Analytical Evaluation of the Ideal Strategy for High-Throughput Flow Injection Analysis by Tandem Mass Spectrometry in Routine Newborn Screening
Abstract
:1. Introduction
2. Results
2.1. Evaluation of the Analytical Performances between Acquity UPLC I-Class and Renatadx Screening Systems: Study 1
2.2. Methods Comparison between ACQUITY UPLC I-Class and RenataDX Systems
2.3. Evaluation of Two Commercial MS/MS Kits Used Rountinely in Newborn Screening on the RenataDX Screening System: Study 2
2.3.1. Linearity of the MassChrom® LC-MS/MS Kit
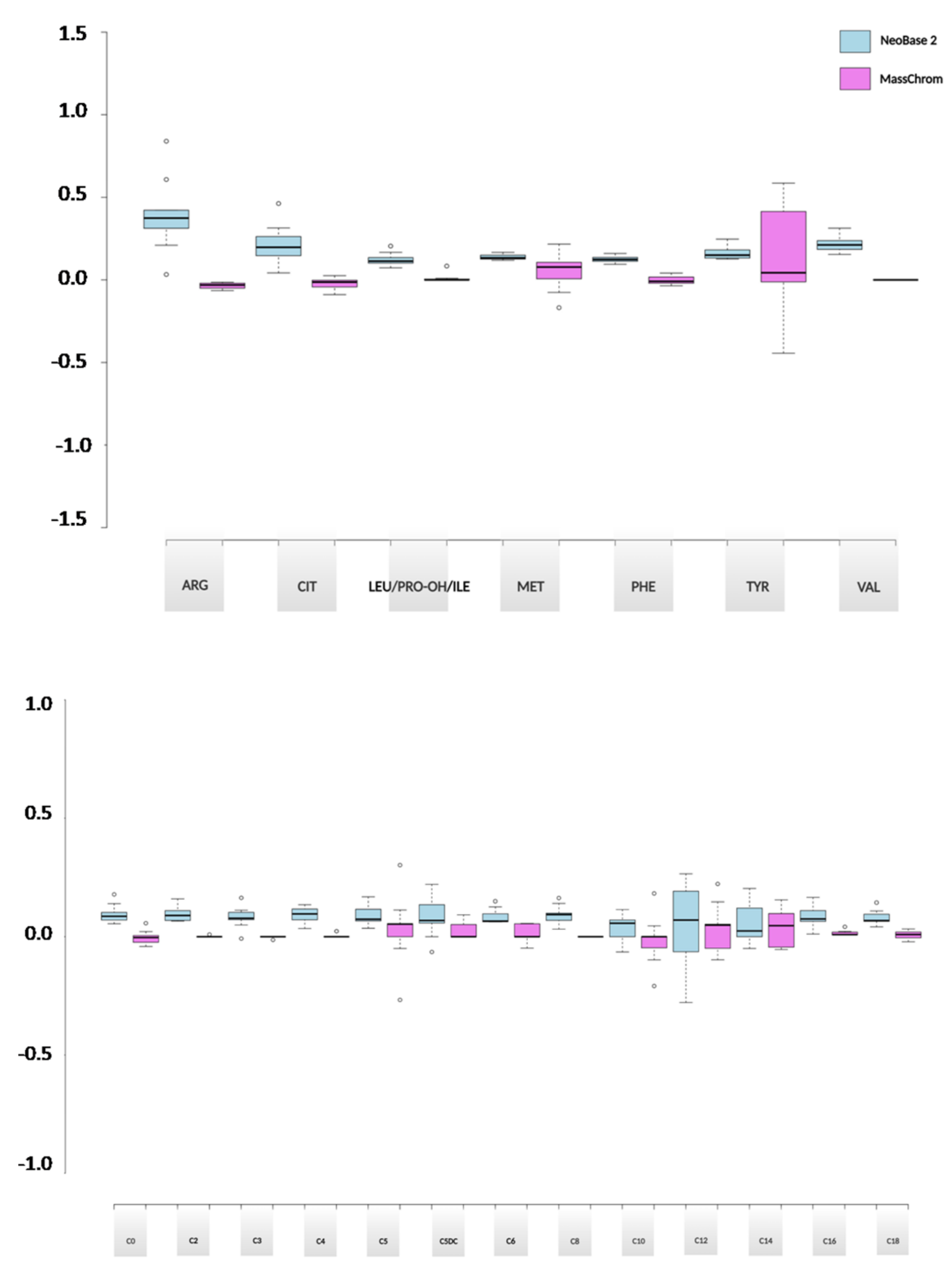
2.3.2. Evaluation of the Analytical Performances between NeoBase™ 2 and MassChrom® Non-Derivatized Kits
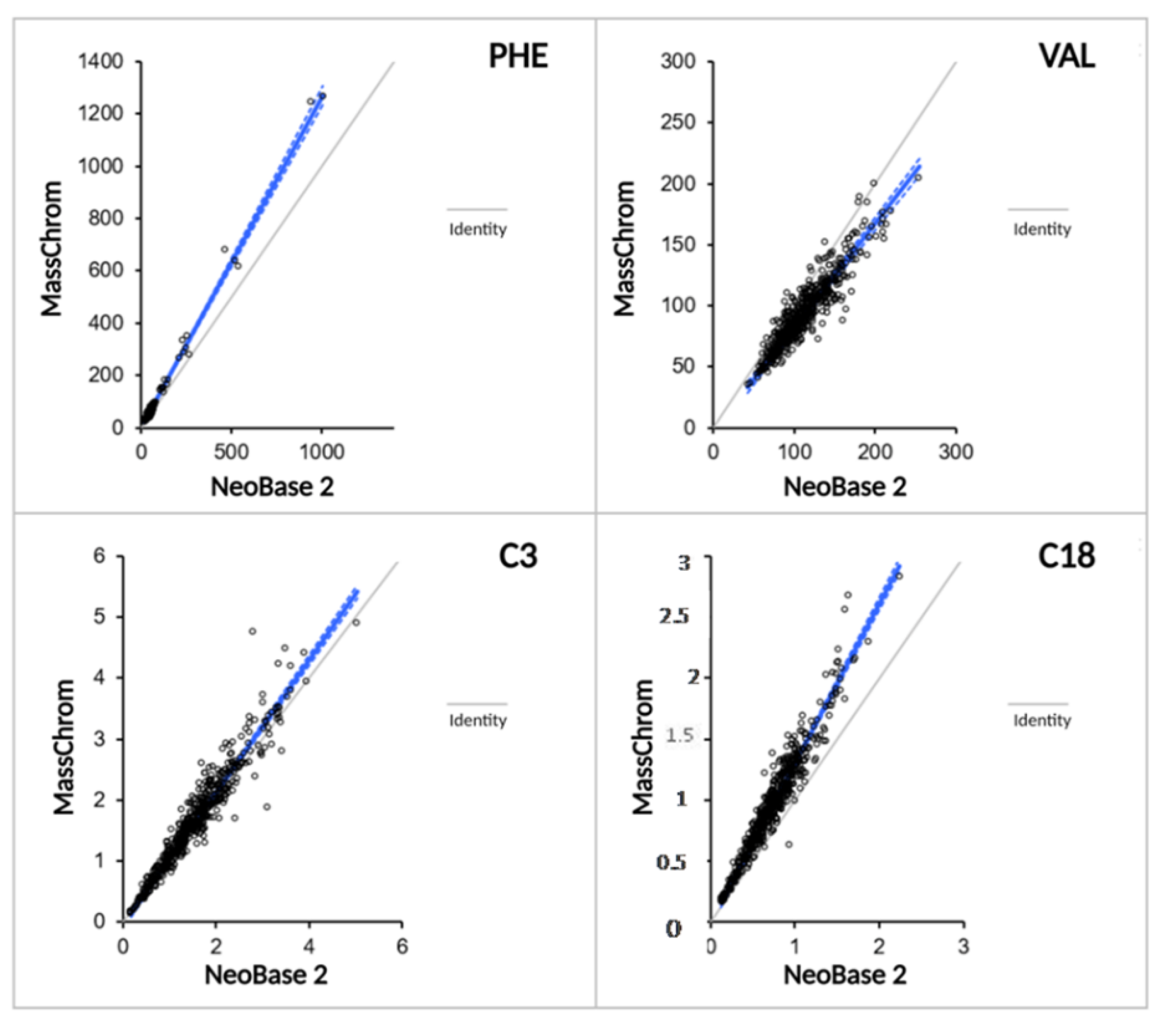
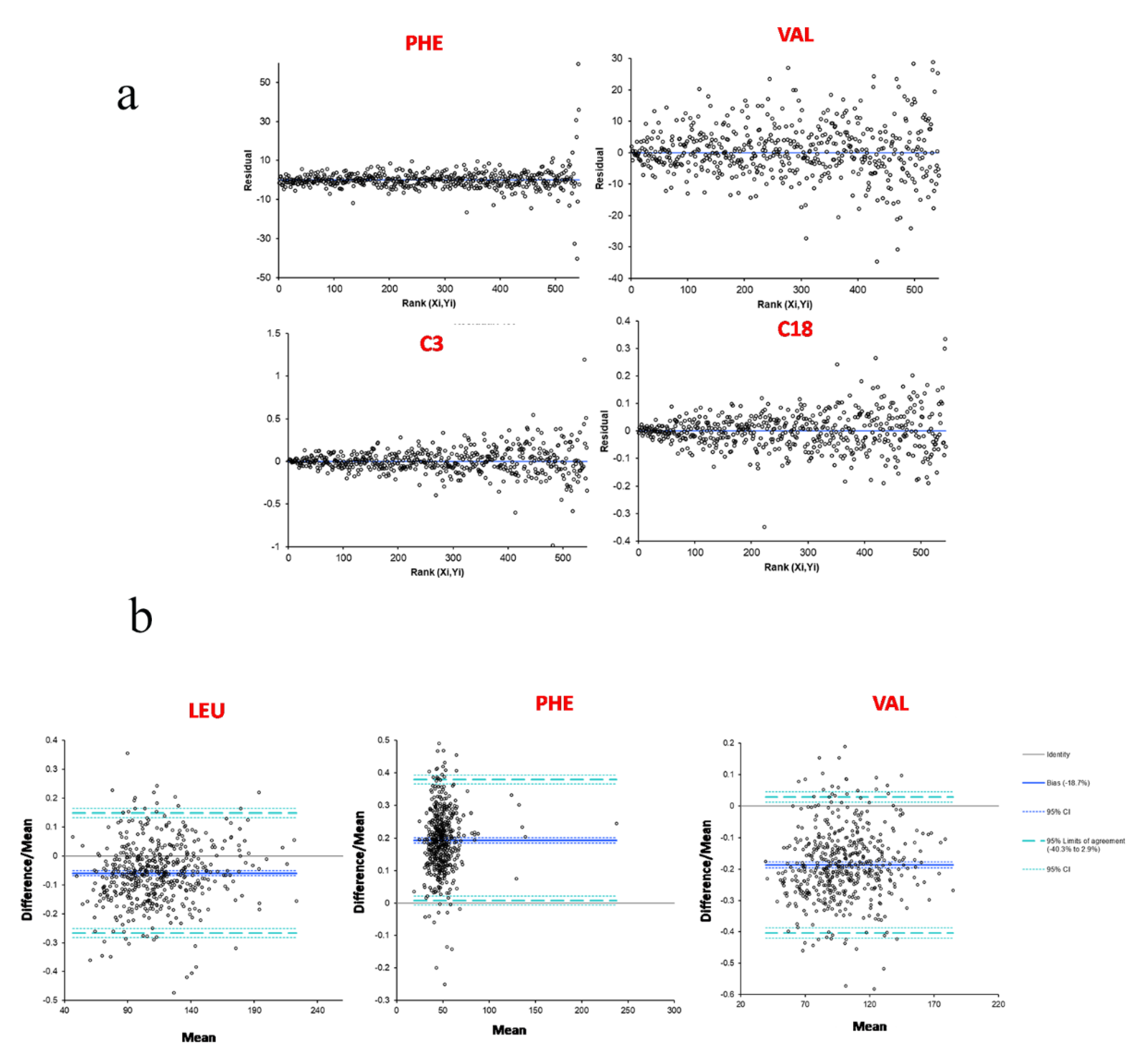
2.3.3. Methods Comparison between NeoBase™ 2 and MassChrom® Non-Derivatized Kits
3. Discussion
4. Materials and Methods
4.1. FIA-MS/MS for the Comparison between Two Analytical Platforms
4.2. Evaluation of the Analytical Performances between ACQUITY UPLC I-Class and RenataDX Systems
4.3. Linearity of the MassChrom® LC-MS/MS Kit
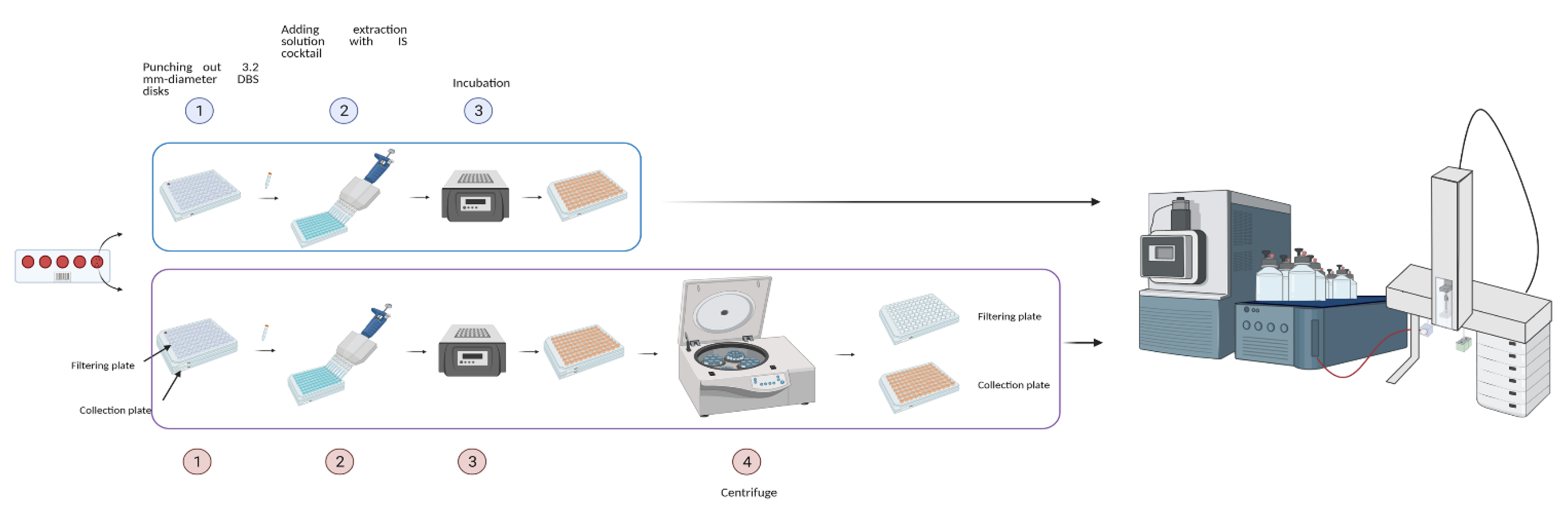
4.4. Extraction of DBS Samples by NeoBase™2 Kit
4.5. Extraction of DBS Samples by MassChrom® Kit
4.6. Evaluation of the Analytical Performances between NeoBase™ 2 and MassChrom® Non-Derivatized Kits
4.7. Methods Comparison
