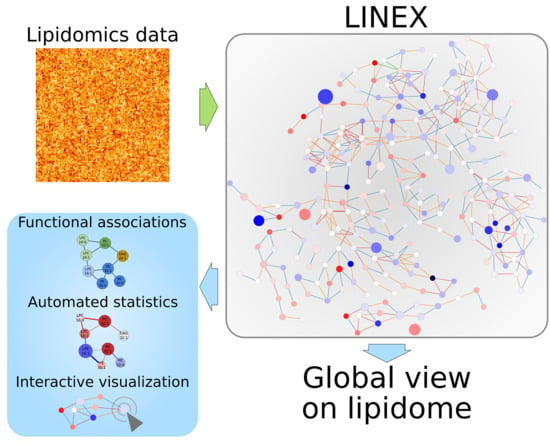
Investigating Global Lipidome Alterations with the Lipid Network Explorer
Abstract
:1. Introduction
2. Results
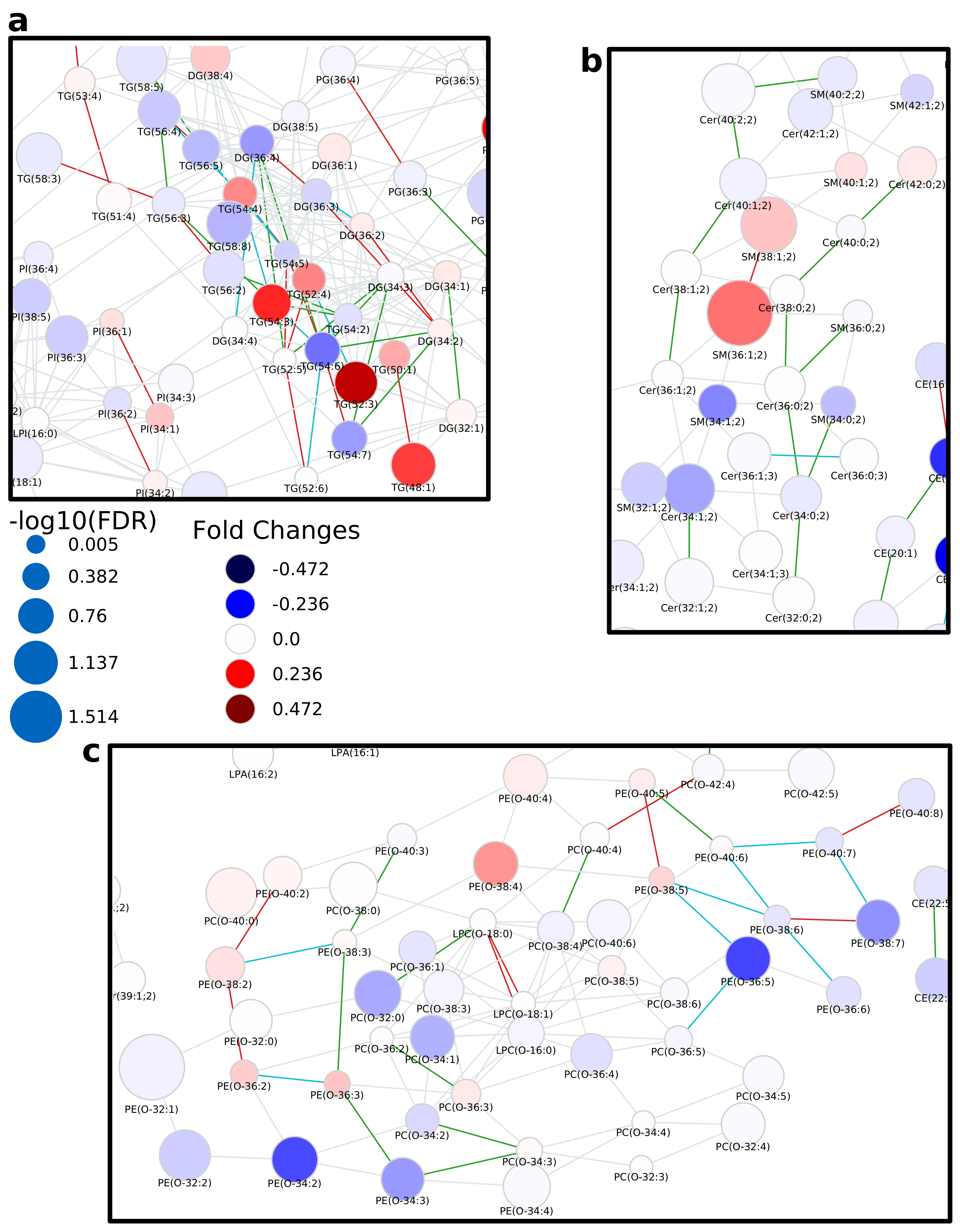
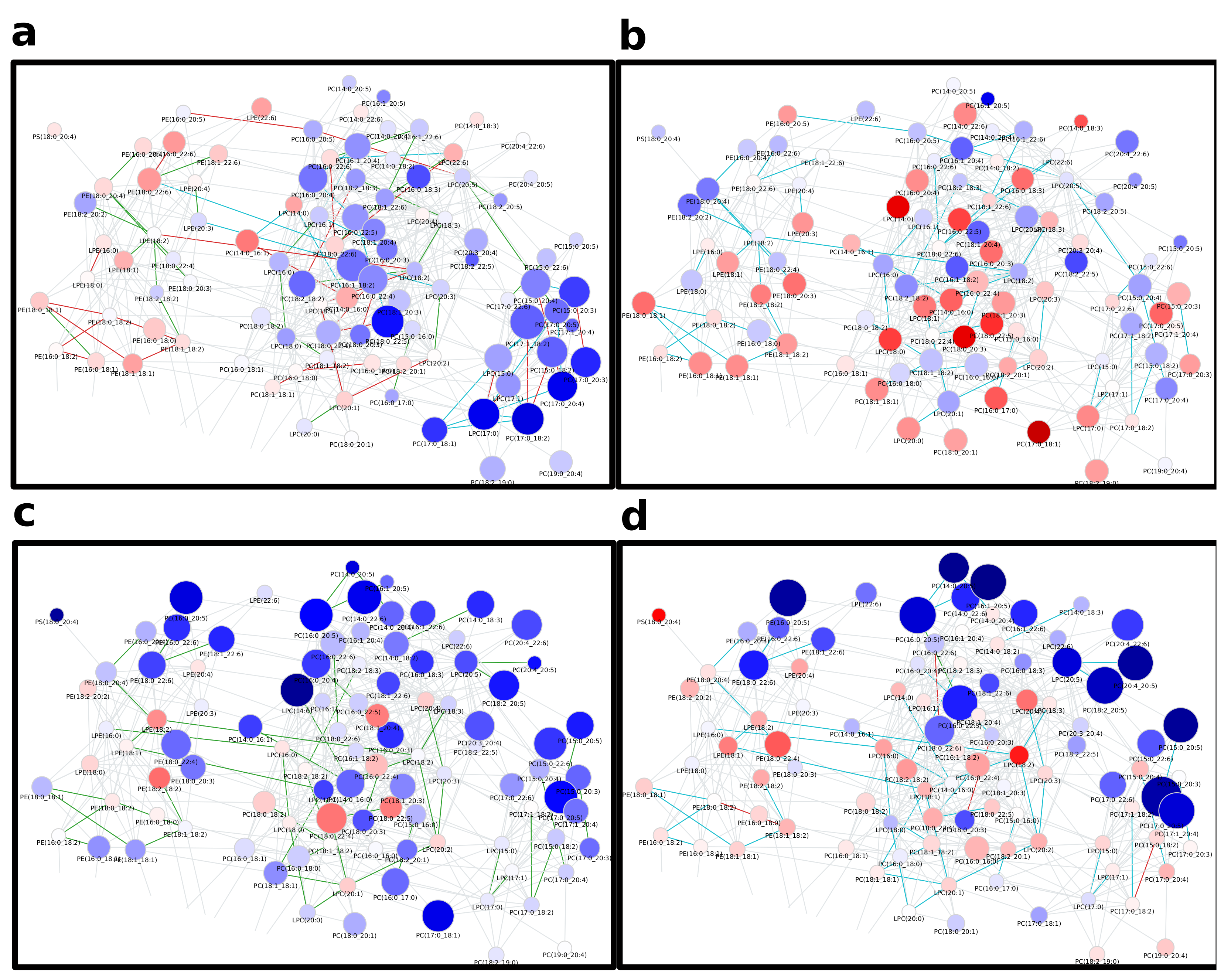
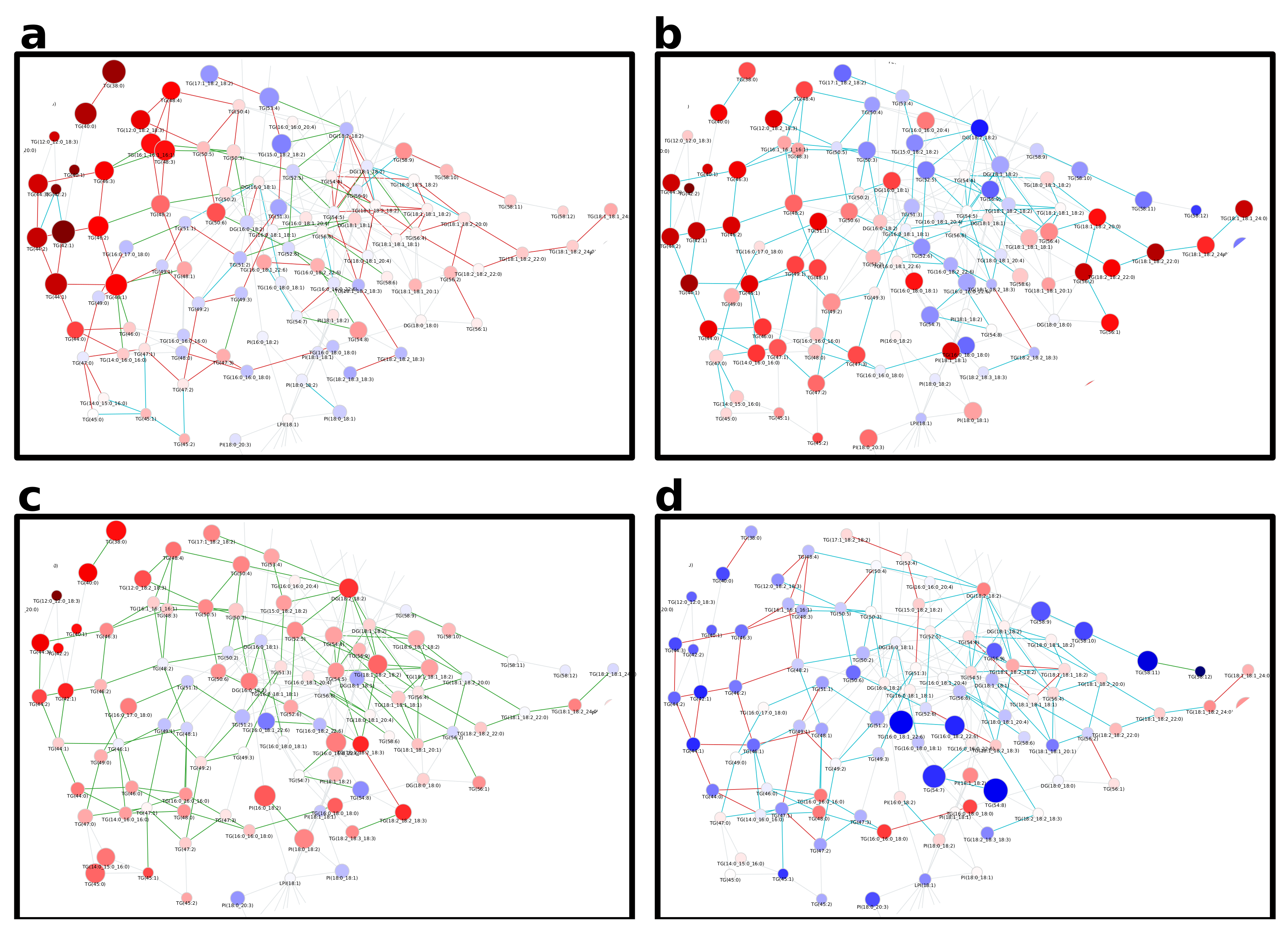
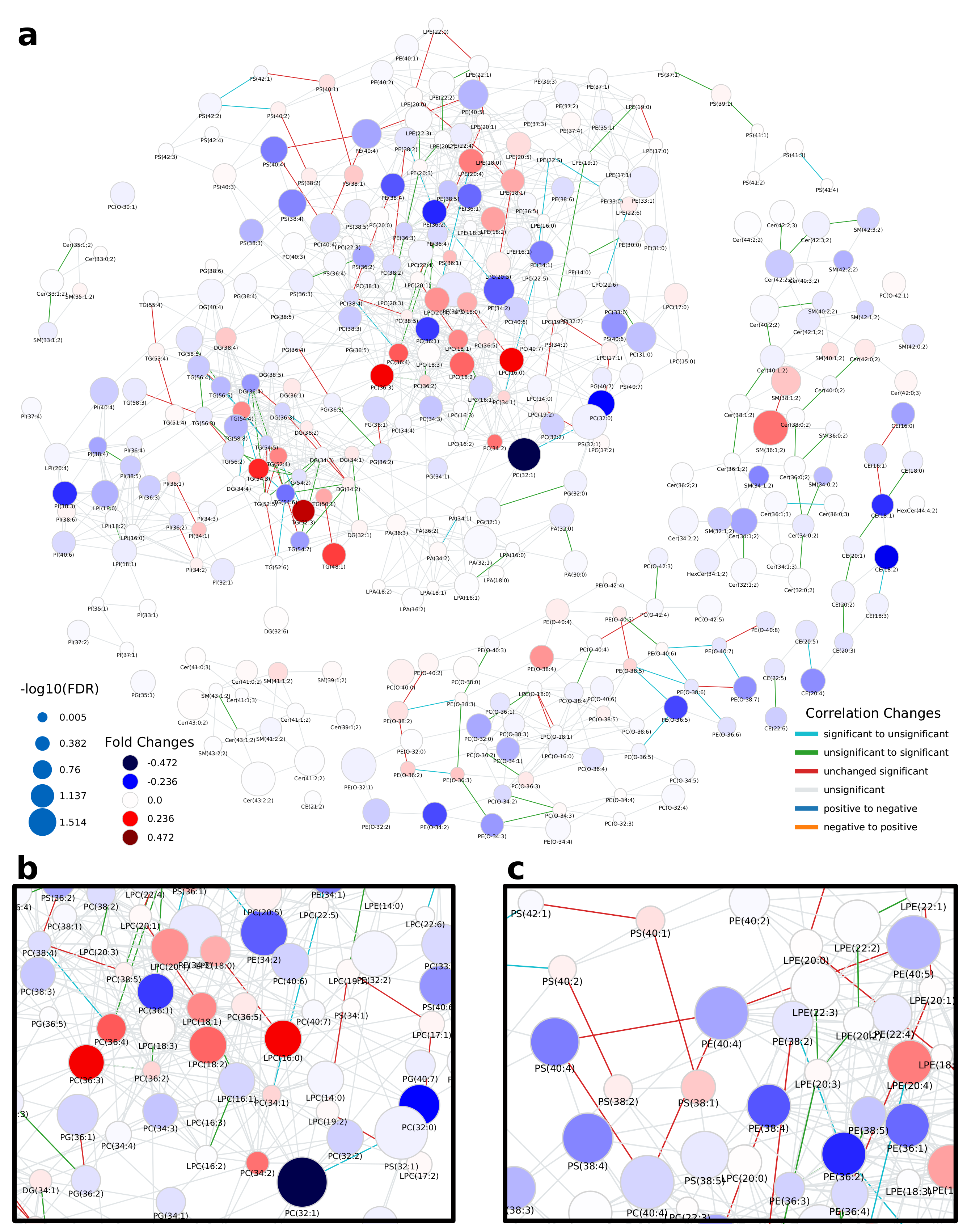
2.1. Lipidomics of Colorectal Cancer
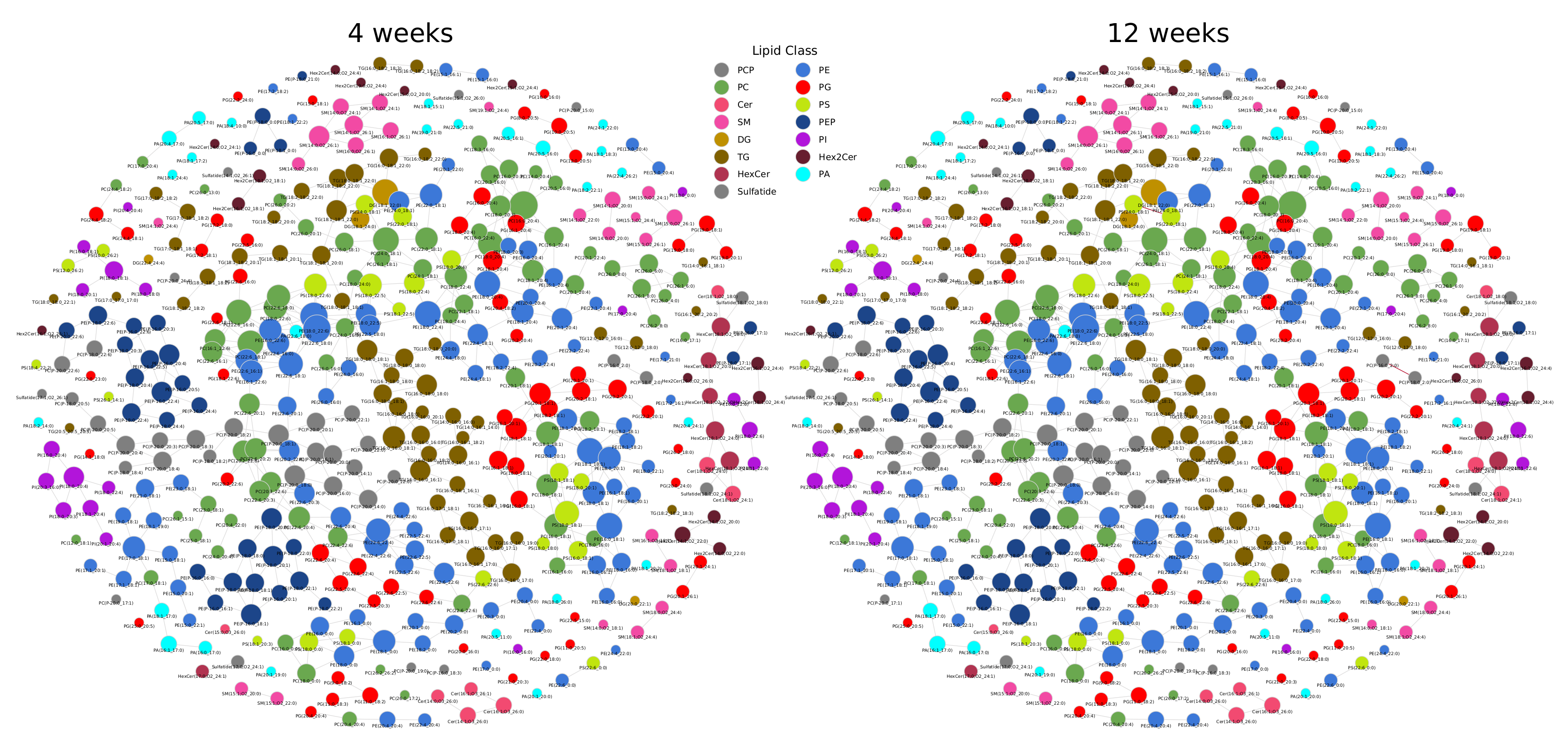
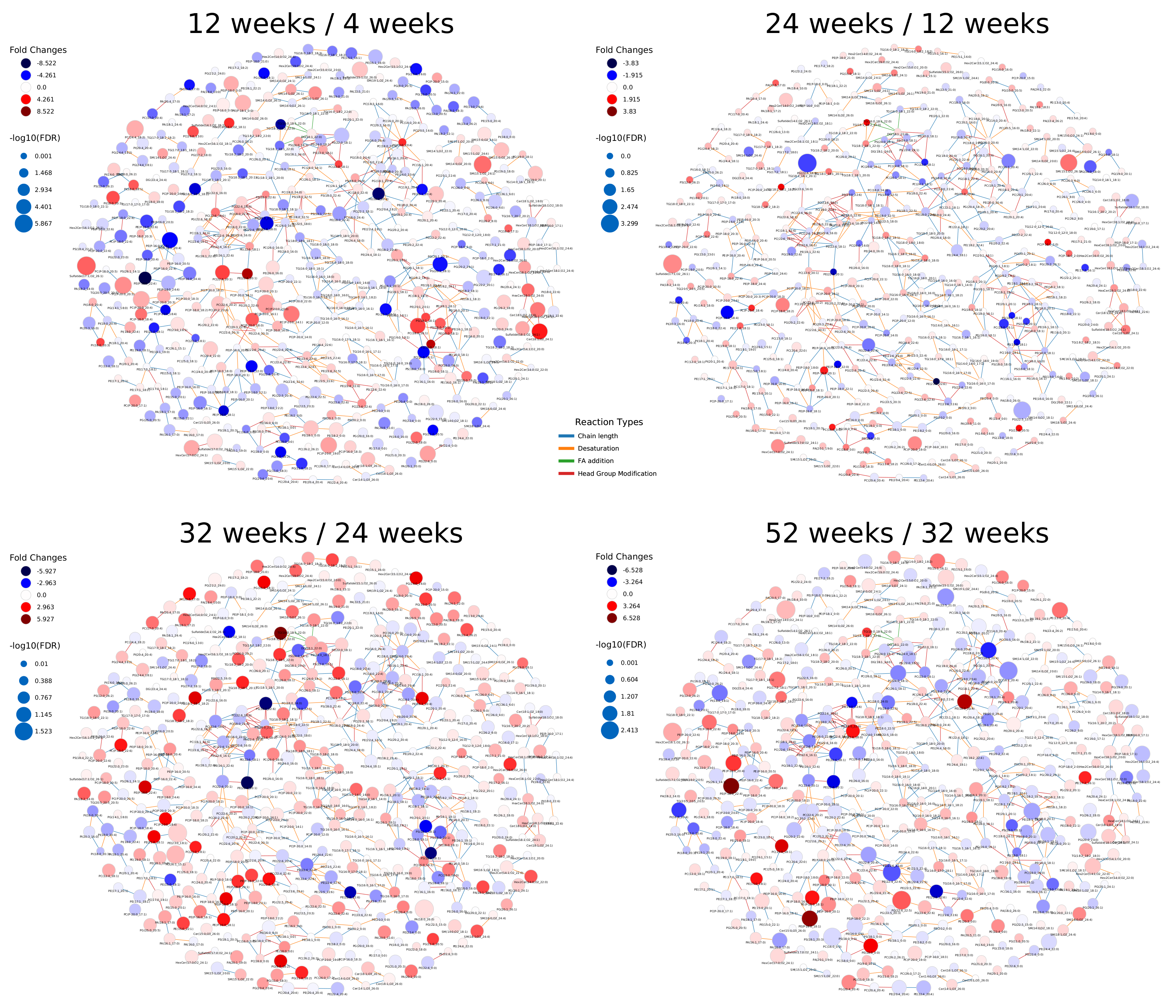
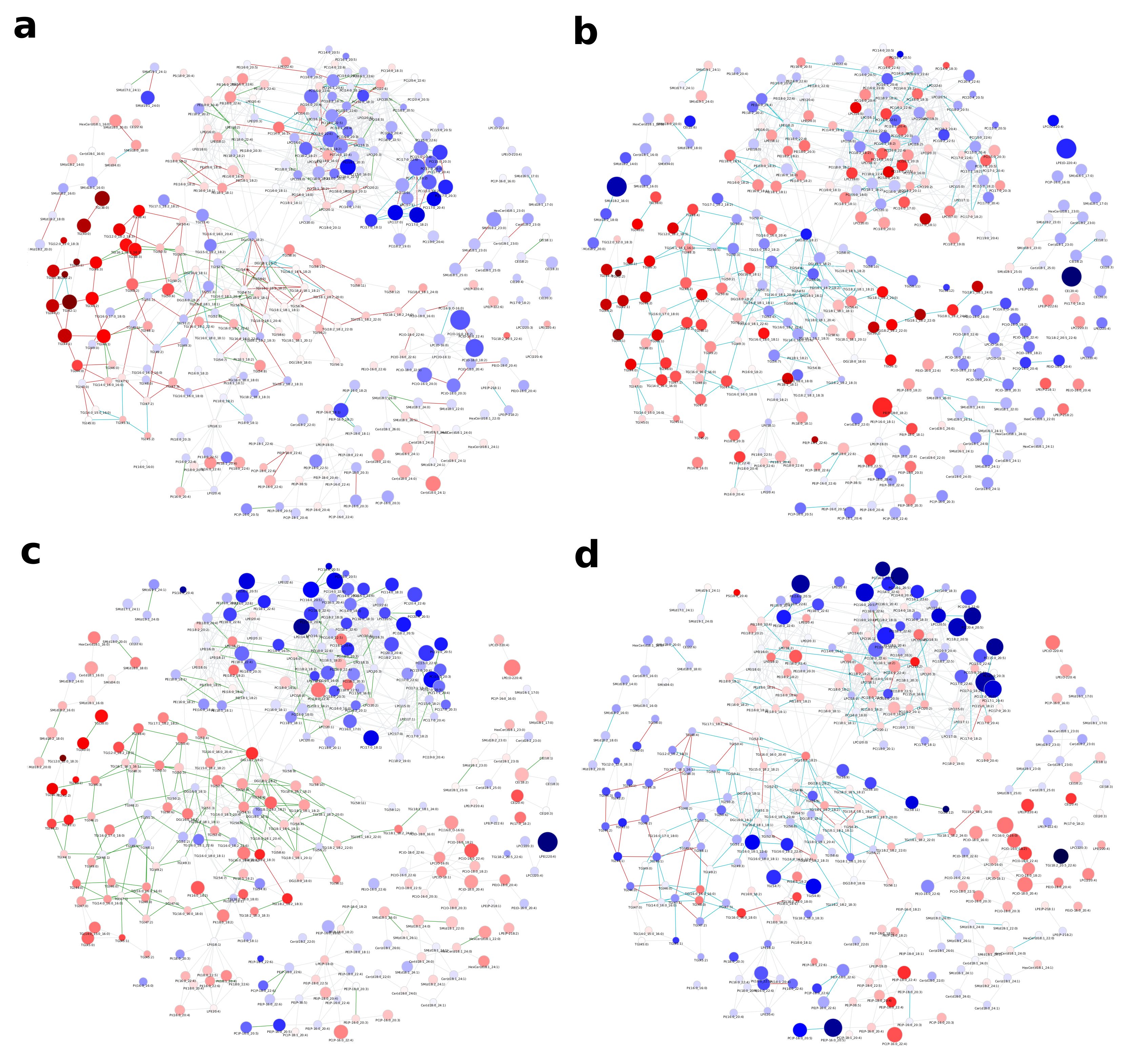
2.2. Lipidome Alterations in Aging Brain of Mice
2.3. Healthy Human Reference Plasma Lipidome in Aging
3. Discussion
4. Materials and Methods
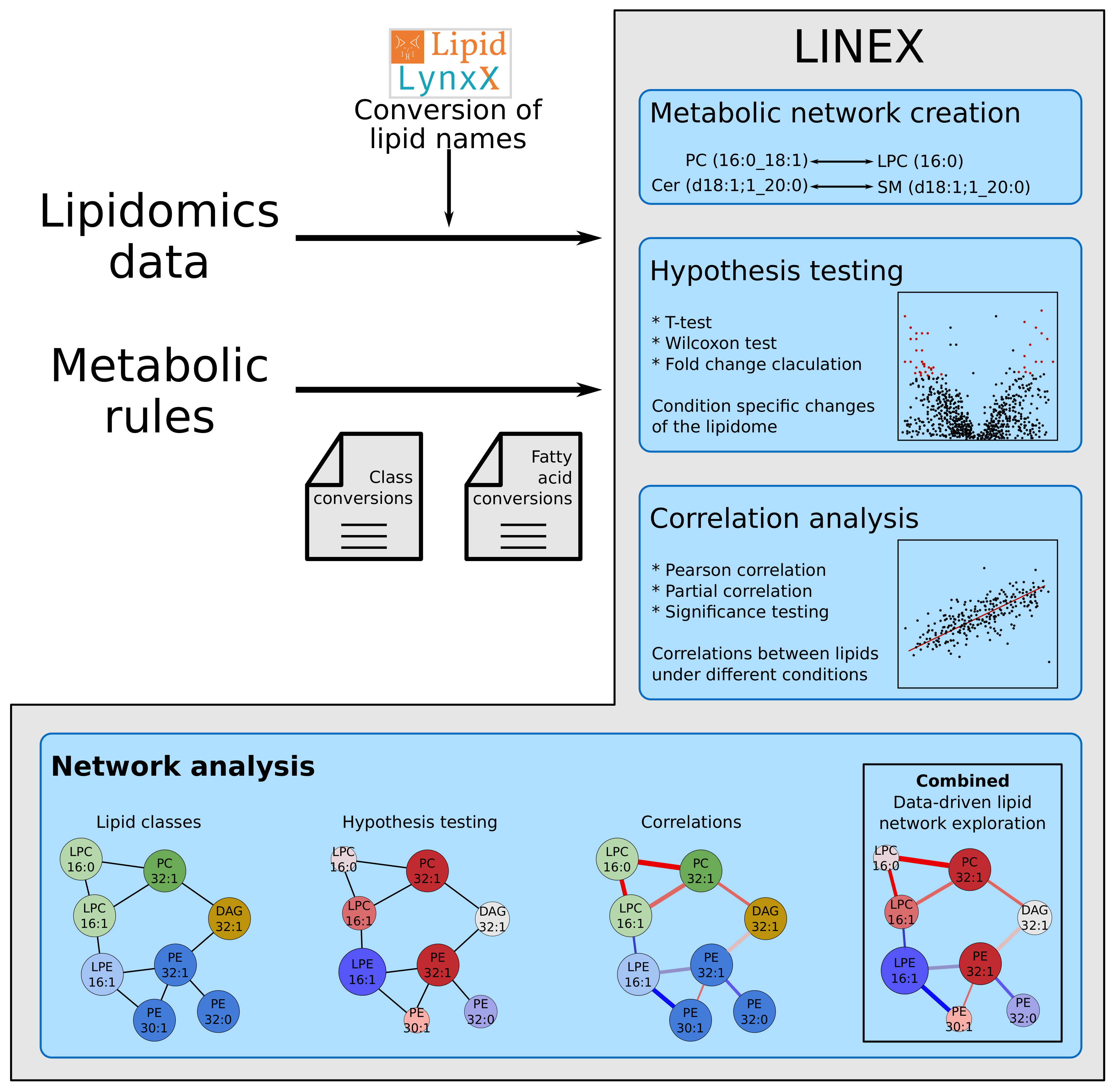
4.1. Webtool
4.2. Lipid Name Conversion
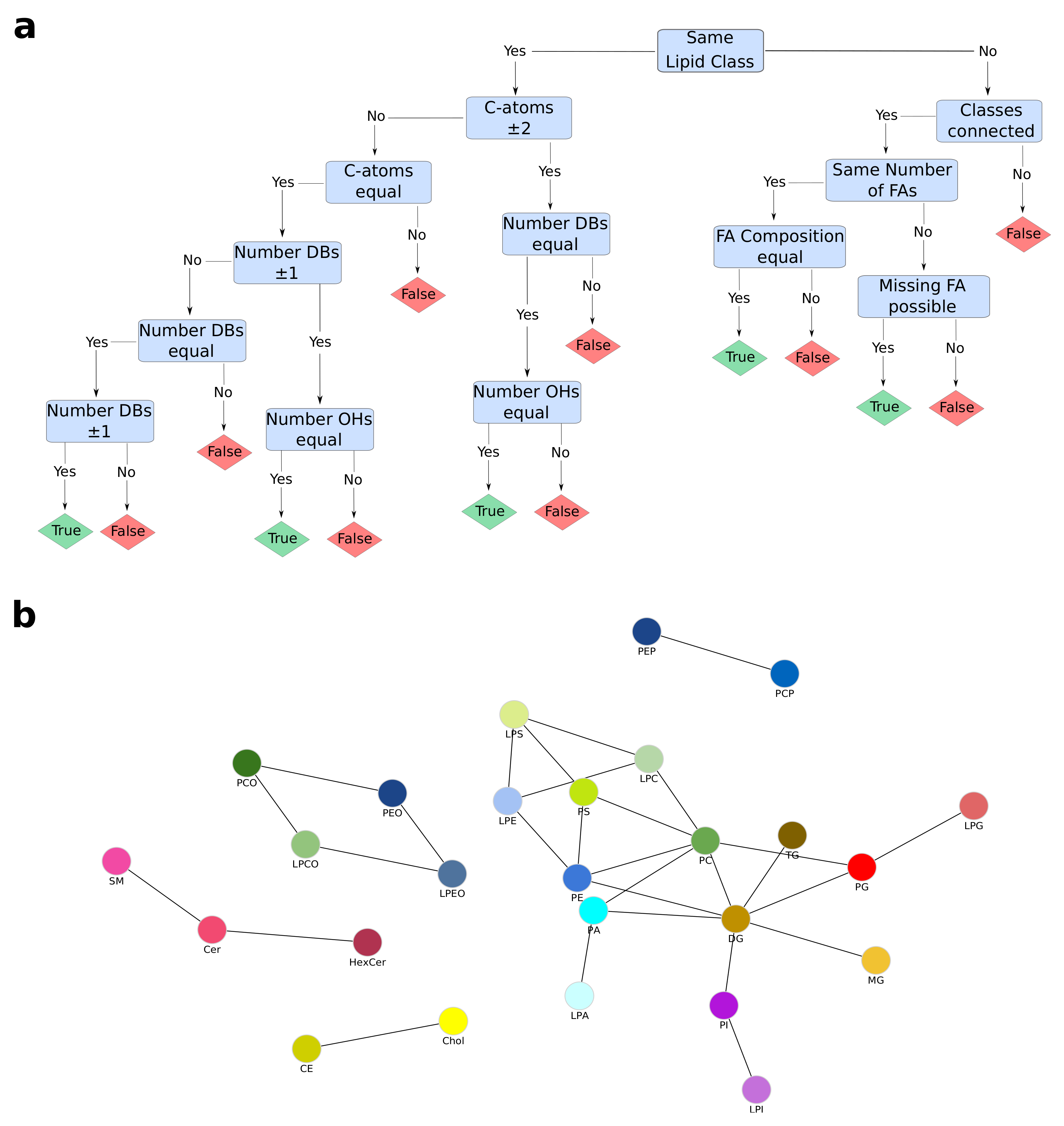
4.3. Dynamic Network Creation
4.4. Lipid Class Color Scheme
4.5. Statistical Methods
4.6. Experimental Data Processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FA | fatty Acid |
| GPL | glycerophospholipid |
| GR | Gene Regulatory |
| LGPL | lyso-glycerophospholipid |
| LPC | lyso-phosphatidylcholine |
| LPE | lyso-phosphatidylethanolamine |
