Maternal Plasma Metabolic Profile Demarcates a Role for Neuroinflammation in Non-Typical Development of Children
Abstract
1. Introduction
2. Results
2.1. Participant Characteristics and Covariate Selection
2.2. Metabolomic Analyses
2.2.1. Full Feature Metabolome Set
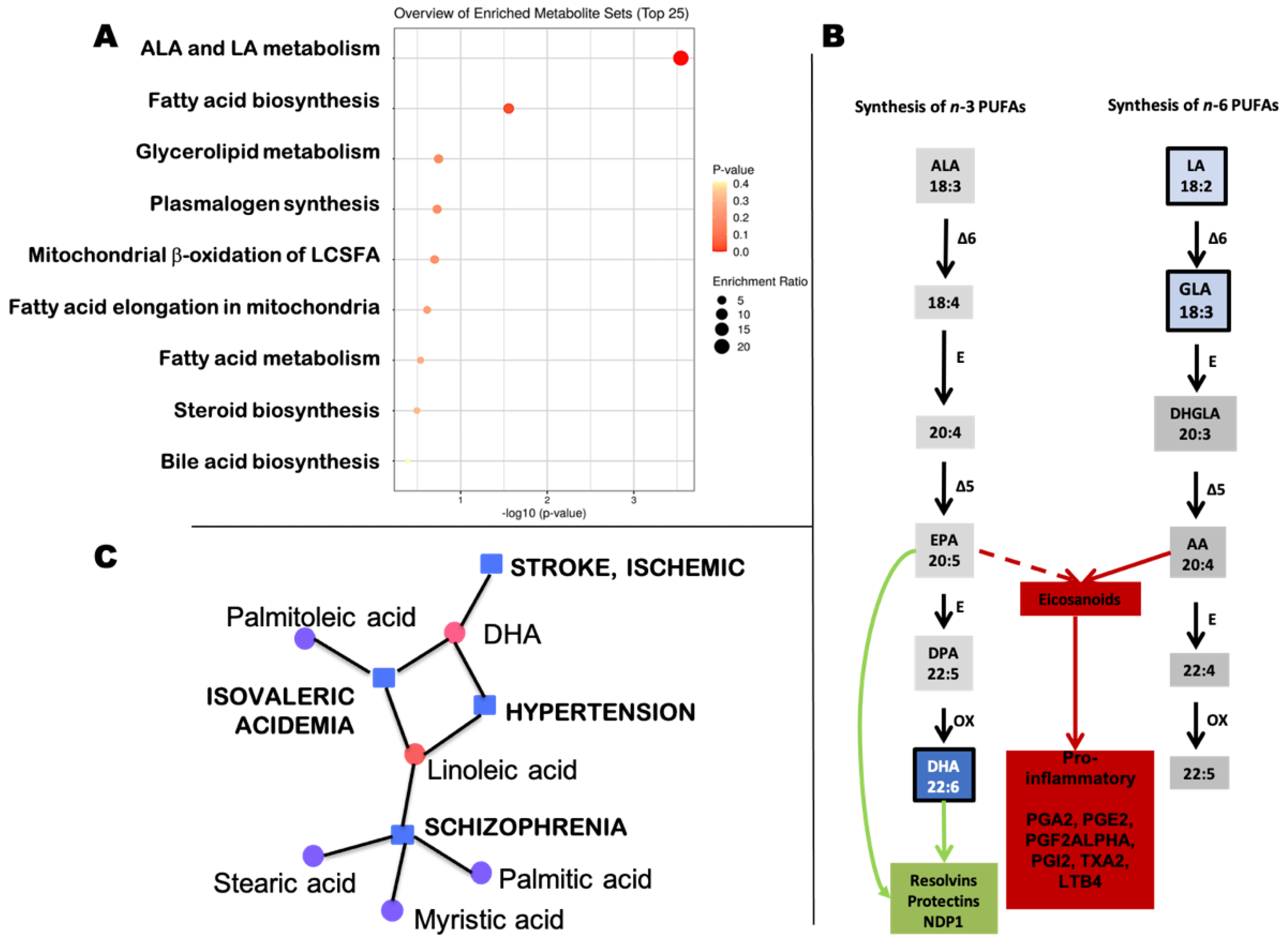
2.2.2. Annotated Metabolome Set
2.3. Fatty Acids in Prostaglandin Pathways
3. Discussion
4. Materials and Methods
4.1. Study Design and Population
4.2. ASD and Non-TD Outcome Classification
4.3. Metabolomics Analysis
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maenner, M.J.; Shaw, K.A.; Baio, J. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chiarotti, F.; Venerosi, A. Epidemiology of Autism Spectrum Disorders: A Review of Worldwide Prevalence Estimates Since 2014. Brain Sci. 2020, 10, 274. [Google Scholar] [CrossRef]
- Ozonoff, S.; Young, G.S.; Carter, A.; Messinger, D.; Yirmiya, N.; Zwaigenbaum, L.; Bryson, S.; Carver, L.J.; Constantino, J.N.; Dobkins, K.; et al. Recurrence risk for autism spectrum disorders: A Baby Siblings Research Consortium study. Pediatrics 2011, 128, e488–e495. [Google Scholar] [CrossRef] [PubMed]
- Werner, E.; Heilier, J.F.; Ducruix, C.; Ezan, E.; Junot, C.; Tabet, J.C. Mass spectrometry for the identification of the discriminating signals from metabolomics: Current status and future trends. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 871, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Werner, E.; Croixmarie, V.; Umbdenstock, T.; Ezan, E.; Chaminade, P.; Tabet, J.C.; Junot, C. Mass spectrometry-based metabolomics: Accelerating the characterization of discriminating signals by combining statistical correlations and ultrahigh resolution. Anal. Chem. 2008, 80, 4918–4932. [Google Scholar] [CrossRef]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef]
- German, J.B.; Hammock, B.D.; Watkins, S.M. Metabolomics: Building on a century of biochemistry to guide human health. Metabolomics 2005, 1, 3–9. [Google Scholar] [CrossRef]
- Wang, C.; Kong, H.; Guan, Y.; Yang, J.; Gu, J.; Yang, S.; Xu, G. Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis. Anal. Chem. 2005, 77, 4108–4116. [Google Scholar] [CrossRef]
- Rozen, S.; Cudkowicz, M.E.; Bogdanov, M.; Matson, W.R.; Kristal, B.S.; Beecher, C.; Harrison, S.; Vouros, P.; Flarakos, J.; Vigneau-Callahan, K.; et al. Metabolomic analysis and signatures in motor neuron disease. Metabolomics 2005, 1, 101–108. [Google Scholar] [CrossRef][Green Version]
- Scherzer, C.R.; Eklund, A.C.; Morse, L.J.; Liao, Z.; Locascio, J.J.; Fefer, D.; Schwarzschild, M.A.; Schlossmacher, M.G.; Hauser, M.A.; Vance, J.M.; et al. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. USA 2007, 104, 955–960. [Google Scholar] [CrossRef]
- Barba, I.; Fernandez-Montesinos, R.; Garcia-Dorado, D.; Pozo, D. Alzheimer’s disease beyond the genomic era: Nuclear magnetic resonance (NMR) spectroscopy-based metabolomics. J. Cell Mol. Med. 2008, 12, 1477–1485. [Google Scholar] [CrossRef]
- Boudonck, K.J.; Mitchell, M.W.; Nemet, L.; Keresztes, L.; Nyska, A.; Shinar, D.; Rosenstock, M. Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol. Pathol. 2009, 37, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Boudonck, K.J.; Rose, D.J.; Karoly, E.D.; Lee, D.P.; Lawton, K.A.; Lapinskas, P.J. Metabolomics for early detection of drug-induced kidney injury: Review of the current status. Bioanalysis 2009, 1, 1645–1663. [Google Scholar] [CrossRef]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the adult human plasma metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Ritz, B.; Yan, Q.; Uppal, K.; Liew, Z.; Cui, X.; Ling, C.; Inoue, K.; von Ehrenstein, O.; Walker, D.I.; Jones, D.P. Untargeted Metabolomics Screen of Mid-pregnancy Maternal Serum and Autism in Offspring. Autism Res. 2020, 13, 1258–1269. [Google Scholar] [CrossRef]
- Frye, R.E.; Slattery, J.C.; Quadros, E.V. Folate metabolism abnormalities in autism: Potential biomarkers. Biomark. Med. 2017, 11, 687–699. [Google Scholar] [CrossRef]
- Jones, K.L.; Croen, L.A.; Yoshida, C.K.; Heuer, L.; Hansen, R.; Zerbo, O.; DeLorenze, G.N.; Kharrazi, M.; Yolken, R.; Ashwood, P.; et al. Autism with intellectual disability is associated with increased levels of maternal cytokines and chemokines during gestation. Mol. Psychiatry 2017, 22, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Curtin, P.; Kellogg, J.; Cech, N.; Gennings, C. A random subset implementation of weighted quantile sum (WQSRS) regression for analysis of high-dimensional mixtures. Commun. Stat. Simul. Comput. 2019, 50, 1119–1134. [Google Scholar] [CrossRef]
- Carrico, C.; Gennings, C.; Wheeler, D.C.; Factor-Litvak, P. Characterization of Weighted Quantile Sum Regression for Highly Correlated Data in a Risk Analysis Setting. J Agric Biol Environ. Stat 2015, 20, 100–120. [Google Scholar] [CrossRef]