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ozben, T. Expanded newborn screening and confirmatory follow-up testing for inborn errors of metabolism detected by tandem mass spectrometry. Clin. Chem. Lab. Med. 2013, 51, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Loeber, J.G.; Platis, D.; Zetterström, R.H.; Almashanu, S.; Boemer, F.; Bonham, J.R.; Borde, P.; Brincat, I.; Cheillan, D.; Dekkers, E.; et al. Neonatal Screening in Europe Revisited: An ISNS Perspective on the Current State and Developments Since 2010. Int. J. Neonatal Screen. 2021, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Blumenfeld, C.M.; Wallace, M.J.; Anderson, R. Phenylketonuria-the guthrie screening test-a method of quantitation, observations on reliability and suggestions for improvement. Calif. Med. 1966, 105, 429–434. [Google Scholar] [PubMed]
- Chace, D.H.; Kalas, T.A.; Naylor, E.W. The application of tandem mass spectrometry to neonatal screening for inherited disorders of intermediary metabolism. Annu. Rev. Genom. Hum. Genet. 2002, 3, 17–45. [Google Scholar] [CrossRef] [Green Version]
- Garg, U.; Dasouki, M. Expanded newborn screening of inherited metabolic disorders by tandem mass spectrometry: Clinical and laboratory aspects. Clin. Biochem. 2006, 39, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Tarini, B.A.; Christakis, D.A.; Welch, H.G. State newborn screening in the tandem mass spectrometry era: More tests, more false-positive results. Pediatrics 2006, 118, 448–456. [Google Scholar] [CrossRef]
- Parini, R.; Corbetta, C. Metabolic screening for the newborn. The journal of maternal-fetal & neonatal medicine: The official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. J. Matern. Fetal. Neonatal Med. 2011, 24 (Suppl. 2), 6–8. [Google Scholar] [CrossRef]
