Knockout of Arabidopsis thaliana VEP1, Encoding a PRISE (Progesterone 5β-Reductase/Iridoid Synthase-Like Enzyme), Leads to Metabolic Changes in Response to Exogenous Methyl Vinyl Ketone (MVK)
Abstract
:1. Introduction
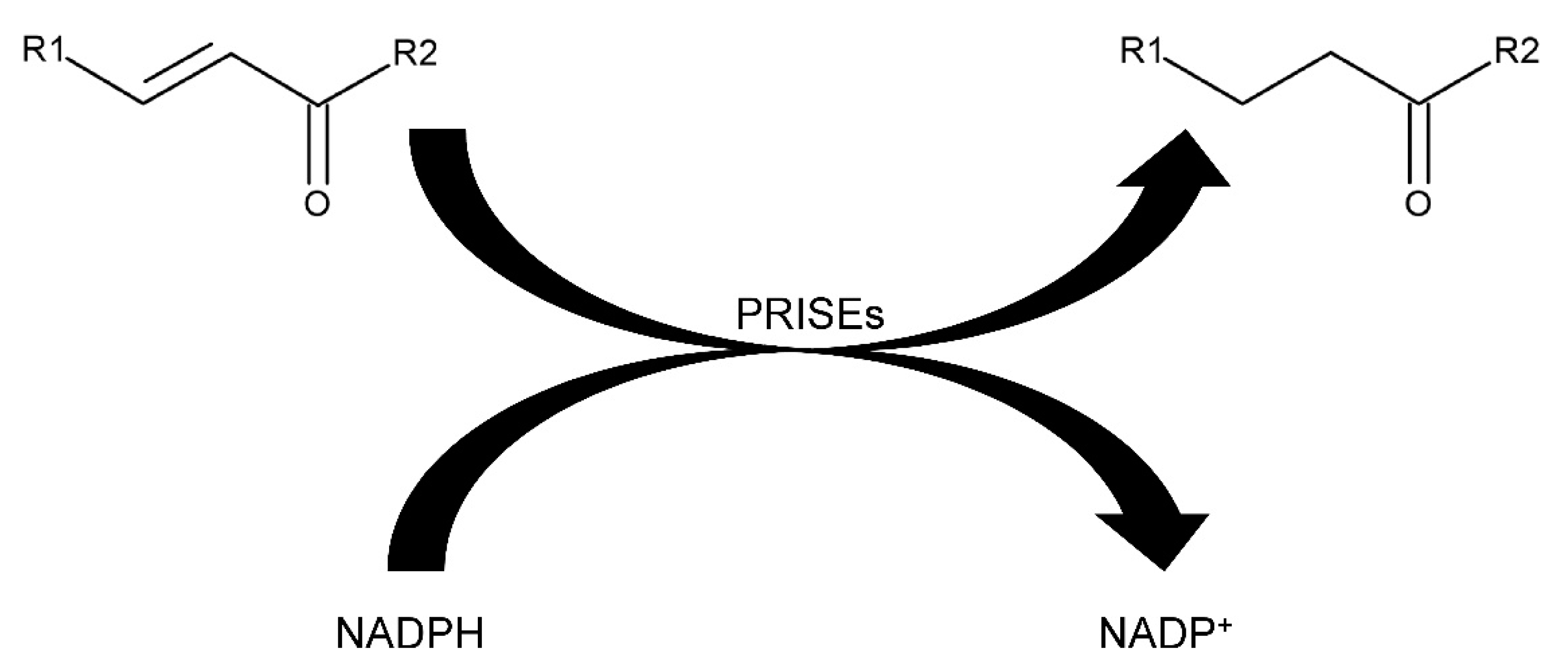
2. Results and Discussion
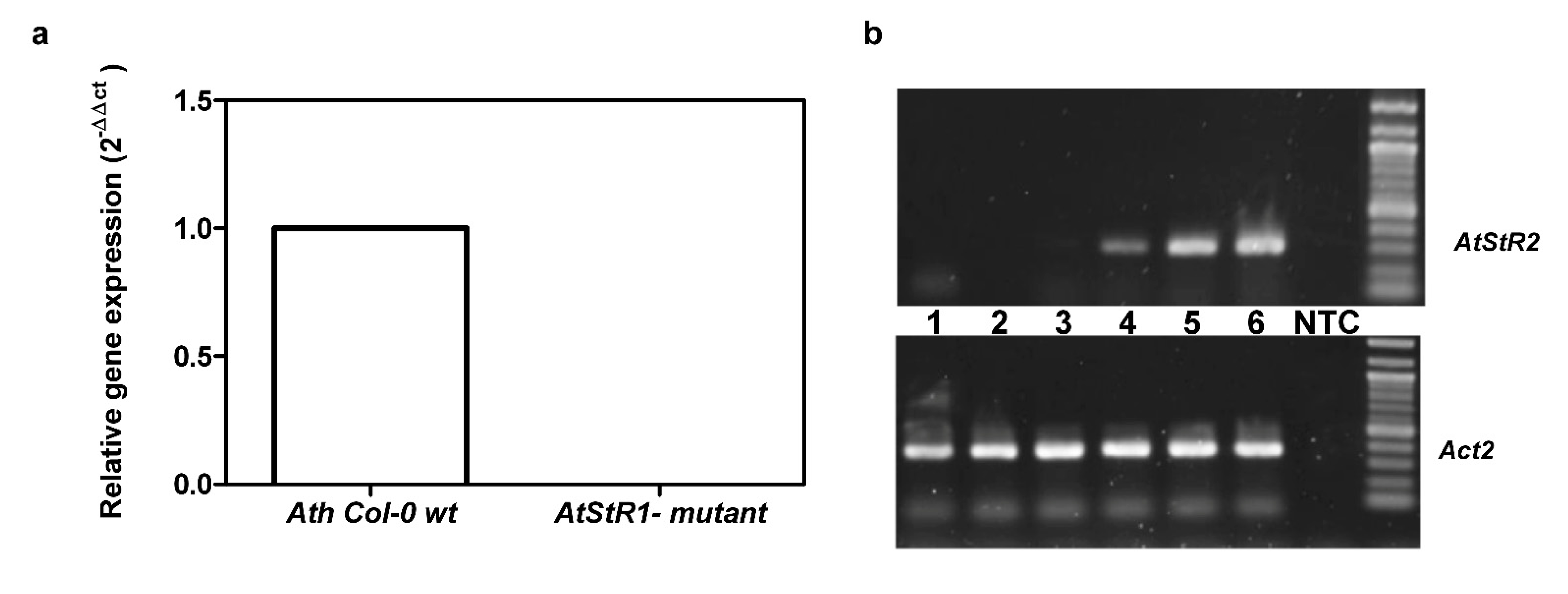
2.1. Characterization of AtStR2
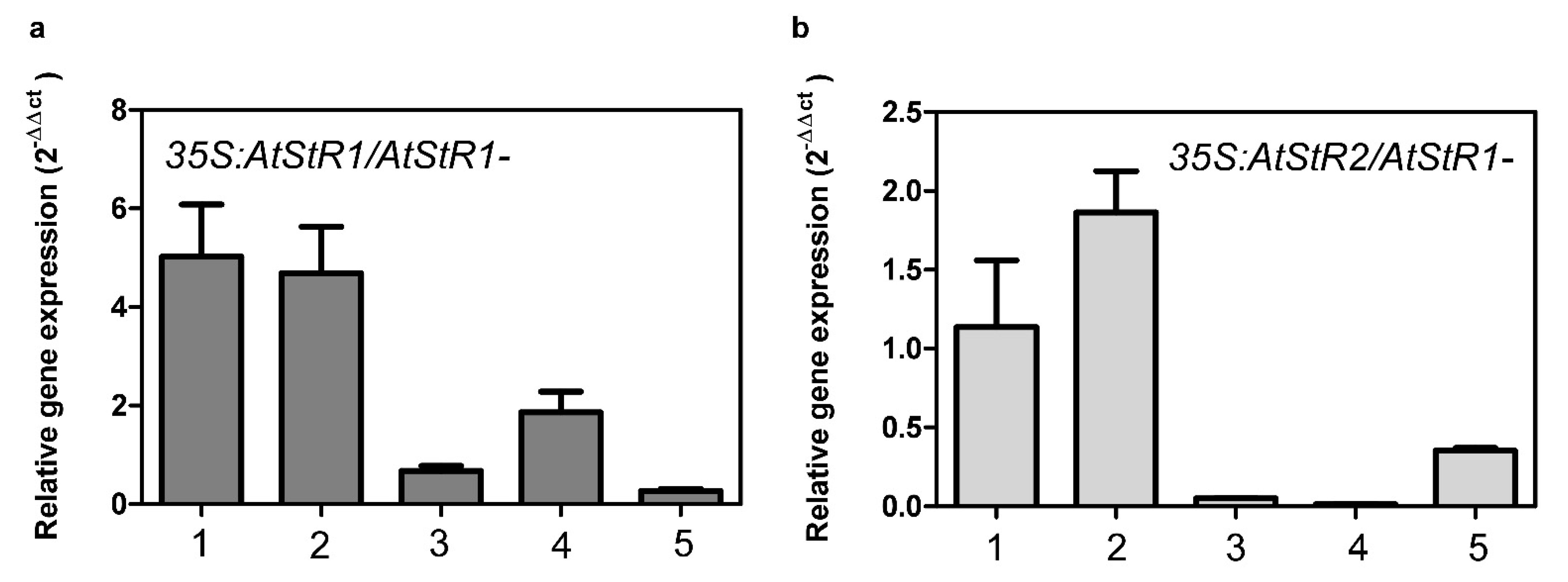
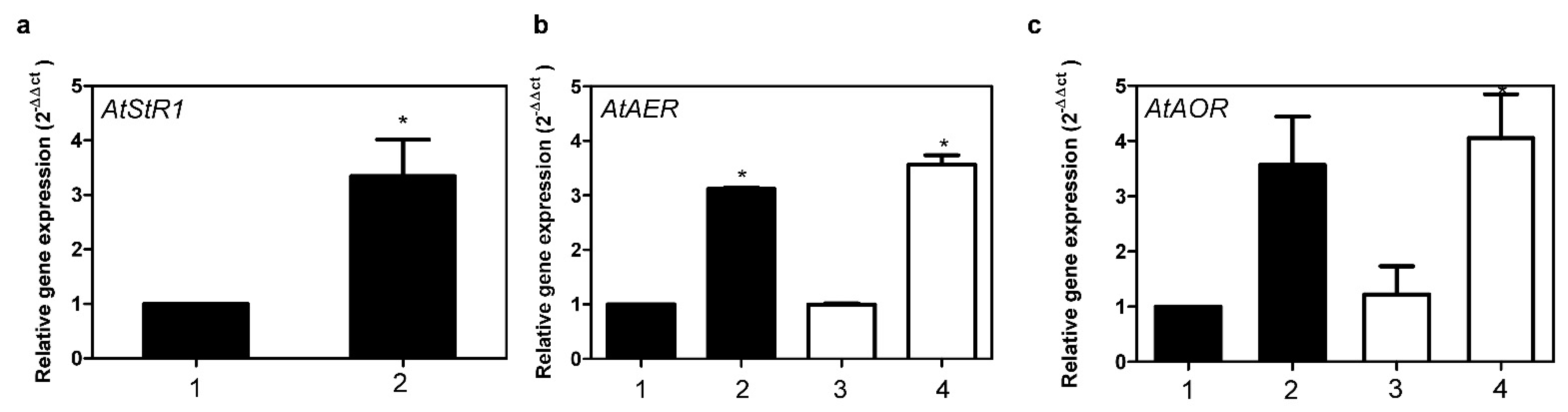
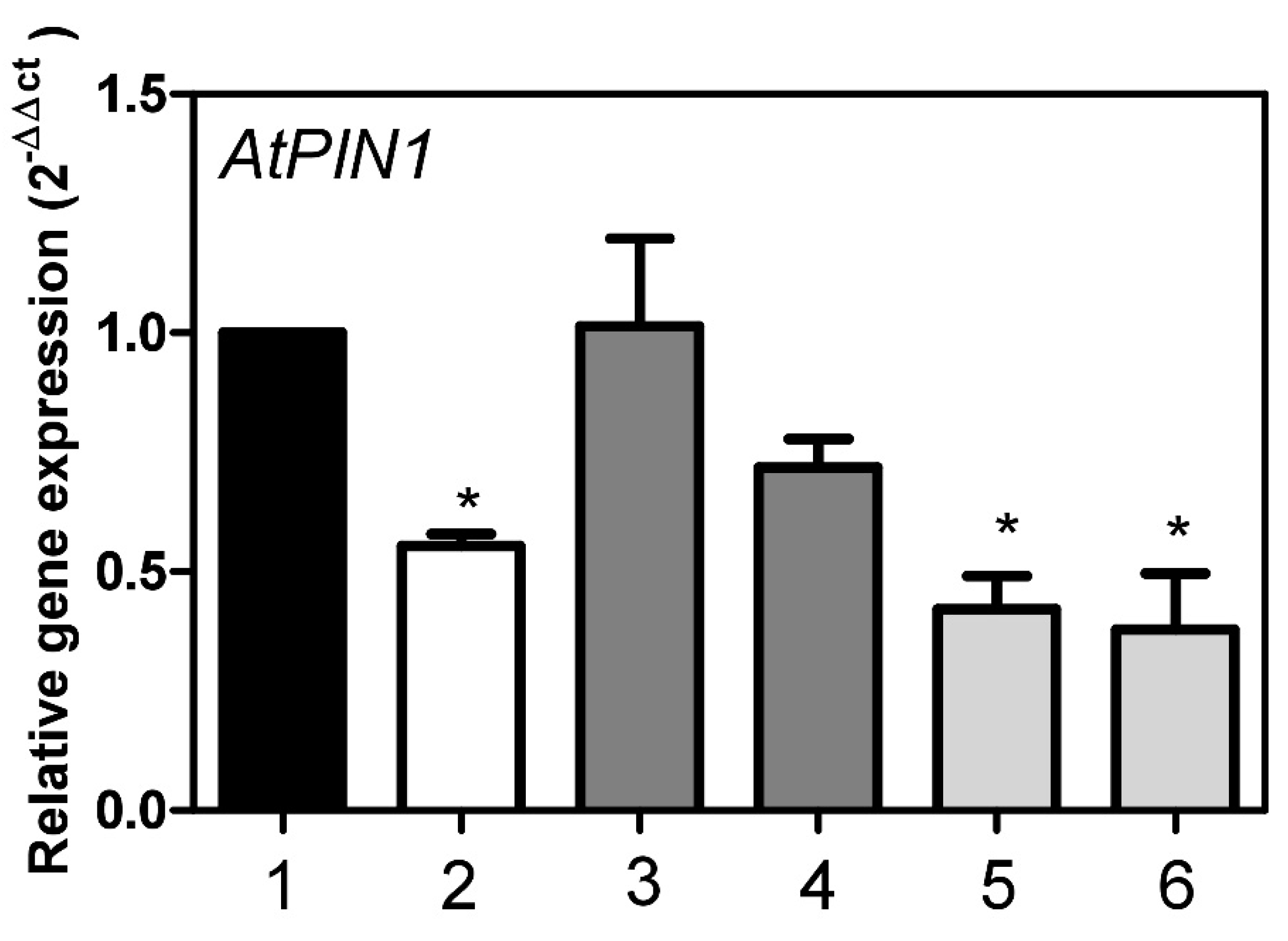
2.2. Expression of AtStR1 and AtStR2
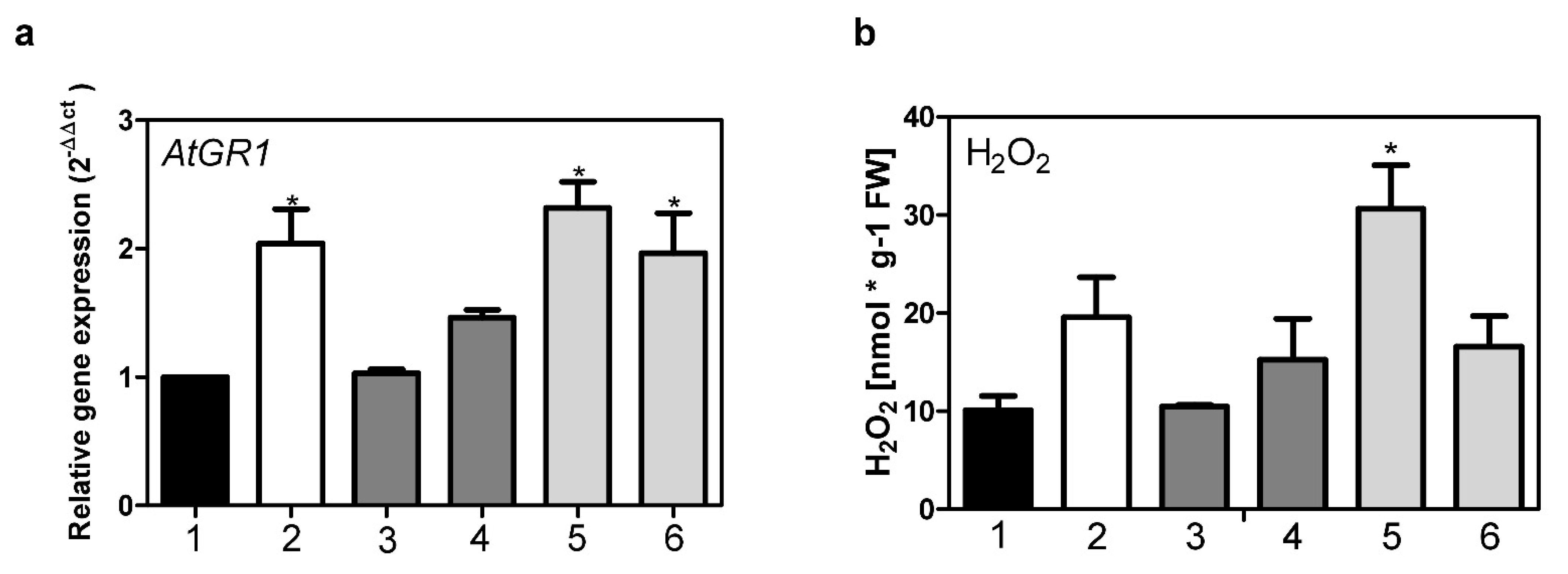
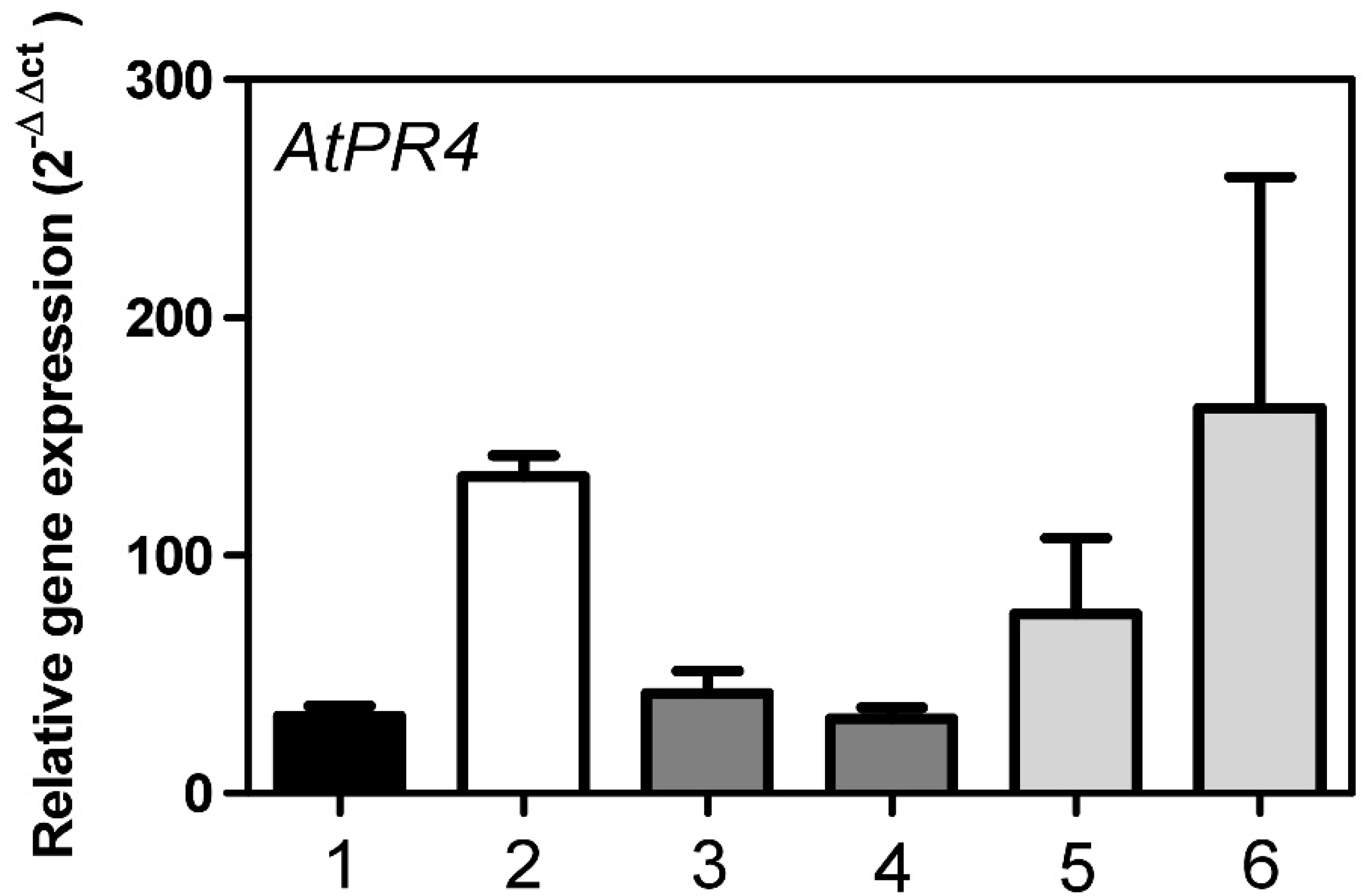
2.3. Roles of AtStR1 and AtStR2 in Plant Reactive Electrophilic Species (RES) Stress
3. Materials and Methods
3.1. Plant Material
3.2. Heterologous Expression of AtStR2
3.3. Enzyme Kinetics
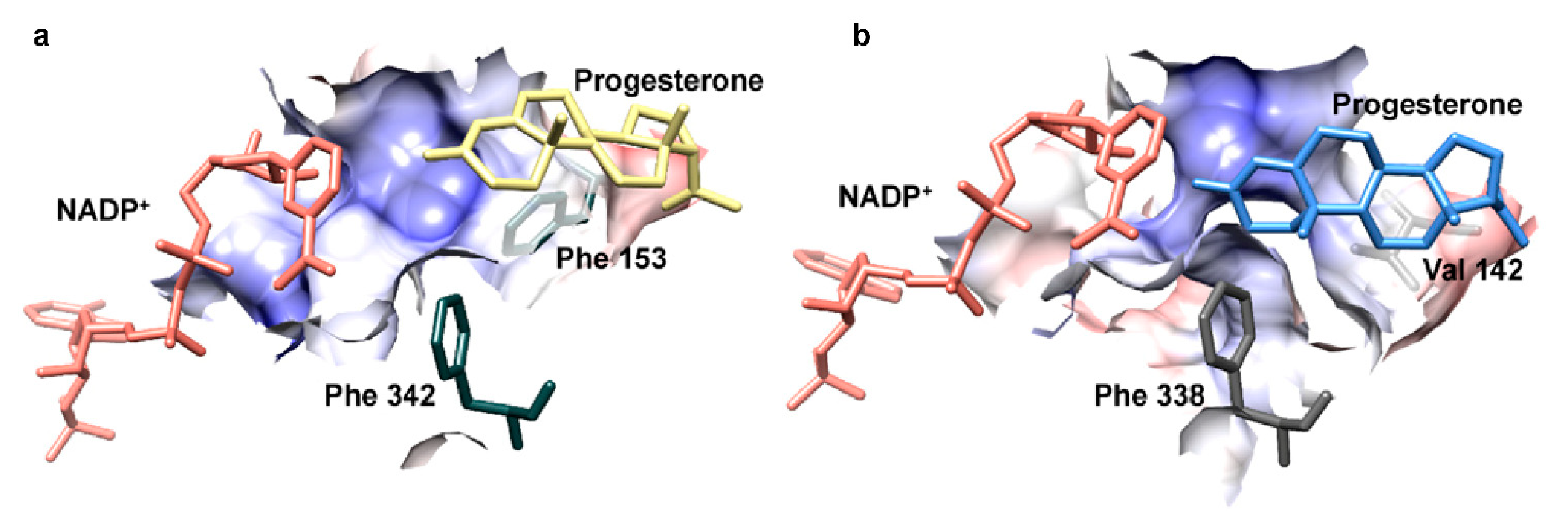
3.4. In Silico Analysis
3.5. MVK Stress Treatment
3.6. Cold Treatment
3.7. RNA Isolation and cDNA-Synthesis
3.8. Quantitative Real-Time PCR and Semiquantitative PCR
3.9. Quantification of Protein Activity in Plant Extracts
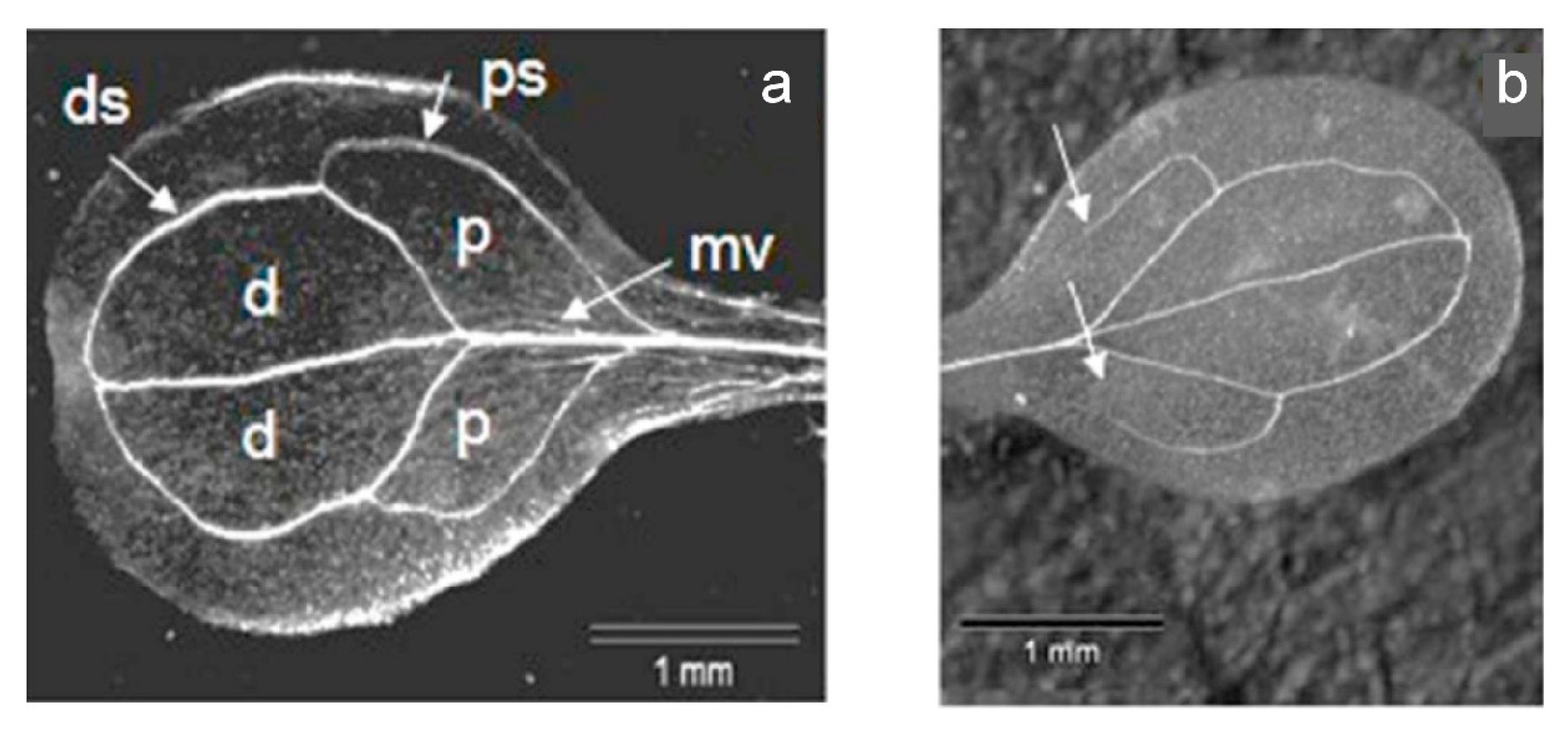
3.10. Analysis of the Vein Patterning in Cotyledons
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kreis, W.; Munkert, J. Exploiting enzyme promiscuity to shape plant specialized metabolism. J. Exp. Bot. 2019, 70, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Noda-Garcia, L.; Liebermeister, W.; Tawfik, D.S. Metabolite-Enzyme Coevolution: From Single Enzymes to Metabolic Pathways and Networks. Annu. Rev. Biochem. 2018, 87, 187–216. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Fiamoncini, J.; Gil-de-la-Fuente, A.; Greiner, R.; Manach, C.; Wishart, D.S. BioTransformer: A comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminform. 2019, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- EFSA Workshop on in vitro comparative metabolism studies in regulatory pesticide risk assessment. EFSA Support. Publ. 2019, 16, 1618E. [CrossRef]