- Akerele, O.A.; Cheema, S.K. Maternal diet high in Omega-3 fatty acids upregulate genes involved in neurotrophin signalling in fetal brain during pregnancy in C57BL/6 mice. Neurochem. Int. 2020, 138, 104778. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, N.; Patil, V.; Joshi, S. Maternal long chain polyunsaturated fatty acid status and pregnancy complications. Prostaglandins Leukot Essent Fat. Acids 2018, 136, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Aldinger, K.A.; Lane, C.J.; Veenstra-VanderWeele, J.; Levitt, P. Patterns of Risk for Multiple Co-Occurring Medical Conditions Replicate Across Distinct Cohorts of Children with Autism Spectrum Disorder. Autism Res. 2015, 8, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.; Le Couteur, A.; Gottesman, I.; Bolton, P.; Simonoff, E.; Yuzda, E.; Rutter, M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol. Med. 1995, 25, 63–77. [Google Scholar] [CrossRef]
- Rosenberg, R.E.; Law, J.K.; Yenokyan, G.; McGready, J.; Kaufmann, W.E.; Law, P.A. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. Pediatrics Adolesc. Med. 2009, 163, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Hallmayer, J.; Cleveland, S.; Torres, A.; Phillips, J.; Cohen, B.; Torigoe, T.; Miller, J.; Fedele, A.; Collins, J.; Smith, K.; et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 2011, 68, 1095–1102. [Google Scholar] [CrossRef]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The familial risk of autism. JAMA 2014, 311, 1770–1777. [Google Scholar] [CrossRef] [PubMed]
- Colvert, E.; Tick, B.; McEwen, F.; Stewart, C.; Curran, S.R.; Woodhouse, E.; Gillan, N.; Hallett, V.; Lietz, S.; Garnett, T.; et al. Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample. JAMA Psychiatry 2015, 72, 415–423. [Google Scholar] [CrossRef]
- Eichler, E.E.; Flint, J.; Gibson, G.; Kong, A.; Leal, S.M.; Moore, J.H.; Nadeau, J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010, 11, 446–450. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 2012, 30, 1095–1106. [Google Scholar] [CrossRef]
- Nebert, D.W.; Zhang, G.; Vesell, E.S. From human genetics and genomics to pharmacogenetics and pharmacogenomics: Past lessons, future directions. Drug Metab. Rev. 2008, 40, 187–224. [Google Scholar] [CrossRef]
- Park, J.H.; Wacholder, S.; Gail, M.H.; Peters, U.; Jacobs, K.B.; Chanock, S.J.; Chatterjee, N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010, 42, 570–575. [Google Scholar] [CrossRef]
- Chaste, P.; Leboyer, M. Autism risk factors: Genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar]
- Meek, S.E.; Lemery-Chalfant, K.; Jahromi, L.B.; Valiente, C. A review of gene-environment correlations and their implications for autism: A conceptual model. Psychol. Rev. 2013, 120, 497–521. [Google Scholar] [CrossRef]
- Tordjman, S.; Somogyi, E.; Coulon, N.; Kermarrec, S.; Cohen, D.; Bronsard, G.; Bonnot, O.; Weismann-Arcache, C.; Botbol, M.; Lauth, B.; et al. Gene x Environment interactions in autism spectrum disorders: Role of epigenetic mechanisms. Front. Psychiatry 2014, 5, 53. [Google Scholar] [CrossRef]
- Zhu, Y.; Mordaunt, C.E.; Durbin-Johnson, B.P.; Caudill, M.A.; Malysheva, O.V.; Miller, J.W.; Green, R.; James, S.J.; Melnyk, S.B.; Fallin, M.D.; et al. Expression Changes in Epigenetic Gene Pathways Associated with One-Carbon Nutritional Metabolites in Maternal Blood From Pregnancies Resulting in Autism and Non-Typical Neurodevelopment. Autism Res. 2021, 14, 11–28. [Google Scholar] [CrossRef]
- Lombardo, M.V.; Pramparo, T.; Gazestani, V.; Warrier, V.; Bethlehem, R.A.I.; Carter Barnes, C.; Lopez, L.; Lewis, N.E.; Eyler, L.; Pierce, K.; et al. Large-scale associations between the leukocyte transcriptome and BOLD responses to speech differ in autism early language outcome subtypes. Nat. Neurosci. 2018, 21, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.; Shin, J.H.; Rajpurohit, A.; Deep-Soboslay, A.; Collado-Torres, L.; Brandon, N.J.; Hyde, T.M.; Kleinman, J.E.; Jaffe, A.E.; Cross, A.J.; et al. Altered expression of histamine signaling genes in autism spectrum disorder. Transl. Psychiatry 2017, 7, e1126. [Google Scholar] [CrossRef] [PubMed]
- Schmunk, G.; Nguyen, R.L.; Ferguson, D.L.; Kumar, K.; Parker, I.; Gargus, J.J. High-throughput screen detects calcium signaling dysfunction in typical sporadic autism spectrum disorder. Sci. Rep. 2017, 7, 40740. [Google Scholar] [CrossRef] [PubMed]
- Pramparo, T.; Pierce, K.; Lombardo, M.V.; Carter Barnes, C.; Marinero, S.; Ahrens-Barbeau, C.; Murray, S.S.; Lopez, L.; Xu, R.; Courchesne, E. Prediction of autism by translation and immune/inflammation coexpressed genes in toddlers from pediatric community practices. JAMA Psychiatry 2015, 72, 386–394. [Google Scholar] [CrossRef]
- Kusenda, M.; Vacic, V.; Malhotra, D.; Rodgers, L.; Pavon, K.; Meth, J.; Kumar, R.A.; Christian, S.L.; Peeters, H.; Cho, S.S.; et al. The Influence of Microdeletions and Microduplications of 16p11.2 on Global Transcription Profiles. J. Child Neurol. 2015, 30, 1947–1953. [Google Scholar] [CrossRef]