- Moat, S.J.; George, R.S.; Carling, R.S. Use of Dried Blood Spot Specimens to Monitor Patients with Inherited Metabolic Disorders. Int. J. Neonatal Screen. 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhondt, J.L. Expanded newborn screening: Social and ethical issues. J. Inherit. Metab. Dis. 2010, 33, S211–S217. [Google Scholar] [CrossRef]
- Campos, H.D. Tandem mass spectrometry as screening for inborn errors of metabolism. Rev. Med. Chile 2011, 139, 1356–1364. [Google Scholar]
- American College of Medical Genetics Newborn Screening Expert Group. Newborn screening: Toward a uniform screening panel and system--executive summary. Pediatrics 2006, 117, S296–S307. [Google Scholar] [CrossRef] [Green Version]
- Kronn, D.; Mofidi, S.; Braverman, N.; Harris, K.; Diagnostics Guidelines Work Group. Diagnostic guidelines for newborns who screen positive in newborn screening. Genet. Med. Off. J. Am. Coll. Med Genet. 2010, 12, S251–S255. [Google Scholar] [CrossRef]
- Chan, K.; Petros, M. Simple Test, Complex System: Multifaceted Views of Newborn Screening Science, Technology, and Policy. Glob. Pediatric Health 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, J.; Shen, B. Newborn Screening in the Era of Precision Medicine. Adv. Exp. Med. Biol. 2017, 1005, 47–61. [Google Scholar] [CrossRef]
- Malvagia, S.; Forni, G.; Ombrone, D.; la Marca, G. Development of Strategies to Decrease False Positive Results in Newborn Screening. Int. J. Neonatal Screen. 