Metabolomic Approaches to Studying the Response to Drought Stress in Corn (Zea mays) Cobs
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Lyophilisation on the Plant Metabolome
2.2. Effect of Drought Conditions upon the Extractable Maize Metabolome
2.2.1. Variation in the Extractable Metabolome of Plants Grown under Different Watering Conditions
2.2.2. Identification of Potential Drought Biomarkers
2.3. Drought-Stress Response Dominates Metabolome in Kernel and Inner Cob Tissues
3. Materials and Methods
3.1. Plant Material Description, Cultivation and Sampling
3.1.1. Extraction Condition Experiment
3.1.2. Drought Condition Experiment
3.2. Metabolite Extraction Method
3.3. Data Collection
3.4. LC–MS-Based Analysis of Metabolome
3.4.1. Liquid Chromatography Separation Method
3.4.2. Mass Spectrometry Methods
3.5. LC–MS–MS
Mass Spectrometry Methods
3.6. Data Handling and Analysis
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lobell, D.B.; Burke, M.B.; Tebaldi, C.; Mastrandrea, M.D.; Falcon, W.P.; Naylor, R.L. Prioritizing climate change adaptation needs for food security in 2030. Science 2008, 319, 607–610. [Google Scholar] [CrossRef]
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidhuber, J.; Tubiello, F.N. Global food security under climate change. Proc. Natl Acad. Sci. USA 2007, 104, 19703–19708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Stork, L.G.; Riordan, S.G.; Ridley, W.P.; MacIsaac, S.; Halls, S.C.; Orth, R.; Rau, D.; Smith, R.G.; Wen, L.; et al. Metabolite analyses of grain from maize hybrids grown in the United States under drought and watered conditions during the 2002 field season. J. Agric. Food Chem. 2007, 55, 6169–6176. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Stork, L.G.; Riordan, S.G.; Reynolds, T.L.; Ridley, W.P.; Masucci, J.D.; MacIsaac, S.; Halls, S.C.; Orth, R.; Smith, R.G.; et al. Impact of genetics and environment on nutritional and metabolite components of maize grain. J. Agric. Food Chem. 2007, 55, 6177–6185. [Google Scholar] [CrossRef]
- Röhlig, R.M.; Eder, J.; Engel, K.H. Metabolite profiling of maize grain: Differentiation due to genetics and environment. Metabolomics 2009, 5, 459. [Google Scholar] [CrossRef]
- Witt, S.; Galicia, L.; Lisec, J.; Cairns, J.; Tiessen, A.; Araus, J.L.; Palacios-Rojas, N.; Fernie, A.R. Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Mol. Plant. 2012, 5, 401–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Fountain, J.C.; Ji, P.; Ni, X.; Chen, S.; Lee, R.D.; Kemerait, R.C.; Guo, B. Deciphering drought-induced metabolic responses and regulation in developing maize kernels. Plant Biotechnol. J. 2018, 16, 1616–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, J.A.; Erban, A.; Kopka, J.; Zörb, C. Metabolic contribution to salt stress in two maize hybrids with contrasting resistance. Plant Sci. 2015, 233, 107–115. [Google Scholar] [CrossRef]
- Sun, C.; Gao, X.; Fu, J.; Zhou, J.; Wu, X. Metabolic response of maize (Zea mays L.) plants to combined drought and salt stress. Plant Soil 2015, 388, 99–117. [Google Scholar] [CrossRef]