| LPI | lyso-phosphatidylinositol |
| MS | Mass Spectrometry |
| PA | phosphatic acid |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PEO | phosphatidylethanolamine Ether |
| PG | phosphatidylglycerol |
| PI | phosphatidylinositol |
| PPI | Protein-Protein Interaction |
| PS | phosphatidylserine |
Appendix A
| Saturated FAs | Monounsaturated FAs | Polyunsaturated FAs |
|---|---|---|
| 14:0 | 16:1 | 18:2 |
| 15:0 | 18:1 | 20:2 |
| 16:0 | 20:1 | 20:3 |
| 17:0 | 20:4 | |
| 15:0 | 20:5 | |
| 20:0 | 22:4 | |
| 22:5 | ||
| 22:6 | ||
| 24:6 |

Appendix B
References
- Mohamed, A.; Molendijk, J.; Hill, M.M. Lipidr: A Software Tool for Data Mining and Analysis of Lipidomics Datasets. J. Proteome Res. 2020, 19, 2890–2897. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Hill, M.M. LipidSuite: Interactive web server for lipidomics differential and enrichment analysis. Nucleic Acids Res. 2021, 49, W346–W351. [Google Scholar] [CrossRef]
- Alcaraz, N.; Pauling, J.; Batra, R.; Barbosa, E.; Junge, A.; Christensen, A.G.L.; Azevedo, V.; Ditzel, H.J.; Baumbach, J. KeyPathwayMiner 4.0: Condition-specific pathway analysis by combining multiple omics studies and networks with Cytoscape. BMC Syst. Biol. 2014, 8, 99. [Google Scholar] [CrossRef] [Green Version]
