count. J. Restor. Dent. 2013, 1, 95. [Google Scholar] [CrossRef]
- Tapiainen, T.; Kontiokari, T.; Sammalkivi, L.; Ikäheimo, I.; Koskela, M.; Uhari, M. Effect of Xylitol on Growth of Streptococcus pneumoniae in the Presence of Fructose and Sorbitol. Antimicrob. Agents Chemother. 2001, 45, 166–169. [Google Scholar] [CrossRef]
- Bellaire, B.H.; Elzer, P.H.; Baldwin, C.L.; Roop, R.M., II. Production of the Siderophore 2,3-Dihydroxybenzoic Acid Is Required for Wild-Type Growth of Brucella abortus in the Presence of Erythritol under Low-Iron Conditions in Vitro. Infect. Immun. 2003, 71, 2927–2932. [Google Scholar] [CrossRef]
- George, S.; Benny, P.J.; Kuriakose, S.; George, C. Antibiotic Activity of 2, 3-Dihydroxybenzoic Acid Isolated from Flacourtia inermis Fruit against Multidrug Resistant Bacteria. Asian J. Pharm. Clin. Res. 2011, 4, 126–130. [Google Scholar]
- Chen, X.; Zhao, X.; Deng, Y.; Bu, X.; Ye, H.; Guo, N. Antimicrobial Potential of Myristic Acid against Listeria monocytogenes in Milk. J. Antibiot. 2019, 72, 298–305. [Google Scholar] [CrossRef]
- Agoramoorthy, G.; Chandrasekaran, M.; Venkatesalu, V.; Hsu, M.J. Antibacterial and Antifungal Activities of Fatty Acid Methyl Esters of the Blind-Your-Eye Mangrove from India. Braz. J. Microbiol. 2007, 38, 739–742. [Google Scholar] [CrossRef]
- Widyowati, R.; Agil, M. Chemical Constituents and Bioactivities of Several Indonesian Plants Typically Used in Jamu. Chem. Pharm. Bull. 2018, 66, 506–518. [Google Scholar] [CrossRef]
- Musa, A.M.; Ibrahim, M.A.; Aliyu, A.B.; Abdullahi, M.S.; Tajuddeen, N.; Ibrahim, H.; Oyewale, A.O. Chemical Composition and Antimicrobial Activity of Hexane Leaf Extract of Anisopus mannii (Asclepiadaceae). J. Intercult. Ethnopharmacol. 2015, 4, 129–133. [Google Scholar] [CrossRef]
- Nguyen, X.H.; Naing, K.W.; Lee, Y.S.; Kim, Y.H.; Moon, J.H.; Kim, K.Y. Antagonism of Antifungal Metabolites from Streptomyces griseus H7602 against Phytophthora capsici. J. Basic Microbiol. 2015, 55, 45–53. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Esmail, G.A.; Ghilan, A.-K.M.; Arasu, M.V.; Duraipandiyan, V.; Ponmurugan, K. Chemical Constituents of Streptomyces sp. Strain Al-Dhabi-97 Isolated from the Marine Region of Saudi Arabia with Antibacterial and Anticancer Properties. J. Infect. Public Health 2020, 13, 235–243. [Google Scholar] [CrossRef]
- Azizan, K.A.; Ghani, N.H.A.; Nawawi, M.F. GC-MS Based Metabolomics and Multivariate Statistical Analysis of Wedelia trilobata Extracts for the Identification of Potential Phytochemical Properties. Plant Omics J. 2015, 8, 537–543. [Google Scholar]
- Purohit, A.; Mohan, A. Antimicrobial Effects of Pyruvic and Succinic Acids on Salmonella Survival in Ground Chicken. LWT 2019, 116, 108596. [Google Scholar] [CrossRef]
- Balachandran, C.; Duraipandiyan, V.; Ignacimuthu, S. Cytotoxic (A549) and Antimicrobial Effects of Methylobacterium sp. Isolate (ERI-135) from Nilgiris Forest Soil, India. Asian Pac. J. Trop. Biomed. 2012, 2, 712–716. [Google Scholar] [CrossRef]