- Kong, S.W.; Shimizu-Motohashi, Y.; Campbell, M.G.; Lee, I.H.; Collins, C.D.; Brewster, S.J.; Holm, I.A.; Rappaport, L.; Kohane, I.S.; Kunkel, L.M. Peripheral blood gene expression signature differentiates children with autism from unaffected siblings. Neurogenetics 2013, 14, 143–152. [Google Scholar] [CrossRef]
- Campbell, M.G.; Kohane, I.S.; Kong, S.W. Pathway-based outlier method reveals heterogeneous genomic structure of autism in blood transcriptome. BMC Med. Genom. 2013, 6, 34. [Google Scholar] [CrossRef]
- Kong, S.W.; Collins, C.D.; Shimizu-Motohashi, Y.; Holm, I.A.; Campbell, M.G.; Lee, I.H.; Brewster, S.J.; Hanson, E.; Harris, H.K.; Lowe, K.R.; et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PLoS ONE 2012, 7, e49475. [Google Scholar] [CrossRef]
- Glatt, S.J.; Tsuang, M.T.; Winn, M.; Chandler, S.D.; Collins, M.; Lopez, L.; Weinfeld, M.; Carter, C.; Schork, N.; Pierce, K.; et al. Blood-based gene expression signatures of infants and toddlers with autism. J. Am. Acad. Child Adolesc. Psychiatry 2012, 51, 934–944.e932. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Guell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed]
- Pascal, L.E.; True, L.D.; Campbell, D.S.; Deutsch, E.W.; Risk, M.; Coleman, I.M.; Eichner, L.J.; Nelson, P.S.; Liu, A.Y. Correlation of mRNA and protein levels: Cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genom. 2008, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Laye, M.J.; Tran, V.; Jones, D.P.; Kapahi, P.; Promislow, D.E. The effects of age and dietary restriction on the tissue-specific metabolome of Drosophila. Aging Cell 2015, 14, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Zolkipli-Cunningham, Z.; Naviaux, J.C.; Nakayama, T.; Hirsch, C.M.; Monk, J.M.; Li, K.; Wang, L.; Le, T.P.; Meinardi, S.; Blake, D.R.; et al. Metabolic and behavioral features of acute hyperpurinergia and the maternal immune activation mouse model of autism spectrum disorder. PLoS ONE 2021, 16, e0248771. [Google Scholar] [CrossRef]
- Xu, X.J.; Cai, X.E.; Meng, F.C.; Song, T.J.; Wang, X.X.; Wei, Y.Z.; Zhai, F.J.; Long, B.; Wang, J.; You, X.; et al. Comparison of the Metabolic Profiles in the Plasma and Urine Samples Between Autistic and Typically Developing Boys: A Preliminary Study. Front. Psychiatry 2021, 12, 657105. [Google Scholar] [CrossRef]
- Xiao, L.; Yan, J.; Yang, T.; Zhu, J.; Li, T.; Wei, H.; Chen, J. Fecal Microbiome Transplantation from Children with Autism Spectrum Disorder Modulates Tryptophan and Serotonergic Synapse Metabolism and Induces Altered Behaviors in Germ-Free Mice. mSystems 2021, 6, e01343-20. [Google Scholar] [CrossRef]
- Needham, B.D.; Adame, M.D.; Serena, G.; Rose, D.R.; Preston, G.M.; Conrad, M.C.; Campbell, A.S.; Donabedian, D.H.; Fasano, A.; Ashwood, P.; et al. Plasma and Fecal Metabolite Profiles in Autism Spectrum Disorder. Biol. Psychiatry 2021, 89, 451–462. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, H.; Li, C.; Zou, X.; Luo, X.; Wu, L.; Li, T.; Chen, X.; Mao, M.; Huang, Y.; et al. Differential Metabolites in Chinese Autistic Children: A Multi-Center Study Based on Urinary (1)H-NMR Metabolomics Analysis. Front. Psychiatry 2021, 12, 624767. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; He, Z.; Zubkov, D.; Huang, S.; Kurochkin, I.; Yang, X.; Halene, T.; Willmitzer, L.; Giavalisco, P.; Akbarian, S.; et al. Lipidome alterations in human prefrontal cortex during development, aging, and cognitive disorders. Mol. Psychiatry 2020, 25, 2952–2969. [Google Scholar] [CrossRef] [PubMed]
- Yadama, A.P.; Kelly, R.S.; Lee-Sarwar, K.; Mirzakhani, H.; Chu, S.H.; Kachroo, P.; Litonjua, A.A.; Lasky-Su, J.; Weiss, S.T. Allergic disease and low ASQ communication score in children. Brain Behav. Immun. 2020, 83, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Ventura, G.; Calvano, C.D.; Porcelli, V.; Palmieri, L.; De Giacomo, A.; Xu, Y.; Goodacre, R.; Palmisano, F.; Cataldi, T.R.I. Phospholipidomics of peripheral blood mononuclear cells (PBMCs): The tricky case of children with autism spectrum disorder (ASD) and their healthy siblings. Anal. Bioanal. Chem. 2020, 412, 6859–6874. [Google Scholar] [CrossRef] [PubMed]
- Usui, N.; Iwata, K.; Miyachi, T.; Takagai, S.; Wakusawa, K.; Nara, T.; Tsuchiya, K.J.; Matsumoto, K.; Kurita, D.; Kameno, Y.; et al. VLDL-specific increases of fatty acids in autism spectrum disorder correlate with social interaction. EBioMedicine 2020, 58, 102917. [Google Scholar] [CrossRef]
- Troisi, J.; Autio, R.; Beopoulos, T.; Bravaccio, C.; Carraturo, F.; Corrivetti, G.; Cunningham, S.; Devane, S.; Fallin, D.; Fetissov, S.; et al. Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach. Brain Sci. 2020, 10, 743. [Google Scholar] [CrossRef]
- Sotelo-Orozco, J.; Abbeduto, L.; Hertz-Picciotto, I.; Slupsky, C.M. Association Between Plasma Metabolites and Psychometric Scores Among Children With Developmental Disabilities: Investigating Sex-Differences. Front. Psychiatry 2020, 11, 579538. [Google Scholar] [CrossRef]
- Smith, A.M.; Natowicz, M.R.; Braas, D.; Ludwig, M.A.; Ney, D.M.; Donley, E.L.R.; Burrier, R.E.; Amaral, D.G. A Metabolomics Approach to Screening for Autism Risk in the Children’s Autism Metabolome Project. Autism Res. 2020, 13, 1270–1285. [Google Scholar] [CrossRef]