- Rai, V. Role of amino acids in plants responses to stresses. Biol. Plant. 2002, 45, 481–487. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Stork, L.G.; Riordan, S.G.; Reynolds, T.L.; Taylor, J.P.; Masucci, J.D.; Cao, Y.; Le Deaux, J.R.; Pandravada, A.; Glenn, K.C. Impact of environmental and genetic factors on expression of maize gene classes: Relevance to grain composition. J. Food Compos. Anal. 2009, 22, 158–164. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, S.; Davies, W.J. ABA-based chemical signalling: The co-ordination of responses to stress in plants. Plant Cell Environ. 2002, 25, 195–210. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jogawat, A.; Yadav, B.; Lakra, N.; Singh, A.K.; Narayan, O.P. Crosstalk between phytohormones and secondary metabolites in the drought stress tolerance of crop plants: A review. Physiol. Plant. 2021, 172, 1106–1132. [Google Scholar] [CrossRef]
- Savchenko, T.; Kolla, V.A.; Wang, C.Q.; Nasafi, Z.; Hicks, D.R.; Phadungchob, B.; Chehab, W.E.; Brandizzi, F.; Froehlich, J.; Dehesh, K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Phys. 2014, 164, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Fountain, J.C.; Bajaj, P.; Pandey, M.; Nayak, S.N.; Yang, L.; Kumar, V.; Jayale, A.S.; Chitikineni, A.; Zhuang, W.; Scully, B.T.; et al. Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep. 2016, 6, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar]