2020, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- van Karnebeek, C.D.M.; Wortmann, S.B.; Tarailo-Graovac, M.; Langeveld, M.; Ferreira, C.R.; van de Kamp, J.M.; Hollak, C.E.; Wasserman, W.W.; Waterham, H.R.; Wevers, R.A.; et al. The role of the clinician in the multi-omics era: Are you ready? J. Inherit. Metab. Dis. 2018, 41, 571–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gucciardi, A.; Pirillo, P.; Di Gangi, I.M.; Naturale, M.; Giordano, G. A rapid UPLC-MS/MS method for simultaneous separation of 48 acylcarnitines in dried blood spots and plasma useful as a second-tier test for expanded newborn screening. Anal. Bioanal. Chem. 2012, 404, 741–751. [Google Scholar] [CrossRef]
- Miller, J.H., IV; Poston, P.A.; Karnes, H.T. A quantitative method for acylcarnitines and amino acids using high resolution chromatography and tandem mass spectrometry in newborn screening dried blood spot analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 903, 142–149. [Google Scholar] [CrossRef]
- Koracin, V.; Mlinaric, M.; Baric, I.; Brincat, I.; Djordjevic, M.; Drole Torkar, A.; Fumic, K.; Kocova, M.; Milenkovic, T.; Moldovanu, F.; et al. Current Status of Newborn Screening in Southeastern Europe. Front. Pediatrics 2021, 9, 648939. [Google Scholar] [CrossRef]
- Pickens, C.A.; Petritis, K. High resolution mass spectrometry newborn screening applications for quantitative analysis of amino acids and acylcarnitines from dried blood spots. Anal. Chim. Acta 2020, 1120, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.A.; Wierenga, A. Quantitation of plasma and urine 3-hydroxyglutaric acid, after separation from 2-hydroxyglutaric acid and other compounds of similar ion transition, by liquid chromatography-tandem mass spectrometry for the confirmation of glutaric aciduria type 1. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1097–1098, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Marzano, V.; Consalvo, A.; Zucchelli, M.; Mortera, S.L.; Casagrande, V.; Mavilio, M.; Sacchetta, P.; Federici, M.; Menghini, R.; et al. Proteomic and metabolomic characterization of streptozotocin-induced diabetic nephropathy in TIMP3-deficient mice. Acta Diabetol. 2018, 55, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; Cicalini, I.; Di Michele, S.; Fusilli, P.; Cotugno, G.; Ferrante, R.; Bucci, I.; Dionisi-Vici, C.; Stuppia, L.; De Laurenzi, V.; et al. A Case of Suspected Hyperphenylalaninemia at Newborn Screening by Tandem Mass Spectrometry during Total Parenteral Nutrition. Metabolites 2020, 10, 44. [Google Scholar] [CrossRef] [Green Version]
- Rossi, C.; Cicalini, I.; Rizzo, C.; Zucchelli, M.; Consalvo, A.; Valentinuzzi, S.; Semeraro, D.; Gasparroni, G.; Brindisino, P.; Gazzolo, D.; et al. A False-Positive Case of Methylmalonic Aciduria by Tandem Mass Spectrometry Newborn Screening Dependent on Maternal Malnutrition in Pregnancy. Int. J. Environ. Res. Public Health 2020, 17, 3601. [Google Scholar] [CrossRef] [PubMed]


| Repeatability | ACQUITY | RenataDX | ||
|---|---|---|---|---|
| LC CV% | HC CV% | LC CV% | HC CV% | |
| ARG | 6.15 | 5.08 | 6.17 | 5.66 |
| CIT | 8.81 | 8.43 | 9.73 | 7.48 |
| LEU\ILE\PRO-OH | 6.47 | 6.90 | 6.02 | 6.85 |
| MET | 7.72 | 9.26 | 9.40 | 9.68 |
| PHE | 7.20 | 7.19 | 6.87 | 7.28 |
| TYR | 7.43 | 6.29 | 7.28 | 6.41 |
| VAL | 5.09 | 5.45 | 4.86 | 4.42 |