- Jun, J.H.; Ha, C.M.; Nam, H.G. Involvement of the VEP1 gene in vascular strand development in Arabidopsis thaliana. Plant Cell Physiol. 2002, 43, 323–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.Y.; Moon, Y.H.; Choi, K.H.; Kim, Y.H.; Eun, M.Y.; Guh, J.O.; Kim, K.C.; Cho, B.H. Structure and expression of the AWI 31 gene specifically induced by wounding in Arabidopsis thaliana. Mol. Cells 1997, 7, 131–135. [Google Scholar] [PubMed]
- Munkert, J.; Pollier, J.; Miettinen, K.; van Moerkercke, A.; Payne, R.; Müller-Uri, F.; Burlat, V.; O’Connor, S.E.; Memelink, J.; Kreis, W.; et al. Iridoid synthase activity is common among the plant progesterone 5β-reductase family. Mol. Plant 2015, 8, 136–152. [Google Scholar] [CrossRef] [Green Version]
- Herl, V.; Fischer, G.; Reva, V.A.; Stiebritz, M.; Muller, Y.A.; Müller-Uri, F.; Kreis, W. The VEP1 gene (At4g24220) encodes a short-chain dehydrogenase/reductase with 3-oxo-Delta4,5-steroid 5beta-reductase activity in Arabidopsis thaliana L. Biochimie 2009, 91, 517–525. [Google Scholar] [CrossRef]
- Schmidt, K.; Petersen, J.; Munkert, J.; Egerer-Sieber, C.; Hornig, M.; Muller, Y.A.; Kreis, W. PRISEs (progesterone 5β-reductase and/or iridoid synthase-like 1,4-enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism. Phytochemistry 2018, 156, 9–19. [Google Scholar] [CrossRef]
- Petersen, J.; Lanig, H.; Munkert, J.; Bauer, P.; Müller-Uri, F.; Kreis, W. Progesterone 5β-reductases/iridoid synthases (PRISE): Gatekeeper role of highly conserved phenylalanines in substrate preference and trapping is supported by molecular dynamics simulations. J. Biomol. Struct. Dyn. 2016, 34, 1667–1680. [Google Scholar] [CrossRef]