- Saadouli, I.; Zendah El Euch, I.; Trabelsi, E.; Mosbah, A.; Redissi, A.; Ferjani, R.; Fhoula, I.; Cherif, A.; Sabatier, J.-M.; Sewald, N.; et al. Isolation, Characterization and Chemical Synthesis of Large Spectrum Antimicrobial Cyclic Dipeptide (l-Leu-L-Pro) from V16R3Y1 Bacteria Extracts. A Novel H NMR Metabolomic Approach. Antibiotics 2020, 9, 270. [Google Scholar] [CrossRef]
- Demirci, M.A.; Ipek, Y.; Gul, F.; Ozen, T.; Demirtas, I. Extraction, Isolation of Heat-Resistance Phenolic Compounds, Antioxidant Properties, Characterization and Purification of 5-Hydroxymaltol from Turkish Apple Pulps. Food Chem. 2018, 269, 111–117. [Google Scholar] [CrossRef]
- Matsumoto, A.; Takahashi, Y. Endophytic Actinomycetes: Promising Source of Novel Bioactive Compounds. J. Antibiot. 2017, 70, 514–519. [Google Scholar] [CrossRef]
- Tyler, T.; Olsson, P.A. Substrate pH Ranges of South Swedish bryophytes—Identifying Critical pH Values and Richness Patterns. Flora 2016, 223, 74–82. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Zhou, X.; Guo, D.; Zhao, J.-H.; Yan, L.; Feng, G.-Z.; Gao, Q.; Yu, H.; Zhao, L.-P. Soil pH Is the Primary Factor Driving the Distribution and Function of Microorganisms in Farmland Soils in Northeastern China. Ann. Microbiol. 2019, 69, 1461–1473. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; Sivasithamparam, K. Non-Streptomycete Actinomycetes as Biocontrol Agents of Soil-Borne Fungal Plant Pathogens and as Plant Growth Promoters. Soil Biol. Biochem. 2006, 38, 1505–1520. [Google Scholar] [CrossRef]
- Fang, B.-Z.; Salam, N.; Han, M.-X.; Jiao, J.-Y.; Cheng, J.; Wei, D.-Q.; Xiao, M.; Li, W.-J. Insights on the Effects of Heat Pretreatment, pH, and Calcium Salts on Isolation of Rare Actinobacteria from Karstic Caves. Front. Microbiol. 2017, 8, 1535. [Google Scholar] [CrossRef] [PubMed]
- Intra, B.; Mungsuntisuk, I.; Nihira, T.; Igarashi, Y.; Panbangred, W. Identification of Actinomycetes from Plant Rhizospheric Soils with Inhibitory Activity against Colletotrichum spp., the Causative Agent of Anthracnose Disease. BMC Res. Notes 2011, 4, 98. [Google Scholar] [CrossRef] [PubMed]
- Masand, M.; Jose, P.A.; Menghani, E.; Jebakumar, S.R.D. Continuing Hunt for Endophytic Actinomycetes as a Source of Novel Biologically Active Metabolites. World J. Microbiol. Biotechnol. 2015, 31, 1863–1875. [Google Scholar] [CrossRef]
- Qin, S.; Bian, G.-K.; Zhang, Y.-J.; Xing, K.; Cao, C.-L.; Liu, C.-H.; Dai, C.-C.; Li, W.-J.; Jiang, J.-H. Modestobacter roseus sp. nov., an Endophytic Actinomycete Isolated from the Coastal Halophyte Salicornia europaea Linn., and Emended Description of the Genus Modestobacter. Int. J. Syst. Evol. Microbiol. 2013, 63((Pt. 6)), 2197–2202. [Google Scholar] [CrossRef]
- Passari, A.K.; Chandra, P.; Zothanpuia; Mishra , V.K.; Leo, V.V.; Gupta, V.K.; Kumar, B.; Singh, B.P. Detection of Biosynthetic Gene and Phytohormone Production by Endophytic Actinobacteria Associated with Solanum lycopersicum and Their Plant-Growth-Promoting Effect. Res. Microbiol. 2016, 167, 692–705. [Google Scholar] [CrossRef]