| C0 | 8.89 | 8.18 | 5.03 | 5.29 |
| C10 | 6.74 | 7.71 | 6.93 | 6.37 |
| C2 | 3.72 | 4.93 | 4.20 | 4.88 |
| C3 | 4.72 | 4.52 | 5.59 | 3.32 |
| C4 | 6.04 | 6.32 | 5.93 | 6.60 |
| C5 | 5.47 | 5.28 | 5.03 | 5.19 |
| C5DC\C6OH | 9.86 | 6.77 | 8.36 | 7.99 |
| C6 | 6.20 | 4.58 | 6.66 | 5.08 |
| C8 | 5.71 | 5.59 | 5.30 | 5.90 |
| C12 | 6.53 | 6.20 | 5.42 | 6.48 |
| C14 | 5.47 | 6.12 | 5.77 | 6.41 |
| C16 | 4.79 | 6.29 | 4.76 | 5.56 |
| C18 | 5.92 | 7.65 | 7.96 | 9.97 |
| R2 | SD | |
|---|---|---|
| ARG | 0.991 | 0.007 |
| CIT | 0.996 | 0.001 |
| LEU\ILE\PRO-OH | 0.996 | 0.002 |
| MET | 0.997 | 0.002 |
| PHE | 0.997 | 0.001 |
| TYR | 0.998 | 0.001 |
| VAL | 0.996 | 0.001 |
| C0 | 0.995 | 0.003 |
| C10 | 0.997 | 0.002 |
| C2 | 0.997 | 0.001 |
| C3 | 0.997 | 0.002 |
| C4 | 0.998 | 0.001 |
| C5 | 0.997 | 0.002 |
| C5DC\C6OH | 0.993 | 0.001 |
| C6 | 0.997 | 0.001 |
| C8 | 0.997 | 0.002 |
| C12 | 0.997 | 0.002 |
| C14 | 0.998 | 0.001 |
| C16 | 0.998 | 0.001 |
| C18 | 0.998 | 0.0007 |
| n = 3 | NeoBase™ 2 | MassChrom® | ||
|---|---|---|---|---|
| LC CV% | HC CV% | LC CV% | HC CV% | |
| ARG | 1.93 | 4.76 | 8.21 | 0.46 |
| CIT | 2.50 | 7.37 | 2.74 | 6.72 |
| LEU\ILE\PRO-OH | 0.85 | 5.28 | 1.49 | 8.65 |
| MET | 3.29 | 4.47 | 2.24 | 9.35 |
| PHE | 1.84 | 4.19 | 1.29 | 8.49 |
| TYR | 3.57 | 5.71 | 1.57 | 7.59 |
| VAL | 0.70 | 6.18 | 1.94 | 8.69 |
| C0 | 2.23 | 5.36 | 1.89 | 8.84 |
| C10 | 5.15 | 8.07 | 0.81 | 5.89 |
| C2 | 1.59 | 5.49 | 0.98 | 8.41 |
| C3 | 2.60 | 4.84 | 1.50 | 10.00 |
| C4 | 4.53 | 6.09 | 2.92 | 10.40 |
| C5 | 2.35 | 3.30 | 3.39 | 8.10 |
| C5DC\C6OH | 3.90 | 11.14 | 9.02 | 15.07 |