- Gärtner, D.E.; Keilholz, W.; Seitz, H.U. Purification, characterization and partial peptide microsequencing of progesterone 5 beta-reductase from shoot cultures of Digitalis purpurea. Eur. J. Biochem. 1994, 225, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Roca-Pérez, L.; Boluda, R.; Gavidia, I.; Pérez-Bermúdez, P. Seasonal cardenolide production and Dop5βr gene expression in natural populations of Digitalis obscura. Phytochemistry 2004, 65, 1869–1878. [Google Scholar] [CrossRef]
- Herl, V.; Fischer, G.; Mueller-Uri, F.; Kreis, W. Molecular cloning and heterologous expression of progesterone 5β-reductase from Digitalis lanata Ehrh. Phytochemistry 2006, 67, 225–231, Erratum in Phytochemistry 2008, 69, 2411. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Horn, E.; Ernst, M.; Leykauf, T.; Leupold, T.; Dorfner, M.; Wolf, L.; Ignatova, A.; Kreis, W.; Munkert, J. RNAi-mediated gene knockdown of progesterone 5β-reductases in Digitalis lanata reduces 5β-cardenolide content. Plant Cell Rep. 2021, 40, 1631–1646. [Google Scholar] [CrossRef] [PubMed]
- Geu-Flores, F.; Sherden, N.H.; Courdavault, V.; Burlat, V.; Glenn, W.S.; Wu, C.; Nims, E.; Cui, Y.; O’Connor, S.E. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 2012, 492, 138–142. [Google Scholar] [CrossRef]
- Nguyen, T.-D.; O’Connor, S.E. The Progesterone 5β-Reductase/Iridoid Synthase Family: A Catalytic Reservoir for Specialized Metabolism across Land Plants. ACS Chem. Biol. 2020, 15, 1780–1787. [Google Scholar] [CrossRef]
- Tarrío, R.; Ayala, F.J.; Rodríguez-Trelles, F. The Vein Patterning 1 (VEP1) gene family laterally spread through an ecological network. PLoS ONE 2011, 6, e22279. [Google Scholar] [CrossRef] [Green Version]
- Burda, E.; Kraußer, M.; Fischer, G.; Hummel, W.; Müller-Uri, F.; Kreis, W.; Gröger, H. Recombinant Δ4,5-Steroid 5 β-Reductases as Biocatalysts for the Reduction of Activated C=C-Double Bonds in Monocyclic and Acyclic Molecules. Adv. Synth. Catal. 2009, 351, 2787–2790. [Google Scholar] [CrossRef]