- Shutsrirung, A.; Chromkaew, Y.; Pathom-Aree, W.; Choonluchanon, S.; Boonkerd, N. Diversity of Endophytic Actinomycetes in Mandarin Grown in Northern Thailand, Their Phytohormone Production Potential and Plant Growth Promoting Activity. Soil Sci. Plant Nutr. 2013, 59, 322–330. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore Production by Actinobacteria. Biometals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Aznar, A.; Dellagi, A. New Insights into the Role of Siderophores as Triggers of Plant Immunity: What Can We Learn from Animals? J. Exp. Bot. 2015, 66, 3001–3010. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Sabiha, Y.; Loss, O.; Eisendle, M.; Wallner, A.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Distinct Roles for Intra- and Extracellular Siderophores during Aspergillus fumigatus Infection. PLoS Pathog. 2007, 3, 1195–1207. [Google Scholar] [CrossRef]
- Nimnoi, P.; Pongsilp, N.; Lumyong, S. Endophytic Actinomycetes Isolated from Aquilaria crassna Pierre Ex Lec and Screening of Plant Growth Promoters Production. World J. Microbiol. Biotechnol. 2010, 26, 193–203. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Hamdali, H. Isolation and Characterization of Rock Phosphate Solubilizing Actinobacteria from a Togolese Phosphate Mine. Afr. J. Biotechnol. 2011, 11, 312–320. [Google Scholar] [CrossRef]
- Passari, A.K.; Mishra, V.K.; Singh, G.; Singh, P.; Kumar, B.; Gupta, V.K.; Sarma, R.K.; Saikia, R.; Donovan, A.O.; Singh, B.P. Insights into the Functionality of Endophytic Actinobacteria with a Focus on Their Biosynthetic Potential and Secondary Metabolites Production. Sci. Rep. 2017, 7, 11809. [Google Scholar] [CrossRef]
- Gulati, A.; Sharma, N.; Vyas, P.; Sood, S.; Rahi, P.; Pathania, V.; Prasad, R. Organic Acid Production and Plant Growth Promotion as a Function of Phosphate Solubilization by Acinetobacter rhizosphaerae Strain BIHB 723 Isolated from the Cold Deserts of the Trans-Himalayas. Arch. Microbiol. 2010, 192, 975–983. [Google Scholar] [CrossRef]
- Ghods, S.; Sims, I.M.; Moradali, M.F.; Rehm, B.H.A. Bactericidal Compounds Controlling Growth of the Plant Pathogen Pseudomonas syringae pv. actinidiae, Which Forms Biofilms Composed of a Novel Exopolysaccharide. Appl. Environ. Microbiol. 2015, 81, 4026–4036. [Google Scholar] [CrossRef]
- Keser, S.; Mutlu, M.B.; Güven, K. Antimicrobial Effect of Halophilic Actinomycetes on Xanthomonas and Pseudomonas Tomato Disease Agents. II Int. Symp. Tomato Dis. 2007, 808, 401–404. [Google Scholar] [CrossRef]
- Nedialkova, D.; Naidenova, M. Screening the Antimicrobial Activity of Actinomycetes Strains Isolated from Antarctica. J. Cult. Collect. 2005, 4, 29–35. [Google Scholar]
- Micalizzi, E.W.; Mack, J.N.; White, G.P.; Avis, T.J.; Smith, M.L. Microbial Inhibitors of the Fungus Pseudogymnoascus destructans, the Causal Agent of White-Nose Syndrome in Bats. PLoS ONE 2017, 12, e0179770. [Google Scholar] [CrossRef]
- Li, C.; Jin, P.; Liu, C.; Ma, Z.; Zhao, J.; Li, J.; Wang, X.; Xiang, W. Streptomyces bryophytorum sp. nov., an Endophytic Actinomycete Isolated from Moss (Bryophyta). Antonie Van Leeuwenhoek 2016, 109, 1209–1215. [Google Scholar] [CrossRef]