- Dhakar, K.; Zarecki, R.; van Bommel, D.; Knossow, N.; Medina, S.; Öztürk, B.; Aly, R.; Eizenberg, H.; Ronen, Z.; Freilich, S. Strategies for Enhancing Degradation of Linuron by sp. Strain SRS 16 Under the Guidance of Metabolic Modeling. Front. Bioeng. Biotechnol. 2021, 9, 602464. [Google Scholar] [CrossRef]
- Levi, H.; Elkon, R.; Shamir, R. DOMINO: A network-based active module identification algorithm with reduced rate of false calls. Mol. Syst. Biol. 2021, 17, e9593. [Google Scholar] [CrossRef]
- Leiserson, M.D.M.; Vandin, F.; Wu, H.T.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Papoutsaki, A.; Kim, Y.; Niu, B.; McLellan, M.; et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015, 47, 106–114. [Google Scholar] [CrossRef]
- Kopczynski, D.; Coman, C.; Zahedi, R.P.; Lorenz, K.; Sickmann, A.; Ahrends, R. Multi-OMICS: A critical technical perspective on integrative lipidomics approaches. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 808–811. [Google Scholar] [CrossRef]
- Poupin, N.; Vinson, F.; Moreau, A.; Batut, A.; Chazalviel, M.; Colsch, B.; Fouillen, L.; Guez, S.; Khoury, S.; Dalloux-Chioccioli, J.; et al. Improving lipid mapping in Genome Scale Metabolic Networks using ontologies. Metabolomics 2020, 16, 44. [Google Scholar] [CrossRef] [Green Version]
- Köberlin, M.S.; Snijder, B.; Heinz, L.X.; Baumann, C.L.; Fauster, A.; Vladimer, G.I.; Gavin, A.C.; Superti-Furga, G. A Conserved Circular Network of Coregulated Lipids Modulates Innate Immune Responses. Cell 2015, 162, 170–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yetukuri, L.; Katajamaa, M.; Medina-Gomez, G.; Seppänen-Laakso, T.; Vidal-Puig, A.; Oresic, M. Bioinformatics strategies for lipidomics analysis: Characterization of obesity related hepatic steatosis. BMC Syst. Biol. 2007, 1, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, G.; Chan, J.; Kingwell, B.A.; Leckie, C.; Meikle, P.J. LICRE: Unsupervised feature correlation reduction for lipidomics. Bioinformatics 2014, 30, 