- Durchschein, K.; Wallner, S.; Macheroux, P.; Schwab, W.; Winkler, T.; Kreis, W.; Faber, K. Nicotinamide-Dependent Ene Reductases as Alternative Biocatalysts for the Reduction of Activated Alkenes. Eur. J. Org. Chem. 2012, 2012, 4963–4968. [Google Scholar] [CrossRef]
- Kai, H.; Hirashima, K.; Matsuda, O.; Ikegami, H.; Winkelmann, T.; Nakahara, T.; Iba, K. Thermotolerant cyclamen with reduced acrolein and methyl vinyl ketone. J. Exp. Bot. 2012, 63, 4143–4150. [Google Scholar] [CrossRef] [Green Version]
- Alméras, E.; Stolz, S.; Vollenweider, S.; Reymond, P.; Mène-Saffrané, L.; Farmer, E.E. Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 2003, 34, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorn, A.; Egerer-Sieber, C.; Jäger, C.M.; Herl, V.; Müller-Uri, F.; Kreis, W.; Muller, Y.A. The crystal structure of progesterone 5β-reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. J. Biol. Chem. 2008, 283, 17260–17269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Bermúdez, P.; García, A.A.M.; Tuñón, I.; Gavidia, I. Digitalis purpurea P5βR2, encoding steroid 5β-reductase, is a novel defense-related gene involved in cardenolide biosynthesis. New Phytol. 2010, 185, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Rieck, C.; Geiger, D.; Munkert, J.; Messerschmidt, K.; Petersen, J.; Strasser, J.; Meitinger, N.; Kreis, W. Biosynthetic approach to combine the first steps of cardenolide formation in Saccharomyces cerevisiae. Microbiologyopen 2019, 8, e925. [Google Scholar] [CrossRef] [Green Version]