- Suriyachadkun, C.; Ngaemthao, W.; Pujchakarn, T.; Chunhametha, S. Cryptosporangium phraense sp. nov., Isolated from Plant Litter. Int. J. Syst. Evol. Microbiol. 2020, 70, 971–976. [Google Scholar] [CrossRef]
- Nurkanto, A.; Lisdiyanti, P.; Hamada, M.; Ratnakomala, S.; Shibata, C.; Tamura, T. Cryptosporangium cibodasense sp. nov., Isolated from Leaf Litter in Indonesia. Int. J. Syst. Evol. Microbiol. 2015, 65, 4632–4637. [Google Scholar] [CrossRef]
- Himaman, W.; Suksaard, P.; Mingma, R.; Matsumoto, A.; Duangmal, K. Cryptosporangium eucalypti sp. nov., an Actinomycete Isolated from Eucalyptus camaldulensis Roots. Int. J. Syst. Evol. Microbiol. 2017, 67, 3077–3082. [Google Scholar] [CrossRef]
- Zitouni, A.; Lamari, L.; Boudjella, H.; Badji, B.; Sabaou, N.; Gaouar, A.; Mathieu, F.; Lebrihi, A.; Labeda, D.P. Saccharothrix algeriensis sp. nov., Isolated from Saharan Soil. Int. J. Syst. Evol. Microbiol 2004, 54((Pt. 4)), 1377–1381. [Google Scholar] [CrossRef]
- Liu, C.; Guan, X.; Wang, S.; Zhao, J.; Wang, H.; He, H.; Xiang, W.; Wang, X. Saccharothrix carnea sp. nov., an Actinobacterium Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 4033–4037. [Google Scholar] [CrossRef]
- Zhang, L.-P.; Jiang, C.-L.; Chen, W.-X. Streptosporangium yunnanense sp. nov. and Streptosporangium purpuratum sp. nov., from Soil in China. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 2, 719–724. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, L.; Li, Z.; Piao, C.; Li, Y.; Li, J.; Liu, C.; Wang, X.; Xiang, W. Streptosporangium jiaoheense sp. nov. and Streptosporangium taraxaci sp. nov., Actinobacteria Isolated from Soil and Dandelion Root (Taraxacum mongolicum Hand.-Mazz.). Int. J. Syst. Evol. Microbiol. 2016, 66, 2370–2376. [Google Scholar] [CrossRef] [PubMed]
- Ara, I.; Yamamura, H.; Tsetseg, B.; Daram, D.; Ando, K. Actinoplanes toevensis sp. nov. and Actinoplanes tereljensis sp. nov., Isolated from Mongolian Soil. Int. J. Syst. Evol. Microbiol. 2010, 60((Pt. 4)), 919–927. [Google Scholar] [CrossRef] [PubMed]
- Sazak, A.; Sahin, N.; Camas, M. Actinoplanes abujensis sp. nov., Isolated from Nigerian Arid Soil. Int. J. Syst. Evol. Microbiol. 2012, 62(Pt. 4), 960–965. [Google Scholar] [CrossRef] [PubMed]
- Borsetto, C.; Amos, G.C.A.; da Rocha, U.N.; Mitchell, A.L.; Finn, R.D.; Laidi, R.F.; Vallin, C.; Pearce, D.A.; Newsham, K.K.; Wellington, E.M.H. Microbial Community Drivers of PK/NRP Gene Diversity in Selected Global Soils. Microbiome 2019, 7, 78. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III Polyketide Synthases in Natural Product Biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef]
- Miller, K.I.; Qing, C.; Sze, D.M.Y.; Neilan, B.A. Investigation of the Biosynthetic Potential of Endophytes in Traditional Chinese Anticancer Herbs. PLoS ONE 2012, 7, e35953. [Google Scholar] [CrossRef]
- Chen, F.; Ludwiczuk, A.; Wei, G.; Chen, X.; Crandall-Stotler, B.; Bowman, J.L. Terpenoid Secondary Metabolites in Bryophytes: Chemical Diversity, Biosynthesis and Biological Functions. CRC Crit. Rev. Plant Sci. 2018, 37, 210–231. [Google Scholar] [CrossRef]