- Panjwani, A.A.; Ji, Y.; Fahey, J.W.; Palmer, A.; Wang, G.; Hong, X.; Zuckerman, B.; Wang, X. Maternal Dyslipidemia, Plasma Branched-Chain Amino Acids, and the Risk of Child Autism Spectrum Disorder: Evidence of Sex Difference. J. Autism Dev. Disord. 2020, 50, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xiao, Z.; Ke, X.; Yao, P.; Chen, Y.; Lin, L.; Lu, J. Urinary Metabonomic Profiling Discriminates Between Children with Autism and Their Healthy Siblings. Med. Sci. Monit. 2020, 26, e926634. [Google Scholar] [CrossRef]
- Liang, Y.; Ke, X.; Xiao, Z.; Zhang, Y.; Chen, Y.; Li, Y.; Wang, Z.; Lin, L.; Yao, P.; Lu, J. Untargeted Metabolomic Profiling Using UHPLC-QTOF/MS Reveals Metabolic Alterations Associated with Autism. Biomed. Res. Int. 2020, 2020, 6105608. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Zhu, J.; Yang, T.; Guo, M.; Li, Q.; Chen, J.; Li, T. The Gut Microbiota and Associated Metabolites Are Altered in Sleep Disorder of Children With Autism Spectrum Disorders. Front. Psychiatry 2020, 11, 855. [Google Scholar] [CrossRef]
- Hollowood-Jones, K.; Adams, J.B.; Coleman, D.M.; Ramamoorthy, S.; Melnyk, S.; James, S.J.; Woodruff, B.K.; Pollard, E.L.; Snozek, C.L.; Kruger, U.; et al. Altered metabolism of mothers of young children with Autism Spectrum Disorder: A case control study. BMC Pediatrics 2020, 20, 557. [Google Scholar] [CrossRef]
- Gevi, F.; Belardo, A.; Zolla, L. A metabolomics approach to investigate urine levels of neurotransmitters and related metabolites in autistic children. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165859. [Google Scholar] [CrossRef] [PubMed]
- Yehia, L.; Ni, Y.; Feng, F.; Seyfi, M.; Sadler, T.; Frazier, T.W.; Eng, C. Distinct Alterations in Tricarboxylic Acid Cycle Metabolites Associate with Cancer and Autism Phenotypes in Cowden Syndrome and Bannayan-Riley-Ruvalcaba Syndrome. Am. J. Hum. Genet. 2019, 105, 813–821. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, D.; He, W.; Sheng, X.; Zhou, W.; Xie, D.; Liang, H.; Zeng, T.; Li, T.; Wang, Y. Identification of gender-related metabolic disturbances in autism spectrum disorders using urinary metabolomics. Int. J. Biochem. Cell Biol. 2019, 115, 105594. [Google Scholar] [CrossRef]
- Smith, A.M.; King, J.J.; West, P.R.; Ludwig, M.A.; Donley, E.L.R.; Burrier, R.E.; Amaral, D.G. Amino Acid Dysregulation Metabotypes: Potential Biomarkers for Diagnosis and Individualized Treatment for Subtypes of Autism Spectrum Disorder. Biol. Psychiatry 2019, 85, 345–354. [Google Scholar] [CrossRef]
- Rangel-Huerta, O.D.; Gomez-Fernandez, A.; de la Torre-Aguilar, M.J.; Gil, A.; Perez-Navero, J.L.; Flores-Rojas, K.; Martin-Borreguero, P.; Gil-Campos, M. Metabolic profiling in children with autism spectrum disorder with and without mental regression: Preliminary results from a cross-sectional case-control study. Metabolomics 2019, 15, 99. [Google Scholar] [CrossRef]
- Panjwani, A.A.; Ji, Y.; Fahey, J.W.; Palmer, A.; Wang, G.; Hong, X.; Zuckerman, B.; Wang, X. Maternal Obesity/Diabetes, Plasma Branched-Chain Amino Acids, and Autism Spectrum Disorder Risk in Urban Low-Income Children: Evidence of Sex Difference. Autism Res. 2019, 12, 1562–1573. [Google Scholar] [CrossRef]
- Orozco, J.S.; Hertz-Picciotto, I.; Abbeduto, L.; Slupsky, C.M. Metabolomics analysis of children with autism, idiopathic-developmental delays, and Down syndrome. Transl. Psychiatry 2019, 9, 243. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.S.; Boulin, A.; Laranjo, N.; Lee-Sarwar, K.; Chu, S.H.; Yadama, A.P.; Carey, V.; Litonjua, A.A.; Lasky-Su, J.; Weiss, S.T. Metabolomics and Communication Skills Development in Children; Evidence from the Ages and Stages Questionnaire. Metabolites 2019, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Qiao, Y.; Xu, X.J.; You, X.; Tao, Y. Urine Organic Acids as Potential Biomarkers for Autism-Spectrum Disorder in Chinese Children. Front. Cell. Neurosci. 2019, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Belardo, A.; Gevi, F.; Zolla, L. The concomitant lower concentrations of vitamins B6, B9 and B12 may cause methylation deficiency in autistic children. J. Nutr. Biochem. 2019, 70, 38–46. [Google Scholar] [CrossRef]
- Kang, D.W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howsmon, D.P.; Shaffer, M.; Lozupone, C.A.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe 2018, 49, 121–131. [Google Scholar] [CrossRef]
- Bitar, T.; Mavel, S.; Emond, P.; Nadal-Desbarats, L.; Lefevre, A.; Mattar, H.; Soufia, M.; Blasco, H.; Vourc’h, P.; Hleihel, W.; et al. Identification of metabolic pathway disturbances using multimodal metabolomics in autistic disorders in a Middle Eastern population. J. Pharm. Biomed. Anal. 2018, 152, 57–65. [Google Scholar] [CrossRef]
- Bent, S.; Lawton, B.; Warren, T.; Widjaja, F.; Dang, K.; Fahey, J.W.; Cornblatt, B.; Kinchen, J.M.; Delucchi, K.; Hendren, R.L. Identification of urinary metabolites that correlate with clinical improvements in children with autism treated with sulforaphane from broccoli. Mol. Autism 2018, 9, 35. [Google Scholar] [CrossRef]