- Mano, J.; Torii, Y.; Hayashi, S.; Takimoto, K.; Matsui, K.; Nakamura, K.; Inzé, D.; Babiychuk, E.; Kushnir, S.; Asada, K. The NADPH:quinone oxidoreductase P1-ζ-crystallin in Arabidopsis catalyzes the α,β-hydrogenation of 2-alkenals: Detoxication of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol. 2002, 43, 1445–1455. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Hasegawa, A.; Taninaka, A.; Mizutani, M.; Sugimoto, Y. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 2011, 286, 6999–7009. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Bauer, P.; Munkert, J.; Brydziun, M.; Burda, E.; Müller-Uri, F.; Gröger, H.; Muller, Y.A.; Kreis, W. Highly conserved progesterone 5β-reductase genes (P5βR) from 5 beta-cardenolide-free and 5β-cardenolide-producing angiosperms. Phytochemistry 2010, 71, 1495–1505. [Google Scholar] [CrossRef]
- Tropper, M.; Höhn, S.; Wolf, L.-S.; Fritsch, J.; Kastner-Detter, N.; Rieck, C.; Munkert, J.; Meitinger, N.; Lanig, H.; Kreis, W. 21-Hydroxypregnane 21-O-malonylation, a crucial step in cardenolide biosynthesis, can be achieved by substrate-promiscuous BAHD-type phenolic glucoside malonyltransferases from Arabidopsis thaliana and homolog proteins from Digitalis lanata. Phytochemistry 2021, 187, 112710. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Mano, J.; Tanaka, K.; Wang, S.; Zhang, M.; Deng, X.; Zhang, S. High level of reduced glutathione contributes to detoxification of lipid peroxide-derived reactive carbonyl species in transgenic Arabidopsis overexpressing glutathione reductase under aluminum stress. Physiol. Plant. 2017, 161, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Lewinsohn, E.; Gijzen, M. Phytochemical diversity: The sounds of silent metabolism. Plant Sci. 2009, 176, 161–169. [Google Scholar] [CrossRef]
- Jardine, K.J.; Meyers, K.; Abrell, L.; Alves, E.G.; Serrano, A.M.Y.; Kesselmeier, J.; Karl, T.; Guenther, A.; Chambers, J.Q.; Vickers, C. Emissions of putative isoprene oxidation products from mango branches under abiotic stress. J. Exp. Bot. 2013, 64, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Cappellin, L.; Loreto, F.; Biasioli, F.; Pastore, P.; McKinney, K. A mechanism for biogenic production and emission of MEK from MVK decoupled from isoprene biosynthesis. Atmos. Chem. Phys. 2019, 19, 3125–3135. [Google Scholar] [CrossRef] [Green Version]