- Vinuchakkaravarthy, T.; Sangeetha, C.K.; Velmurugan, D. Tris(2,4-Di-Tert-Butyl-Phen-Yl) Phosphate. Acta Crystallogr. Sect. E Struct. Rep. Online 2010, 66(Pt. 9), o2207–o2208. [Google Scholar] [CrossRef]
- Janso, J.E.; Carter, G.T. Biosynthetic Potential of Phylogenetically Unique Endophytic Actinomycetes from Tropical Plants. Appl. Environ. Microbiol. 2010, 76, 4377–4386. [Google Scholar] [CrossRef]
- Jones, S.E.; Pham, C.A.; Zambri, M.P.; McKillip, J.; Carlson, E.E.; Elliot, M.A. Streptomyces Volatile Compounds Influence Exploration and Microbial Community Dynamics by Altering Iron Availability. mBio 2019, 10, e00171-19. [Google Scholar] [CrossRef]
- Wu, P.C.; Wang, M.Z. Polytrichaceae. In Moss Flora of China; He, S., Ed.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2005; Volume 8, pp. 306–366. [Google Scholar]
- Eddy, A. A Handbook of Malesian Mosses; British Museum (Natural History): London, UK, 1988. [Google Scholar]
- Zhang, M.X.; He, S. Hypnaceae. In Moss Flora of China; He, S., Ed.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2005; Volume 8, pp. 80–260. [Google Scholar]
- Gangulee, H.C. Mosses of Eastern India and Adjacent Regions; University of Culcutta: Culcutta, India, 1969. [Google Scholar]
- Touw, A. A Taxonomic Revision of the Thuidiaceae (Musci) of Tropical Asia, the Western Pacific, and Hawaii. J. Hattori Bot. Lab. 2001, 91, 1–136. [Google Scholar]
- Li, X.J.; He, S.; Zang, M.; Missouri Botanical Garden Press:, St. Moss Flora of China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2007; Volume 4. [Google Scholar]
- Noguchi, A. A Revision of the Genus Macrothamnium M. Fleisch. (Musci). Kumamoto J. Sci. Biol. 1972, 11, 1–12. [Google Scholar]
- Noguchi, A. Illustrated Moses Flora of Japan, Part 3. J. Hattori Bot. Lab. 1989, 3, 493–742. [Google Scholar]
- Ignatov, M.S.; Huttunen, S.; Koponen, T. Bryophyte Flora of Hunan Province, China. 5. Brachytheciaceae (Musci), with an Overview of Eurhynchiadelphus and Rhynchostegiella in South-East Asia. Acta Bot. Fenn. 2005, 178, 1–56. [Google Scholar]
- Wang, Y.F.; Hu, R.L. Brachytheciaceae. In Moss Flora of China; He, S., Ed.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008; Volume 7, pp. 71–167. [Google Scholar]
- Dabek-Szremiawska, M.; Hattori, T. Winogradsky’s Salts Solution as a Diluting Medium for Plate Count of Oligotrophic Bacteria in Soil. J. Gen. Appl. Microbiol. 1981, 27, 517–518. [Google Scholar] [CrossRef]
- Inter-Society Color Council; Kelly, K.L. ISCC-NBS Color-Name Charts Illustrated with Centroid Colors; National Bureau of Standards: Washington, DC, USA, 1965. [Google Scholar]
- Glickmann, E.; Dessaux, Y. A Critical Examination of the Specificity of the Salkowski Reagent for Indolic Compounds Produced by Phytopathogenic Bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Rangey, D.E. Two Simple Media for Demonstrate of Pyocyanin and Fluorescei. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Atkin, C.L.; Neilands, J.B.; Phaff, H.J. Rhodotorulic Acid from Species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a New Alanine-Containing Ferrichrome from Cryptococcus melibiosum. J. Bacteriol. 1970, 103, 722–733. [Google Scholar] [CrossRef]