| C6 | 6.00 | 4.82 | 3.73 | 6.57 |
| C8 | 1.39 | 3.56 | 3.88 | 10.56 |
| C12 | 5.07 | 8.18 | 2.70 | 8.98 |
| C14 | 1.02 | 5.05 | 1.31 | 11.01 |
| C16 | 1.63 | 5.74 | 1.88 | 12.65 |
| C18 | 0.30 | 7.15 | 1.76 | 14.08 |
| n = 9 | NeoBase™ 2 | MassChrom® | ||
|---|---|---|---|---|
| LC CV% | HC CV% | LC CV% | HC CV% | |
| ARG | 4.75 | 3.57 | 7.04 | 6.49 |
| CIT | 9.29 | 8.22 | 7.09 | 6.14 |
| LEU\ILE\PRO-OH | 5.31 | 4.29 | 4.24 | 4.98 |
| MET | 4.43 | 4.75 | 5.61 | 5.47 |
| PHE | 4.09 | 3.74 | 4.02 | 5.22 |
| TYR | 5.33 | 6.28 | 3.05 | 5.76 |
| VAL | 6.93 | 4.77 | 4.36 | 5.36 |
| C0 | 5.84 | 5.60 | 5.20 | 5.03 |
| C10 | 8.92 | 7.25 | 5.41 | 5.43 |
| C2 | 7.11 | 6.39 | 4.17 | 5.48 |
| C3 | 9.61 | 6.38 | 5.10 | 5.33 |
| C4 | 6.45 | 6.27 | 6.01 | 5.99 |
| C5 | 7.62 | 5.25 | 4.79 | 5.13 |
| C5DC\C6OH | 12.15 | 7.44 | 6.59 | 5.97 |
| C6 | 4.21 | 5.34 | 4.37 | 4.73 |
| C8 | 7.43 | 4.89 | 5.10 | 4.97 |
| C12 | 8.40 | 5.69 | 3.47 | 7.02 |
| C14 | 4.92 | 2.54 | 5.45 | 5.99 |
| C16 | 9.78 | 8.19 | 4.94s | 7.46 |
| C18 | 6.56 | 4.22 | 6.65 | 8.63 |
| Passing-Bablok Fit | Slope | Intercept |
|---|---|---|
| ARG | 0.95 | −0.92 |
| CIT | 1.08 | −0.92 |
| LEU\ILE\PRO-OH | 1 | −6.05 |
| MET | 0.94 | 0.17 |
| PHE | 1.26 | −2.04 |
| TYR | 1.10 | 0.62 |
| VAL | 0.86 | −3.37 |
| C0 | 0.98 | 0.16 |
| C2 | 1.03 | −0.48 |
| C3 | 1.08 | −0.04 |
| C4 | 1.13 | −0.01 |
| C5 | 0.89 | 0 |
| C5DC\C6OH | 1.84 | −0.01 |
| C6 | 1.03 | −0.01 |
| C8 | 1.09 | −0.02 |
| C10 | 1.52 | −0.01 |
| C12 | 1.11 | 0 |
| C14 | 1.02 | 0 |
| C16 | 1.12 | 0 |