- Vollenweider, S.; Weber, H.; Stolz, S.; Chételat, A.; Farmer, E.E. Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 2000, 24, 467–476. [Google Scholar] [CrossRef]
- Horiyama, S.; Hatai, M.; Ichikawa, A.; Yoshikawa, N.; Nakamura, K.; Kunitomo, M. Detoxification Mechanism of α,β-Unsaturated Carbonyl Compounds in Cigarette Smoke Observed in Sheep Erythrocytes. Chem. Pharm. Bull. 2018, 66, 721–726. [Google Scholar] [CrossRef] [Green Version]
- Ernst, M. Untersuchungen zur physiologischen Funktion der Progesteron-5β-Reduktase in Arabidopsis thaliana und Vitis vinifera. Ph.D. Thesis, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany, 2015. [Google Scholar]
- Harvey, C.M.; Sharkey, T.D. Exogenous isoprene modulates gene expression in unstressed Arabidopsis thaliana plants. Plant Cell Environ. 2016, 39, 1251–1263. [Google Scholar] [CrossRef] [Green Version]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.-P. Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 2010, 22, 376–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, M.S.; Fukaki, H.; Mori, I.C.; Nakahara, K.; Mano, J. Reactive oxygen species and reactive carbonyl species constitute a feed-forward loop in auxin signaling for lateral root formation. Plant J. 2019, 100, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.; Biswas, M.S.; Sugimoto, K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants 2019, 8, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusano, H.; Li, H.; Minami, H.; Kato, Y.; Tabata, H.; Yazaki, K. Evolutionary Developments in Plant Specialized Metabolism, Exemplified by Two Transferase Families. Front. Plant Sci. 2019, 10, 794. [Google Scholar] [CrossRef]