- Lussu, M.; Noto, A.; Masili, A.; Rinaldi, A.C.; Dessi, A.; De Angelis, M.; De Giacomo, A.; Fanos, V.; Atzori, L.; Francavilla, R. The urinary (1) H-NMR metabolomics profile of an italian autistic children population and their unaffected siblings. Autism Res. 2017, 10, 1058–1066. [Google Scholar] [CrossRef]
- Gevi, F.; Zolla, L.; Gabriele, S.; Persico, A.M. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Mol. Autism 2016, 7, 47. [Google Scholar] [CrossRef]
- Dieme, B.; Mavel, S.; Blasco, H.; Tripi, G.; Bonnet-Brilhault, F.; Malvy, J.; Bocca, C.; Andres, C.R.; Nadal-Desbarats, L.; Emond, P. Metabolomics Study of Urine in Autism Spectrum Disorders Using a Multiplatform Analytical Methodology. J. Proteome Res. 2015, 14, 5273–5282. [Google Scholar] [CrossRef]
- West, P.R.; Amaral, D.G.; Bais, P.; Smith, A.M.; Egnash, L.A.; Ross, M.E.; Palmer, J.A.; Fontaine, B.R.; Conard, K.R.; Corbett, B.A.; et al. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS ONE 2014, 9, e112445. [Google Scholar] [CrossRef] [PubMed]
- Noto, A.; Fanos, V.; Barberini, L.; Grapov, D.; Fattuoni, C.; Zaffanello, M.; Casanova, A.; Fenu, G.; De Giacomo, A.; De Angelis, M.; et al. The urinary metabolomics profile of an Italian autistic children population and their unaffected siblings. J Matern Fetal Neonatal Med. 2014, 27 (Suppl 2), 46–52. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Desbarats, L.; Aidoud, N.; Emond, P.; Blasco, H.; Filipiak, I.; Sarda, P.; Bonnet-Brilhault, F.; Mavel, S.; Andres, C.R. Combined 1H-NMR and 1H-13C HSQC-NMR to improve urinary screening in autism spectrum disorders. Analyst 2014, 139, 3460–3468. [Google Scholar] [CrossRef]
- Mavel, S.; Nadal-Desbarats, L.; Blasco, H.; Bonnet-Brilhault, F.; Barthelemy, C.; Montigny, F.; Sarda, P.; Laumonnier, F.; Vourc’h, P.; Andres, C.R.; et al. 1H-13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta 2013, 114, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Emond, P.; Mavel, S.; Aidoud, N.; Nadal-Desbarats, L.; Montigny, F.; Bonnet-Brilhault, F.; Barthelemy, C.; Merten, M.; Sarda, P.; Laumonnier, F.; et al. GC-MS-based urine metabolic profiling of autism spectrum disorders. Anal. Bioanal. Chem. 2013, 405, 5291–5300. [Google Scholar] [CrossRef]
- Ming, X.; Stein, T.P.; Barnes, V.; Rhodes, N.; Guo, L. Metabolic perturbance in autism spectrum disorders: A metabolomics study. J. Proteome Res. 2012, 11, 5856–5862. [Google Scholar] [CrossRef]
- Nolin, S.L.; Napoli, E.; Flores, A.; Hagerman, R.J.; Giulivi, C. Deficits in Prenatal Serine Biosynthesis Underlie the Mitochondrial Dysfunction Associated with the Autism-Linked FMR1 Gene. Int. J. Mol. Sci. 2021, 22, 5886. [Google Scholar] [CrossRef]
- Bala, K.A.; Dogan, M.; Mutluer, T.; Kaba, S.; Aslan, O.; Balahoroglu, R.; Cokluk, E.; Ustyol, L.; Kocaman, S. Plasma amino acid profile in autism spectrum disorder (ASD). Eur. Rev. Med. Pharm. Sci. 2016, 20, 923–929. [Google Scholar]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef]
- Hamlin, J.; Crook, T.; James, J.; Gonzales, D.; Hakkak, R. Correlation between dietary intake and plasma levels of choline and betaine in children with autism. In Proceedings of the Experimental Biology Meeting, San Diego, CA, USA, 26–30 April 2014; p. 827.821. [Google Scholar]
- Yu, X.; Qian-Qian, L.; Cong, Y.; Xiao-Bing, Z.; Hong-Zhu, D. Reduction of essential amino acid levels and sex-specific alterations in serum amino acid concentration profiles in children with autism spectrum disorder. Psychiatry Res. 2021, 297, 113675. [Google Scholar] [CrossRef]
- Vargason, T.; Kruger, U.; McGuinness, D.L.; Adams, J.B.; Geis, E.; Gehn, E.; Coleman, D.; Hahn, J. Investigating Plasma Amino Acids for Differentiating Individuals with Autism Spectrum Disorder and Typically Developing Peers. Res. Autism Spectr. Disord. 2018, 50, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.F.; Wang, W.Q.; Li, X.M.; Rauw, G.; Baker, G.B. Body fluid levels of neuroactive amino acids in autism spectrum disorders: A review of the literature. Amino Acids 2017, 49, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, S.; Wang, M.; Gao, J.; Sun, C.; Wang, J.; Xia, W.; Wu, S.; Sumner, S.J.; Zhang, F.; et al. Potential serum biomarkers from a metabolomics study of autism. J. Psychiatry Neurosci. 2016, 41, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, M.; Emond, V.; Chen, C.T.; Julien, C.; Bourasset, F.; Oddo, S.; LaFerla, F.; Bazinet, R.P.; Calon, F. Diffusion of docosahexaenoic and eicosapentaenoic acids through the blood-brain barrier: An in situ cerebral perfusion study. Neurochem. Int. 2009, 55, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.D.; Duran, M.; Vlieger, E.J.; Majoie, C.B.; den Heeten, G.J.; Linszen, D.H.; de Haan, L. Polyunsaturated fatty acids and brain white matter anisotropy in recent-onset schizophrenia: A preliminary study. Prostaglandins Leukot Essent Fat. Acids 2009, 81, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Conklin, S.M.; Runyan, C.A.; Leonard, S.; Reddy, R.D.; Muldoon, M.F.; Yao, J.K. Age-related changes of n-3 and n-6 polyunsaturated fatty acids in the anterior cingulate cortex of individuals with major depressive disorder. Prostaglandins Leukot Essent Fat. Acids 2010, 82, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Fabelo, N.; Santpere, G.; Puig, B.; Marin, R.; Ferrer, I.; Diaz, M. Lipid alterations in lipid rafts from Alzheimer’s disease human brain cortex. J. Alzheimers Dis. 2010, 19, 489–502. [Google Scholar] [CrossRef]