- Fu, J.; Sun, X.; Wang, J.; Chu, J.; Yan, C. Progress in quantitative analysis of plant hormones. Sci. Bull. 2011, 56, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2010, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Hamid, S.S.; Wakayama, M.; Soga, T.; Tomita, M. Drying and extraction effects on three edible brown seaweeds for metabolomics. J. Appl. Phycol. 2018, 30, 3335–3350. [Google Scholar] [CrossRef]
- Oikawa, A.; Otsuka, T.; Jikumaru, Y.; Yamaguchi, S.; Matsuda, F.; Nakabayashi, R.; Takashina, T.; Isuzugawa, K.; Saito, K.; Shiratake, K. Effects of freeze-drying of samples on metabolite levels in metabolome analyses. J. Sep. Sci. 2011, 34, 3561–3567. [Google Scholar] [CrossRef]
- Marti, G.; Erb, M.; Boccard, J.; Glauser, G.; Doyen, G.R.; Villard, N.; Robert, C.A.; Turlings, T.C.; Rudaz, S.; Wolfender, J.L. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant Cell Environ. 2013, 36, 621–639. [Google Scholar] [CrossRef]
- Röhlig, R.M.; Engel, K.H. Influence of the input system (conventional versus organic farming) on metabolite profiles of maize (Zea mays) kernels. J. Agric. Food Chem. 2010, 58, 3022–3030. [Google Scholar] [CrossRef]
- Zhu, D.; Kebede, B.; Chen, G.; McComb, K.; Frew, R. Impact of freeze-drying and subsequent storage on milk metabolites based on 1H NMR and UHPLC-QToF/MS. Food Control. 2020, 116, 107017. [Google Scholar] [CrossRef]
- Cheng, K.; Brunius, C.; Fristedt, R.; Landberg, R. An LC-QToF MS based method for untargeted metabolomics of human fecal samples. Metabolomics. 2020, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Cutler, A.J.; Ambrose, S.J.; Galka, M.M.; Nelson, K.M.; Squires, T.M.; Loewen, M.K.; Jadhav, A.S.; Ross, A.R.; Taylor, D.C.; et al. A new abscisic acid catabolic pathway. Plant Physiol. 