2832–2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benedetti, E.; Pučić-Baković, M.; Keser, T.; Gerstner, N.; Büyüközkan, M.; Štambuk, T.; Selman, M.H.J.; Rudan, I.; Polašek, O.; Hayward, C.; et al. A strategy to incorporate prior knowledge into correlation network cutoff selection. Nat. Commun. 2020, 11, 5153. [Google Scholar] [CrossRef]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; van de Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/web: A web-based ontology enrichment tool for lipidomic data analysis. Gigascience 2019, 8, giz061. [Google Scholar] [CrossRef] [Green Version]
- Gaud, C.; Sousa, B.C.; Nguyen, A.; Fedorova, M.; Ni, Z.; O’Donnell, V.B.; Wakelam, M.J.O.; Andrews, S.; Lopez-Clavijo, A.F. BioPAN: A web-based tool to explore mammalian lipidome metabolic pathways on LIPID MAPS. F1000Res 2021, 10, 4. [Google Scholar] [CrossRef]
- Marella, C.; Torda, A.E.; Schwudke, D. The LUX Score: A Metric for Lipidome Homology. PLoS Comput. Biol. 2015, 11, e1004511. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hinz, S.; Uckermann, O.; Hönscheid, P.; von Schönfels, W.; Burmeister, G.; Hendricks, A.; Ackerman, J.M.; Baretton, G.B.; Hampe, J.; et al. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158579. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, S.K.M.; Hess, T.; Becker, J.; Hamann, L.; Vashist, Y.K.; Butterbach, K.; Schmidt, T.; Alakus, H.; Krasniuk, I.; Höblinger, A.; et al. Evidence for PTGER4, PSCA, and MBOAT7 as risk genes for gastric cancer on the genome and transcriptome level. Cancer Med. 2018, 7, 5057–5065. [Google Scholar] [CrossRef]
- Thangapandi, V.R.; Knittelfelder, O.; Brosch, M.; Patsenker, E.; Vvedenskaya, O.; Buch, S.; Hinz, S.; Hendricks, A.; Nati, M.; Herrmann, A.; et al. Loss of hepatic Mboat7 leads to liver fibrosis. Gut 2021, 70, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Yin, Y.; Xu, M.; Wang, R.; Zhu, Z.J. Absolute quantitative lipidomics reveals lipidome-wide alterations in aging brain. Metabolomics 2017, 14, 5. [Google Scholar] [CrossRef]
- Ni, Z.; Fedorova, M. LipidLynxX: Lipid annotations converter for large scale lipidomics and epilipidomics datasets. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Balgoma, D.; Pettersson, C.; Hedeland, M. Common Fatty Markers in Diseases with Dysregulated Lipogenesis. Trends Endocrinol. Metab. 2019, 30, 283–285. [Google Scholar] [CrossRef]