- Arnow, L.E. Colorimetric Determination of the Components of 3,4-Dihydroxyphenylalaninetyrosine Mixtures. J. Biol. Chem. 1937, 118, 531–537. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- McArthur, R.L.; Ghosh, S.; Cheeptham, N. Improvement of Protocols for the Screening of Biological Control Agents against White-Nose Syndrome. J. Exp. Microbiol. Immunol. 2017, 2, 1–7. [Google Scholar]
- Ghosh, S.; Persad, E.; Shiue, T.-Y.; Lam, C.; Islam, A.; Mascibroda, L.G.; Sherman, M.B.; Smith, T.; Cheeptham, N. Explorative Study on Isolation and Characterization of a Microviridae G4 Bacteriophage, EMCL318, against Multi-Drug-Resistant Escherichia coli 15-318. Antibiotics 2018, 7, 92. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for Characterization of Streptomyces Species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Alzohairy, A.M. BioEdit: An Important Software for Molecular Biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evolution. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.; Fiedler, H.-P. A Guide to Successful Bioprospecting: Informed by Actinobacterial Systematics. Antonie Van Leeuwenhoek 2010, 98, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Khattab, A.I.; Babiker, E.H.; Saeed, H.A. Streptomyces: Isolation, Optimization of Culture Conditions and Extraction of Secondary Metabolites. Int. Curr. Pharm. J. 2016, 5, 27–32. [Google Scholar] [CrossRef]
- Pang, Z.Q.; Chen, J.; Wang, T.H.; Gao, C.S.; Li, Z.M.; Guo, L.T.; Xu, J.; Cheng, Y. Linking Plant Secondary Metabolites and Plant Microbiomes: A Review. Front. Plant Sci. 2021, 12, 300. [Google Scholar] [CrossRef] [PubMed]
| Method | Host Moss Species | Location | Isolation Media | Average Value CFU/g | No. Isolates | |
|---|---|---|---|---|---|---|
| Rhizosphere Soil | Inside Moss | |||||
| Method 1 | P. microstomum | 8°35′19′’ N98°29′12′’ E | WPA | 1.3 × 104 | 0 | 15 |
| V8 juice agar | 4.6 × 104 | 0 | 36 | |||
| Hypnum sp. | 8°35′19′’ N98°29′12′’ E | WPA | 5.9 × 104 | 0 | 23 | |
| V8 juice agar | 9.5 × 103 | 0 | 6 | |||
| P. maximoviczii | 8°35′19′’ N98°29′12′’ E | WPA | 1.5 × 103 | 2 × 103 | 3 | |
| V8 juice agar | 4.4 × 104 | 2 × 103 | 0 | |||
| Method 2 | B. buchananii | 18°33′14′’ N98°46′52′’ E | WPA | 1.1 × 104 | 4.5 × 103 | 30 |
| V8 juice agar | 7.5 × 103 | 0 | 45 | |||
| D. maschalogena | 18°40′44′’ N98°50′25′’ E | WPA | 0 | 2.9 × 105 | 0 | |
| V8 juice agar | 0 | 3 × 105 | 2 | |||
| B. recurvulum | 18°40′44′’ N98°50′25′’ E | WPA | 0 | 3.5 × 104 | 11 | |
| V8 juice agar | 5 × 103 | 2.3 × 104 | 14 | |||
| T. cymbifolium | 8°35′19′’ N98°29′12′’ E | WPA | 1.1 × 104 | 0 | 2 | |
| V8 juice agar | 7.5 × 103 | 0 | 0 | |||
| M. submacrocarpum | 8°35′19′’ N98°29′12′’ E | WPA | 0 | 4.9 × 104 | 4 | |
| V8 juice agar | 0 | 6.7 × 104 | 5 | |||
| C. purpureus | 18°40′44′’ N98°50′25′’ E | WPA | 0 | 0 | 0 | |