- Fabelo, N.; Martin, V.; Santpere, G.; Marin, R.; Torrent, L.; Ferrer, I.; Diaz, M. Severe alterations in lipid composition of frontal cortex lipid rafts from Parkinson’s disease and incidental Parkinson’s disease. Mol. Med. 2011, 17, 1107–1118. [Google Scholar] [CrossRef]
- Parletta, N.; Niyonsenga, T.; Duff, J. Omega-3 and Omega-6 Polyunsaturated Fatty Acid Levels and Correlations with Symptoms in Children with Attention Deficit Hyperactivity Disorder, Autistic Spectrum Disorder and Typically Developing Controls. PLoS ONE 2016, 11, e0156432. [Google Scholar] [CrossRef]
- Robson, L.G.; Dyall, S.; Sidloff, D.; Michael-Titus, A.T. Omega-3 polyunsaturated fatty acids increase the neurite outgrowth of rat sensory neurones throughout development and in aged animals. Neurobiol. Aging 2010, 31, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Labrousse, V.F.; Nadjar, A.; Joffre, C.; Costes, L.; Aubert, A.; Gregoire, S.; Bretillon, L.; Laye, S. Short-term long chain omega3 diet protects from neuroinflammatory processes and memory impairment in aged mice. PLoS ONE 2012, 7, e36861. [Google Scholar] [CrossRef]
- Harbeby, E.; Jouin, M.; Alessandri, J.M.; Lallemand, M.S.; Linard, A.; Lavialle, M.; Huertas, A.; Cunnane, S.C.; Guesnet, P. n-3 PUFA status affects expression of genes involved in neuroenergetics differently in the fronto-parietal cortex compared to the CA1 area of the hippocampus: Effect of rest and neuronal activation in the rat. Prostaglandins Leukot Essent Fat. Acids 2012, 86, 211–220. [Google Scholar] [CrossRef]
- Blondeau, N.; Nguemeni, C.; Debruyne, D.N.; Piens, M.; Wu, X.; Pan, H.; Hu, X.; Gandin, C.; Lipsky, R.H.; Plumier, J.C.; et al. Subchronic alpha-linolenic acid treatment enhances brain plasticity and exerts an antidepressant effect: A versatile potential therapy for stroke. Neuropsychopharmacology 2009, 34, 2548–2559. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.P.; Zhang, H.Q.; Wei, W.; Guo, Y.F.; Na, X.; Cao, X.H.; Liu, L.J. Some subtypes of endocannabinoid/endovanilloid receptors mediate docosahexaenoic acid-induced enhanced spatial memory in rats. Brain Res. 2011, 1412, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.C.; Michael, G.J.; Michael-Titus, A.T. Omega-3 fatty acids reverse age-related decreases in nuclear receptors and increase neurogenesis in old rats. J. Neurosci. Res. 2010, 88, 2091–2102. [Google Scholar] [CrossRef] [PubMed]
- Martins, B.P.; Bandarra, N.M.; Figueiredo-Braga, M. The role of marine omega-3 in human neurodevelopment, including Autism Spectrum Disorders and Attention-Deficit/Hyperactivity Disorder—A review. Crit. Rev. Food. Sci. Nutr. 2020, 60, 1431–1446. [Google Scholar] [CrossRef]
- Lebbadi, M.; Julien, C.; Phivilay, A.; Tremblay, C.; Emond, V.; Kang, J.X.; Calon, F. Endogenous conversion of omega-6 into omega-3 fatty acids improves neuropathology in an animal model of Alzheimer’s disease. J. Alzheimers Dis. 2011, 27, 853–869. [Google Scholar] [CrossRef]
- Refolo, V.; Stefanova, N. Neuroinflammation and Glial Phenotypic Changes in Alpha-Synucleinopathies. Front. Cell. Neurosci. 2019, 13, 263. [Google Scholar] [CrossRef]
- Yuan, J.; Amin, P.; Ofengeim, D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 2019, 20, 19–33. [Google Scholar] [CrossRef]
- Das, U.N. A defect in the activity of Delta6 and Delta5 desaturases may be a factor predisposing to the development of insulin resistance syndrome. Prostaglandins Leukot Essent Fat. Acids 2005, 72, 343–350. [Google Scholar] [CrossRef]
- Bazan, N.G.; Colangelo, V.; Lukiw, W.J. Prostaglandins and other lipid mediators in Alzheimer’s disease. Prostaglandins Other Lipid Mediat 2002, 68-69, 197–210. [Google Scholar] [CrossRef]
- Trushina, E.; Dutta, T.; Persson, X.M.; Mielke, M.M.; Petersen, R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 2013, 8, e63644. [Google Scholar] [CrossRef]
- Combrinck, M.; Williams, J.; De Berardinis, M.A.; Warden, D.; Puopolo, M.; Smith, A.D.; Minghetti, L. Levels of CSF prostaglandin E2, cognitive decline, and survival in Alzheimer’s disease. J. Neurol. Neurosurg Psychiatry 2006, 77, 85–88. [Google Scholar] [CrossRef]
- Panisi, C.; Guerini, F.R.; Abruzzo, P.M.; Balzola, F.; Biava, P.M.; Bolotta, A.; Brunero, M.; Burgio, E.; Chiara, A.; Clerici, M.; et al. Autism Spectrum Disorder from the Womb to Adulthood: Suggestions for a Paradigm Shift. J. Pers. Med. 2021, 11, 70. [Google Scholar] [CrossRef]