- Moghe, G.D.; Last, R.L. Something Old, Something New: Conserved Enzymes and the Evolution of Novelty in Plant Specialized Metabolism. Plant Physiol. 2015, 169, 1512–1523. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Bond, S.R.; Naus, C.C. RF-Cloning.org: An online tool for the design of restriction-free cloning projects. Nucleic Acids Res. 2012, 40, W209–W213. [Google Scholar] [CrossRef] [Green Version]
- Unger, T.; Jacobovitch, Y.; Dantes, A.; Bernheim, R.; Peleg, Y. Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 2010, 172, 34–44. [Google Scholar] [CrossRef]
- Stevens, R.C. Design of high-throughput methods of protein production for structural biology. Structure 2000, 8, R177–R185. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ernst, M.; de Padua, R.M.; Herl, V.; Müller-Uri, F.; Kreis, W. Expression of 3β-HSD and P5βR, genes respectively coding for Δ5-3β-hydroxysteroid dehydrogenase and progesterone 5β-reductase, in leaves and cell cultures of Digitalis lanata EHRH. Planta Med. 2010, 76, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Enzyme (Substrate) | Km (µM) | kcat (s−1) | Catalytic Efficiency (s−1 M−1) |
|---|---|---|---|
| rAtStR2 | |||
| rAtStR2 (progesterone) | 64.7 | 0.15 | 2314 |
| rAtStR2 (MVK) | 157.5 | 0.03 | 163 |
| rAtStR2 (NADPH2) | 41.0 | 0.12 | 2927 |
| Other PRISEs of cluster II | |||
| rEcP5βR2 (progesterone) a | 82 | 0.05 | 552 |
| rEcP5βR2 (MVK) a | 224 | 0.05 | 245 |
| rCrP5βR6 (progesterone) a | 75.5 | 0.40 | 5530 |
| rCrP5βR6 (MVK) a | 118 | 0.03 | 261 |
| rAtStR1 and rAtSt5βR1_F153A_F342A | |||
| rAtStR1 (progesterone) b | 124.8 | 0.28 | 2244 |
| rAtStR1 (MVK) b | 75.5 | 0.20 | 7299 |
| rAtStR1_F153A_F342A (progesterone) c | 217 | 2.00 | 9218 |
| rAtStR1_F153A_F342A (MVK) c | - | n.a. | - |