| V8 juice agar | 5 × 103 | 0 | 0 | |||
| Total | 196 | |||||
| Source | Host Moss Species | Isolate | Clear Zone on PVK Agar (cm) |
|---|---|---|---|
| Plant tissue | B. buchananii | P32-20 | 0.71 ± 0.11 ab |
| D. maschalogena | P33-17 | 0.96 ± 0.13 ab | |
| M. submacrocarpum | P49-10 | 0.71 ± 0.05 ab | |
| M. submacrocarpum | P49-11 | 0.85 ± 0.08 ab | |
| M. submacrocarpum | P49-14 | 1.21 ± 0.74 bc | |
| Soil | P. microstomum | S3-11 | 0.86 ± 0.06 ab |
| P. microstomum | S3-16 | 0.95 ± 0.01 ab | |
| P. microstomum | S3-17 | 0.62 ± 0.03 ab | |
| P. microstomum | S3-31 | 0.54 ± 0.05 a | |
| Hypnum sp. | S6-3 | 1.04 ± 0.04 ab | |
| Hypnum sp. | S6-6 | 0.63 ± 0.04 ab | |
| Hypnum sp. | S6-14 | 0.51 ± 0.33 a | |
| Hypnum sp. | S6-17 | 0.6 ± 0 ab | |
| Hypnum sp. | S6-28 | 1.75 ± 0.12 d | |
| Hypnum sp. | S6-31 | 1.36 ± 0.08 ab | |
| P. maximoviczii | S13-2 | 0.75 ± 0.14 ab | |
| B. buchananii | S32-30 | 0.77 ± 0.21 ab | |
| B. buchananii | S54-2 | 0.91 ± 0.02 ab | |
| B. buchananii | S54-18 | 0.92 ± 0.1 ab | |
| B. buchananii | S54-19 | 0.87 ± 0.06 ab |
| Genera | Isolate | DDBJ Accession Number | Top Hit Taxon | % Similarity | Length (bp) |
|---|---|---|---|---|---|
| Streptomyces | P32-2 | LC551864 | Streptomyces althioticus NRRL B-3981T | 100 | 1221 |
| P32-15 | LC551863 | Streptomyces althioticus NRRL B-3981T | 100 | 1221 | |
| P32-21 | LC551860 | Streptomyces althioticus NRRL B-3981T | 100 | 1221 | |
| P54-7 | LC551871 | Streptomyces althioticus NRRL B-3981T | 100 | 1328 | |
| P54-15 | LC551870 | Streptomyces althioticus NRRL B-3981T | 100 | 1329 | |
| P49-13 | LC551869 | Streptomyces violaceolatus DSM 40438T | 100 | 1344 | |
| P49-18 | LC551868 | Streptomyces griseoincarnatus LMG 19316T | 100 | 1303 | |
| S3-11 | LC551876 | Streptomyces rhizosphaerihabitans JR-35T | 99.85 | 1324 | |
| S3-16 | LC551877 | Streptomyces mirabilis NBRC 13450T | 100 | 1338 | |
| S3-17 | LC551878 | Streptomyces aureus NBRC 100912T | 100 | 1355 | |
| S3-26 | LC551872 | Streptomyces wedmorensis NRRL 3426T | 99.55 | 1324 | |
| S3-30 | LC551874 | Streptomyces camponoticapitis 2H-TWYE14T | 99.62 | 1329 | |
| S6-17 | LC551893 | Streptomyces setonii NRRL ISP-5322T | 100 | 1329 | |
| S32-79 | LC551890 | Streptomyces setonii NRRL ISP-5322T | 100 | 1335 | |
| S6-31 | LC551895 | Streptomyces sporoverrucosus NBRC 15458T | 100 | 1360 | |
| S32-5 | LC551886 | Streptomyces fulvissimus DSM 40593T | 99.93 | 1359 | |
| S32-27 | LC551891 | Streptomyces fulvissimus DSM 40593T | 99.93 | 1360 | |
| S32-55 | LC551884 | Streptomyces fulvissimus DSM 40593T | 99.93 | 1356 | |
| S32-76 | LC551888 | Streptomyces fulvissimus DSM 40593T | 99.93 | 1353 | |
| S32-10 | LC551885 | Streptosporangium oxazolinicum K07-0460T | 99.25 | 1343 | |
| S32-29 | LC551887 | Streptomyces badius NRRL B-2567T | 100 | 1300 | |
| S32-43 | LC551880 | Streptomyces badius NRRL B-2567T | 100 | 1329 | |
| S32-52 | LC551882 | Streptomyces omiyaensis NBRC 13449T | 99.55 | 1343 | |
| S32-65 | LC551889 | Streptomyces globisporus NBRC 12867T | 100 | 1337 | |
| S32-74 | LC551883 | Streptomyces wedmorensis NRRL 3426T | 99.62 | 1330 | |