- Sotgiu, S.; Manca, S.; Gagliano, A.; Minutolo, A.; Melis, M.C.; Pisuttu, G.; Scoppola, C.; Bolognesi, E.; Clerici, M.; Guerini, F.R.; et al. Immune regulation of neurodevelopment at the mother-foetus interface: The case of autism. Clin. Transl. Immunol. 2020, 9, e1211. [Google Scholar] [CrossRef]
- Theoharides, T.C.; Zhang, B. Neuro-inflammation, blood-brain barrier, seizures and autism. J. Neuroinflammation 2011, 8, 168. [Google Scholar] [CrossRef]
- Cohen-Cory, S.; Fraser, S.E. Effects of brain-derived neurotrophic factor on optic axon branching and remodelling in vivo. Nature 1995, 378, 192–196. [Google Scholar] [CrossRef]
- Suzuki, S.; Hongli, Q.; Okada, A.; Kasama, T.; Ohta, K.; Warita, K.; Tanaka, K.; Miki, T.; Takeuchi, Y. BDNF-dependent accumulation of palmitoleic acid in CNS neurons. Cell. Mol. Neurobiol. 2012, 32, 1367–1373. [Google Scholar] [CrossRef]
- Breuer, S.; Pech, K.; Buss, A.; Spitzer, C.; Ozols, J.; Hol, E.M.; Heussen, N.; Noth, J.; Schwaiger, F.W.; Schmitt, A.B. Regulation of stearoyl-CoA desaturase-1 after central and peripheral nerve lesions. BMC Neurosci. 2004, 5, 15. [Google Scholar] [CrossRef][Green Version]
- LaBarre, J.L.; Miller, A.L.; Bauer, K.W.; Burant, C.F.; Lumeng, J.C. Early life stress exposure associated with reduced polyunsaturated-containing lipids in low-income children. Pediatr. Res. 2021, 89, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Kessler, K.; Gerl, M.J.; Hornemann, S.; Damm, M.; Klose, C.; Petzke, K.J.; Kemper, M.; Weber, D.; Rudovich, N.; Grune, T.; et al. Shotgun Lipidomics Discovered Diurnal Regulation of Lipid Metabolism Linked to Insulin Sensitivity in Nondiabetic Men. J. Clin. Endocrinol. Metab. 2020, 105, 1501–1514. [Google Scholar] [CrossRef]
- Skene, D.J.; Skornyakov, E.; Chowdhury, N.R.; Gajula, R.P.; Middleton, B.; Satterfield, B.C.; Porter, K.I.; Van Dongen, H.P.A.; Gaddameedhi, S. Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc. Natl. Acad. Sci. USA 2018, 115, 7825–7830. [Google Scholar] [CrossRef]
- Chua, E.C.; Shui, G.; Lee, I.T.; Lau, P.; Tan, L.C.; Yeo, S.C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473. [Google Scholar] [CrossRef]
- Honma, A.; Revell, V.L.; Gunn, P.J.; Davies, S.K.; Middleton, B.; Raynaud, F.I.; Skene, D.J. Effect of acute total sleep deprivation on plasma melatonin, cortisol and metabolite rhythms in females. Eur. J. Neurosci. 2020, 51, 366–378. [Google Scholar] [CrossRef]
- Ang, J.E.; Revell, V.; Mann, A.; Mantele, S.; Otway, D.T.; Johnston, J.D.; Thumser, A.E.; Skene, D.J.; Raynaud, F. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol. Int. 2012, 29, 868–881. [Google Scholar] [CrossRef] [PubMed]
- Hertz-Picciotto, I.; Schmidt, R.J.; Walker, C.K.; Bennett, D.H.; Oliver, M.; Shedd-Wise, K.M.; LaSalle, J.M.; Giulivi, C.; Puschner, B.; Thomas, J.; et al. A Prospective Study of Environmental Exposures and Early Biomarkers in Autism Spectrum Disorder: Design, Protocols, and Preliminary Data from the MARBLES Study. Environ. Health Perspect. 2018, 126, 117004. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Risi, S.; Lambrecht, L.; Cook, E.H., Jr.; Leventhal, B.L.; DiLavore, P.C.; Pickles, A.; Rutter, M. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000, 30, 205–223. [Google Scholar] [CrossRef]
- Lord, C.; Rutter, M.; DiLavore, P.C.; Risi, S. The Autism Diagnostic Observation Schedule (ADOS); Western Psychological Services: Los Angeles, CA, USA, 2000. [Google Scholar]
- Mullen, E.M. Scales of Early Learning; American Guidance Services Inc.: Circle Pines, MN, USA, 1995. [Google Scholar]
- Chawarska, K.; Shic, F.; Macari, S.; Campbell, D.J.; Brian, J.; Landa, R.; Hutman, T.; Nelson, C.A.; Ozonoff, S.; Tager-Flusberg, H.; et al. 18-month predictors of later outcomes in younger siblings of children with autism spectrum disorder: A baby siblings research consortium study. J. Am. Acad. Child Adolesc. Psychiatry 2014, 53, 1317–1327.e1311. [Google Scholar] [CrossRef]
- Ozonoff, S.; Young, G.S.; Belding, A.; Hill, M.; Hill, A.; Hutman, T.; Johnson, S.; Miller, M.; Rogers, S.J.; Schwichtenberg, A.J.; et al. The broader autism phenotype in infancy: When does it emerge? J. Am. Acad. Child Adolesc. Psychiatry 2014, 53, 398–407.e392. [Google Scholar] [CrossRef]
- Charman, T.; Young, G.S.; Brian, J.; Carter, A.; Carver, L.J.; Chawarska, K.; Curtin, S.; Dobkins, K.; Elsabbagh, M.; Georgiades, S.; et al. Non-ASD outcomes at 36 months in siblings at familial risk for autism spectrum disorder (ASD): A baby siblings research consortium (BSRC) study. Autism Res. 2017, 10, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Ozonoff, S.; Young, G.S.; Landa, R.J.; Brian, J.; Bryson, S.; Charman, T.; Chawarska, K.; Macari, S.L.; Messinger, D.; Stone, W.L.; et al. Diagnostic stability in young children at risk for autism spectrum disorder: A baby siblings research consortium study. J. Child Psychol. Psychiatry Allied Discip. 2015, 56, 988–998. [Google Scholar] [CrossRef]