| Other PRISEs of cluster I | |||
| rEcP5βR1 (progesterone) b | 77 | 0.01 | 31 |
| rEcP5βR1 (MVK) b | 344 | 0.72 | 2143 |
| rCrP5βR4 (progesterone) b | 153 | 0.02 | 123 |
| rCrP5βR4 (MVK) b | 123 | 0.20 | 1562 |
| Other MVK-converting enzymes | |||
| AtAER (MVK) d | 55.0 | 83.0 | 1,500,000 |
| AtAOR (MVK) e | 2880 | 74.0 | 25,700 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klein, J.; Ernst, M.; Christmann, A.; Tropper, M.; Leykauf, T.; Kreis, W.; Munkert, J. Knockout of Arabidopsis thaliana VEP1, Encoding a PRISE (Progesterone 5β-Reductase/Iridoid Synthase-Like Enzyme), Leads to Metabolic Changes in Response to Exogenous Methyl Vinyl Ketone (MVK). Metabolites 2022, 12, 11. https://doi.org/10.3390/metabo12010011
Klein J, Ernst M, Christmann A, Tropper M, Leykauf T, Kreis W, Munkert J. Knockout of Arabidopsis thaliana VEP1, Encoding a PRISE (Progesterone 5β-Reductase/Iridoid Synthase-Like Enzyme), Leads to Metabolic Changes in Response to Exogenous Methyl Vinyl Ketone (MVK). Metabolites. 2022; 12(1):11. https://doi.org/10.3390/metabo12010011
Chicago/Turabian StyleKlein, Jan, Mona Ernst, Alexander Christmann, Marina Tropper, Tim Leykauf, Wolfgang Kreis, and Jennifer Munkert. 2022. "Knockout of Arabidopsis thaliana VEP1, Encoding a PRISE (Progesterone 5β-Reductase/Iridoid Synthase-Like Enzyme), Leads to Metabolic Changes in Response to Exogenous Methyl Vinyl Ketone (MVK)" Metabolites 12, no. 1: 11. https://doi.org/10.3390/metabo12010011
APA StyleKlein, J., Ernst, M., Christmann, A., Tropper, M., Leykauf, T., Kreis, W., & Munkert, J. (2022). Knockout of Arabidopsis thaliana VEP1, Encoding a PRISE (Progesterone 5β-Reductase/Iridoid Synthase-Like Enzyme), Leads to Metabolic Changes in Response to Exogenous Methyl Vinyl Ketone (MVK). Metabolites, 12(1), 11. https://doi.org/10.3390/metabo12010011