- Kyle, J.E.; Stratton, K.G.; Zink, E.M.; Kim, Y.M.; Bloodsworth, K.J.; Monroe, M.E.; Waters, K.M.; Webb-Robertson, B.J.M.; Koeller, D.M.; Metz, T.O. A resource of lipidomics and metabolomics data from individuals with undiagnosed diseases. Sci. Data 2021, 8, 114. [Google Scholar] [CrossRef]
- Perrone, G.; Unpingco, J.; Lu, H.M. Network visualizations with Pyvis and VisJS. arXiv 2020, arXiv:2006.04951. [Google Scholar]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Freeman, L.C. A Set of Measures of Centrality Based on Betweenness. Sociometry 1977, 40, 35. [Google Scholar] [CrossRef]
- Bavelas, A. Communication Patterns in Task-Oriented Groups. J. Acoust. Soc. Am. 1950, 22, 725. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hagberg, A.; Schult, D.; Swart, P. Exploring Network Structure, Dynamics, and Function Using Networkx. In Proceedings of the 7th Python in Science Conference (SciPy 2008), Pasadena, CA, USA, 19–24 August 2008; pp. 11–15. [Google Scholar]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [Green Version]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Demographic Information for Reference Population. Available online: https://doi.org/10.6084/m9.figshare.12440342 (accessed on 11 May 2021).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Köhler, N.; Rose, T.D.; Falk, L.; Pauling, J.K. Investigating Global Lipidome Alterations with the Lipid Network Explorer. Metabolites 2021, 11, 488. https://doi.org/10.3390/metabo11080488
Köhler N, Rose TD, Falk L, Pauling JK. Investigating Global Lipidome Alterations with the Lipid Network Explorer. Metabolites. 2021; 11(8):488. https://doi.org/10.3390/metabo11080488
Chicago/Turabian StyleKöhler, Nikolai, Tim Daniel Rose, Lisa Falk, and Josch Konstantin Pauling. 2021. "Investigating Global Lipidome Alterations with the Lipid Network Explorer" Metabolites 11, no. 8: 488. https://doi.org/10.3390/metabo11080488
APA StyleKöhler, N., Rose, T. D., Falk, L., & Pauling, J. K. (2021). Investigating Global Lipidome Alterations with the Lipid Network Explorer. Metabolites, 11(8), 488. https://doi.org/10.3390/metabo11080488