| S32-77 | LC551881 | Streptomyces dioscori A217T | 99.63 | 1341 | |
| Micromonospora | P32-13 | LC551861 | Micromonospora maritima D10-9-5T | 99.77 | 1324 |
| P32-19 | LC551862 | Micromonospora marina DSM 45555T | 99.85 | 1331 | |
| P33-11 | LC551867 | Micromonospora aurantiaca ATCC 27029T | 100 | 1296 | |
| S32-33 | LC551892 | Micromonospora tulbaghiae DSM 45142T | 100 | 1326 | |
| Actinoplanes | S3-33 | LC551879 | Actinoplanes lutulentus NEAU-GRX6T | 98.57 | 1327 |
| S6-21 | LC551896 | Actinoplanes consettensis JCM 7624T | 99.93 | 1338 | |
| Saccharothrix | S3-21 | LC551875 | Saccharothrix yanglingensis Hhs.015T | 98.86 | 1328 |
| S3-31 | LC551873 | Saccharothrix yanglingensis Hhs.015T | 98.86 | 1332 | |
| Nocardia | P33-17 | LC551866 | Nocardia fluminea S1T | 99.55 | 1332 |
| S6-27 | LC551894 | Nocardia salmonicida subsp. cummidelens R89T | 100 | 1335 | |
| Cryptosporangium | P33-8 | LC551865 | Cryptosporangium minutisporangium IFO 15962T | 99.92 | 1330 |
| Condition | Compound Nature | Estimated Compound Content (%) |
|---|---|---|
| Pre-derivatization | Alkane | 46.75 |
| Alcohol | 12.99 | |
| Ketone | 3.90 | |
| Carboxylic acid | 2.60 | |
| Flavonoid | 2.60 | |
| Amide | 2.60 | |
| Alkaloid derivative | 2.60 | |
| Ether | 2.60 | |
| Imide | 1.30 | |
| Ester | 1.30 | |
| Lactone | 1.30 | |
| Others | 19.48 | |
| Post-derivatization | Alkanes | 15.29 |
| Carboxylic acid | 10.59 | |
| Sugar | 8.24 | |
| Alcohol | 7.06 | |
| Sugar alcohol | 5.88 | |
| Fatty acid | 4.71 | |
| Ether | 3.53 | |
| Ester | 2.35 | |
| Amide | 2.35 | |
| Imide | 2.35 | |
| Terpene | 2.35 | |
| Others | 30.59 |
| Isolates | Activity of Crude Extract against MRSA (mm, ± Standard Error) |
|---|---|
| Absolute Ethanol (negative control) | 0 (±0) |
| Tetracycline (positive control) | 8 (±0.3) |
| S32-76 | 10 (±0.3) |
| S32-77 | 13 (±0.4) |
| S32-79 | 14 (±0.4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Insuk, C.; Pongpamorn, P.; Forsythe, A.; Matsumoto, A.; Ōmura, S.; Pathom-aree, W.; Cheeptham, N.; Xu, J. Taxonomic and Metabolite Diversities of Moss-Associated Actinobacteria from Thailand. Metabolites 2022, 12, 22. https://doi.org/10.3390/metabo12010022
Insuk C, Pongpamorn P, Forsythe A, Matsumoto A, Ōmura S, Pathom-aree W, Cheeptham N, Xu J. Taxonomic and Metabolite Diversities of Moss-Associated Actinobacteria from Thailand. Metabolites. 2022; 12(1):22. https://doi.org/10.3390/metabo12010022
Chicago/Turabian StyleInsuk, Chadabhorn, Pornkanok Pongpamorn, Adrian Forsythe, Atsuko Matsumoto, Satoshi Ōmura, Wasu Pathom-aree, Naowarat Cheeptham, and Jianping Xu. 2022. "Taxonomic and Metabolite Diversities of Moss-Associated Actinobacteria from Thailand" Metabolites 12, no. 1: 22. https://doi.org/10.3390/metabo12010022
APA StyleInsuk, C., Pongpamorn, P., Forsythe, A., Matsumoto, A., Ōmura, S., Pathom-aree, W., Cheeptham, N., & Xu, J. (2022). Taxonomic and Metabolite Diversities of Moss-Associated Actinobacteria from Thailand. Metabolites, 12(1), 22. https://doi.org/10.3390/metabo12010022