- Gotham, K.; Pickles, A.; Lord, C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J. Autism Dev. Disord. 2009, 39, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Moutinho, J.L.; Golan, R.; Yu, T.; Ladva, C.N.; Niedzwiecki, M.; Walker, D.I.; Sarnat, S.E.; Chang, H.H.; Greenwald, R.; et al. Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environ. Int. 2018, 120, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.M.; Walker, D.I.; Liang, Y.; Uppal, K.; Soltow, Q.A.; Tran, V.; Strobel, F.; Quyyumi, A.A.; Ziegler, T.R.; Pennell, K.D.; et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol. Sci. 2015, 148, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Simon-Manso, Y.; Lowenthal, M.S.; Kilpatrick, L.E.; Sampson, M.L.; Telu, K.H.; Rudnick, P.A.; Mallard, W.G.; Bearden, D.W.; Schock, T.B.; Tchekhovskoi, D.V.; et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem. 2013, 85, 11725–11731. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Park, Y.; Johnson, J.M.; Jones, D.P. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009, 25, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
| TD ** (n = 115) | Non-TD (n = 27) | ASD (n = 57) | p-Value from Covariate-Only Model # | |
|---|---|---|---|---|
| Child sex: n (%) | 0.04 * | |||
| Female | 56 (48.7) | 12 (44.4) | 17 (29.8) | |
| Male | 59 (51.3) | 15 (55.6) | 40 (70.2) | |
| Gestational age at delivery (weeks) Mean (SD) | 38.84 (1.39) | 39.29 (1.09) | 39.33 (1.18) | 0.06 * |
| Parity: n (%) | 0.80 | |||
| 1 or less | 53 (46.1) | 12 (44.4) | 22 (38.6) | |
| 2 or more | 62 (53.9) | 15 (55.6) | 35 (61.4) | |
| Child race: n (%) | 0.50 † | |||
| White | 71 (61.7) | 15 (55.6) | 37 (64.9) | |
| Black/African American | 0 (0.0) | 2 (7.4) | 5 (8.8) | |
| Asian | 18 (15.7) | 3 (11.1) | 4 (7.0) | |
| Pacific Islander | 1 (0.9) | 0 (0.0) | 0 (0.0) | |
| Multi-racial | 25 (21.7) | 7 (25.9) | 11 (19.3) | |
| Child ethnicity: n (%) | 0.93 | |||
| Hispanic/Latinx | 38 (33.0) | 10 (37.0) | 21 (36.8) | |
| Not Hispanic/Latinx | 77 (67.0) | 17 (63.0) | 36 (63.2) | |
| Child’s year of birth: mean (SD) | 2011.83 (2.20) | 2012.44 (2.06) | 2012.02 (1.99) | 0.41 |
| Mother’s age at time of birth: mean (SD) | 34.41 (4.91) | 33.44 (4.91) | 34.18 (5.16) | 0.66 |
| Maternal BMI before pregnancy: mean (SD) | 26.12 (5.63) | 29.31 (9.71) | 28.67 (7.83) | 0.01 * |
| Maximum parental education: n (%) | 0.69 | |||
| High school or less | 7 (6.1) | 1 (3.7) | 4 (7.1) | |
| Some college/2 y vocational degree | 29 (25.2) | 12 (44.4) | 20 (35.7) | |
| Bachelor’s degree | 43 (37.4) | 7 (25.9) | 20 (35.7) | |
| MS’s/Doctorate/Prof. degree | 36 (31.3) | 7 (25.9) | 12 (21.4) | |
| Missing | 0 | 0 | 1 | |
| Homeowner status: n (%) | 0.32 | |||
| No | 47 (41.2) | 12 (46.2) | 30 (54.5) | |
| Yes | 67 (58.8) | 14 (53.8) | 25 (45.5) | |
| Missing | 1 | 1 | 2 | |
| Maternal vitamin use at 1 m of pregnancy: n (%) | 0.33 | |||
| No | 53 (46.5) | 12 (46.2) | 34 (60.7) | |
| Yes | 61 (53.5) | 14 (53.8) | 22 (39.3) | |
| Missing | 1 | 1 | 1 |
| Metabolite | Outcome ** | OR (95% CI) | Raw p-Value | FDR |
|---|---|---|---|---|
| Norvaline | ASD | 1.53 (1.07, 2.26) | 0.025 | 0.512 |
| Mevalonic acid | ASD | 1.68 (1.09, 2.76) | 0.027 | 0.512 |
| Docosahexaenoic acid | Non-TD | 0.28 (0.14, 0.50) | <0.001 | 0.003 * |
| Palmitic acid | Non-TD | 0.38 (0.20, 0.65) | 0.001 | 0.020 * |
| Palmitoleic acid | Non-TD | 0.46 (0.27, 0.73) | 0.002 | 0.023 * |
| Gamma-linolenic acid | Non-TD | 0.46 (0.27, 0.74) | 0.003 | 0.023 * |
| Myristic acid | Non-TD | 0.49 (0.29, 0.78) | 0.004 | 0.023 * |
| Stearic acid | Non-TD | 0.42 (0.22, 0.73) | 0.004 | 0.023 * |
| Heptadecanoic acid | Non-TD | 0.47 (0.26,0.77) | 0.005 | 0.023 * |
| Linoleic acid | Non-TD | 0.48 (0.27, 0.78) | 0.005 | 0.023 * |
| Gulonolactone | Non-TD | 2.70 (1.18, 7.57) | 0.038 | 0.143 |
| Citrulline | Non-TD | 0.64 (0.41, 0.97) | 0.04 | 0.143 |
| Hypoxanthine | Non-TD | 0.64 (0.40, 0.99) | 0.046 | 0.143 |
| Glyceric acid | Non-TD | 0.63 (0.40, 0.99) | 0.046 | 0.143 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, R.J.; Liang, D.; Busgang, S.A.; Curtin, P.; Giulivi, C. Maternal Plasma Metabolic Profile Demarcates a Role for Neuroinflammation in Non-Typical Development of Children. Metabolites 2021, 11, 545. https://doi.org/10.3390/metabo11080545
Schmidt RJ, Liang D, Busgang SA, Curtin P, Giulivi C. Maternal Plasma Metabolic Profile Demarcates a Role for Neuroinflammation in Non-Typical Development of Children. Metabolites. 2021; 11(8):545. https://doi.org/10.3390/metabo11080545
Chicago/Turabian StyleSchmidt, Rebecca J., Donghai Liang, Stefanie A. Busgang, Paul Curtin, and Cecilia Giulivi. 2021. "Maternal Plasma Metabolic Profile Demarcates a Role for Neuroinflammation in Non-Typical Development of Children" Metabolites 11, no. 8: 545. https://doi.org/10.3390/metabo11080545
APA StyleSchmidt, R. J., Liang, D., Busgang, S. A., Curtin, P., & Giulivi, C. (2021). Maternal Plasma Metabolic Profile Demarcates a Role for Neuroinflammation in Non-Typical Development of Children. Metabolites, 11(8), 545. https://doi.org/10.3390/metabo11080545