| C18 | 1.31 | −0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cicalini, I.; Valentinuzzi, S.; Pieragostino, D.; Consalvo, A.; Zucchelli, M.; Donzelli, S.; Ambrogi, D.; Brown, H.A.; Calton, L.J.; Stuppia, L.; et al. Analytical Evaluation of the Ideal Strategy for High-Throughput Flow Injection Analysis by Tandem Mass Spectrometry in Routine Newborn Screening. Metabolites 2021, 11, 473. https://doi.org/10.3390/metabo11080473
Cicalini I, Valentinuzzi S, Pieragostino D, Consalvo A, Zucchelli M, Donzelli S, Ambrogi D, Brown HA, Calton LJ, Stuppia L, et al. Analytical Evaluation of the Ideal Strategy for High-Throughput Flow Injection Analysis by Tandem Mass Spectrometry in Routine Newborn Screening. Metabolites. 2021; 11(8):473. https://doi.org/10.3390/metabo11080473
Chicago/Turabian StyleCicalini, Ilaria, Silvia Valentinuzzi, Damiana Pieragostino, Ada Consalvo, Mirco Zucchelli, Simone Donzelli, Davide Ambrogi, Heather A. Brown, Lisa J. Calton, Liborio Stuppia, and et al. 2021. "Analytical Evaluation of the Ideal Strategy for High-Throughput Flow Injection Analysis by Tandem Mass Spectrometry in Routine Newborn Screening" Metabolites 11, no. 8: 473. https://doi.org/10.3390/metabo11080473
APA StyleCicalini, I., Valentinuzzi, S., Pieragostino, D., Consalvo, A., Zucchelli, M., Donzelli, S., Ambrogi, D., Brown, H. A., Calton, L. J., Stuppia, L., De Laurenzi, V., Del Boccio, P., & Rossi, C. (2021). Analytical Evaluation of the Ideal Strategy for High-Throughput Flow Injection Analysis by Tandem Mass Spectrometry in Routine Newborn Screening. Metabolites, 11(8), 473. https://doi.org/10.3390/metabo11080473