2004, 134, 361–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.M.; Habermann, G.; Marchi, M.R.; Zocolo, G.J. The role of matrix effects on the quantification of abscisic acid and its metabolites in the leaves of Bauhinia variegata L. using liquid chromatography combined with tandem mass spectrometry. Braz. J. Plant Physiol. 2012, 24, 223–232. [Google Scholar] [CrossRef] [Green Version]
- Korovetska, H.; Novák, O.; Turečková, V.; Hájíčková, M.; Gloser, V. Signalling mechanisms involved in the response of two varieties of Humulus lupulus L. to soil drying: II. changes in the concentration of abscisic acid catabolites and stress-induced phytohormones. Plant Growth Regul. 2016, 78, 13–20. [Google Scholar] [CrossRef]
- Gigon, A.; Matos, A.R.; Laffray, D.; Zuily-Fodil, Y.; Pham-Thi, A.T. Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann. Bot. Lond. 2004, 94, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Wang, H.; Xin, H.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013, 9, e1003790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laxalt, A.M.; Munnik, T. Phospholipid signaling in plant defense. Curr. Opin. Plant Biol. 2002, 5, 332–338. [Google Scholar] [CrossRef]
- Xue, H.W.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem. J. 2009, 421, 145–156. [Google Scholar] [CrossRef]
- Lancashire, P.D.; Bleiholder, H.; Boom, T.V.; Langelüddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
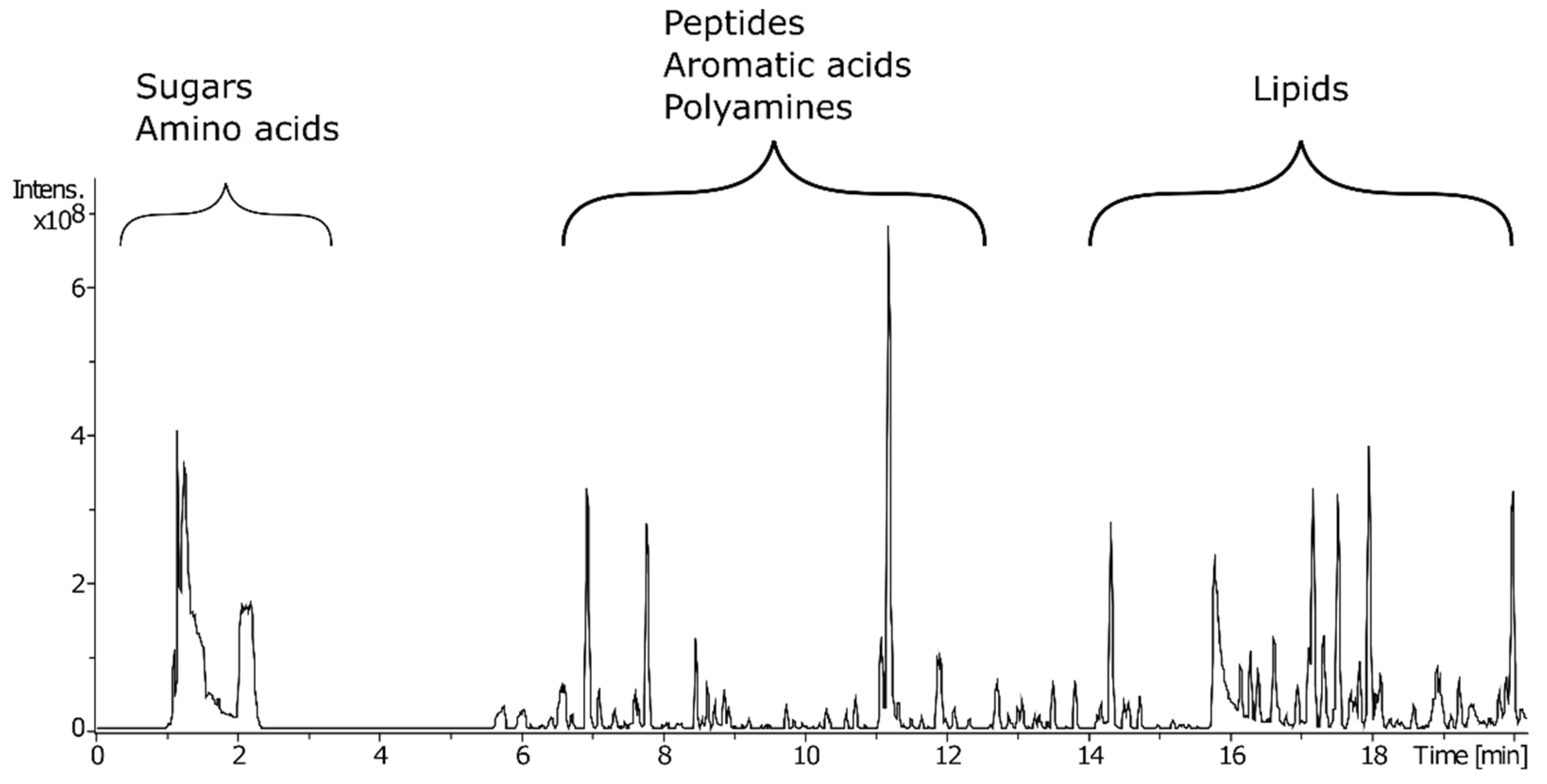
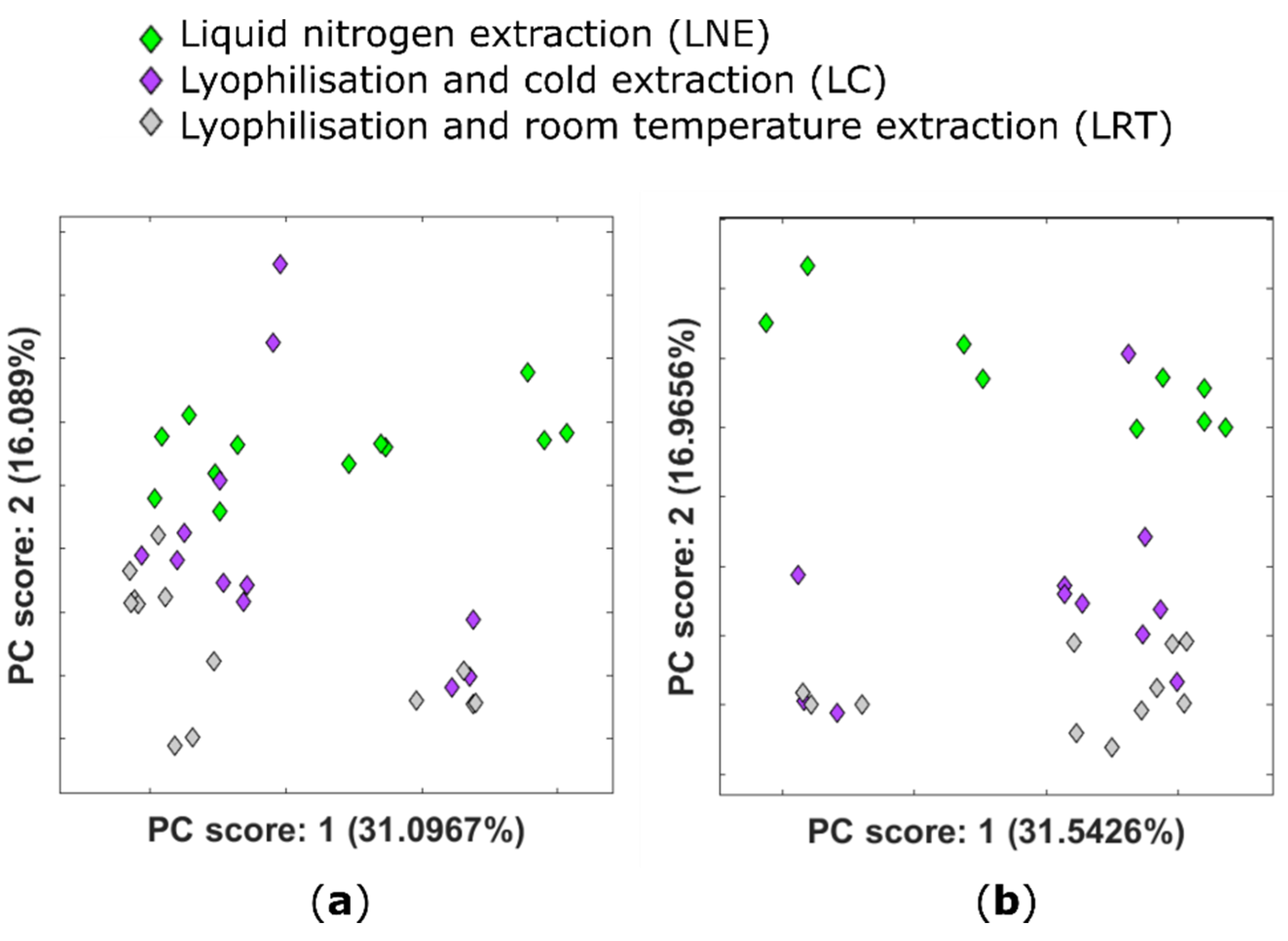

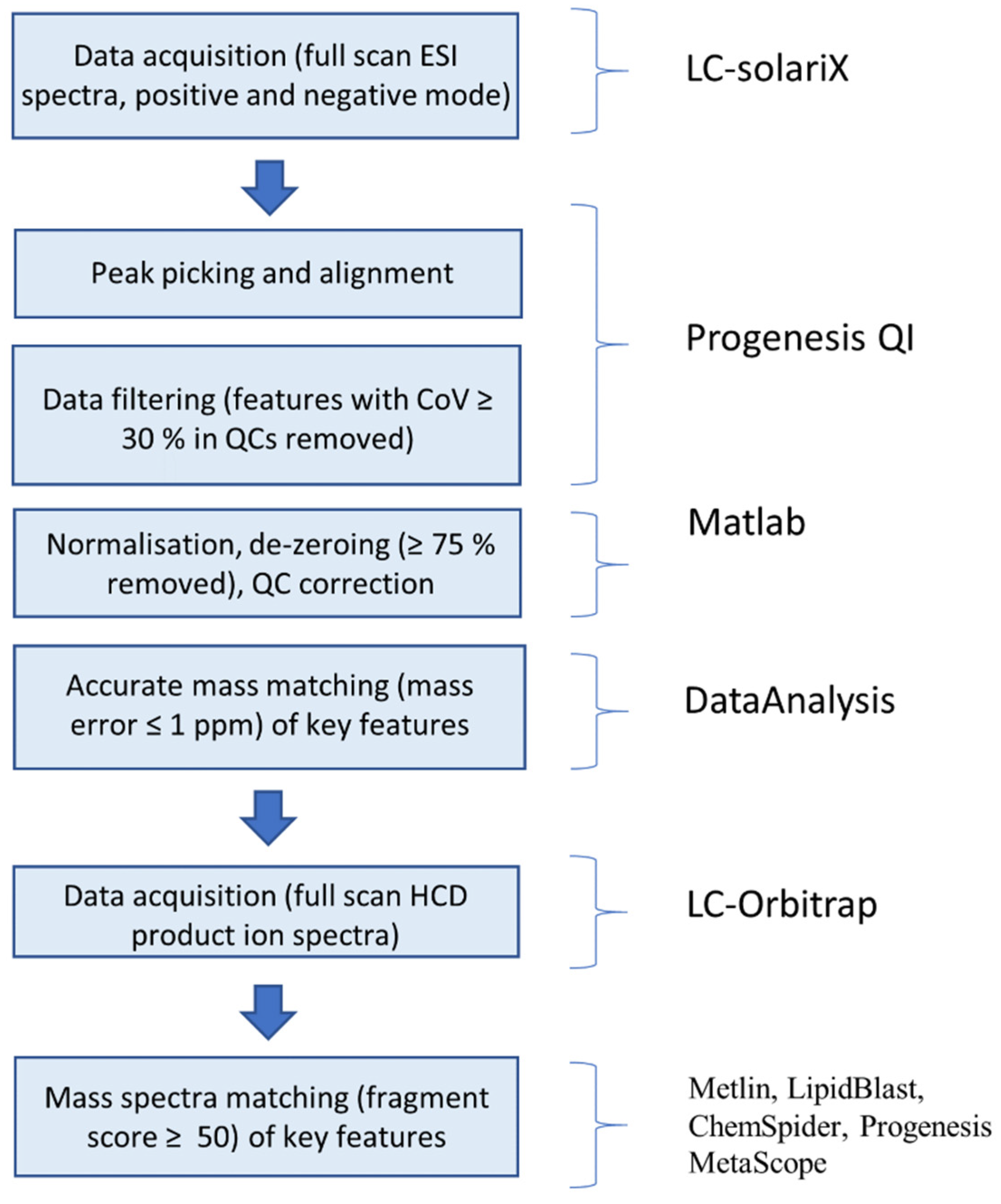

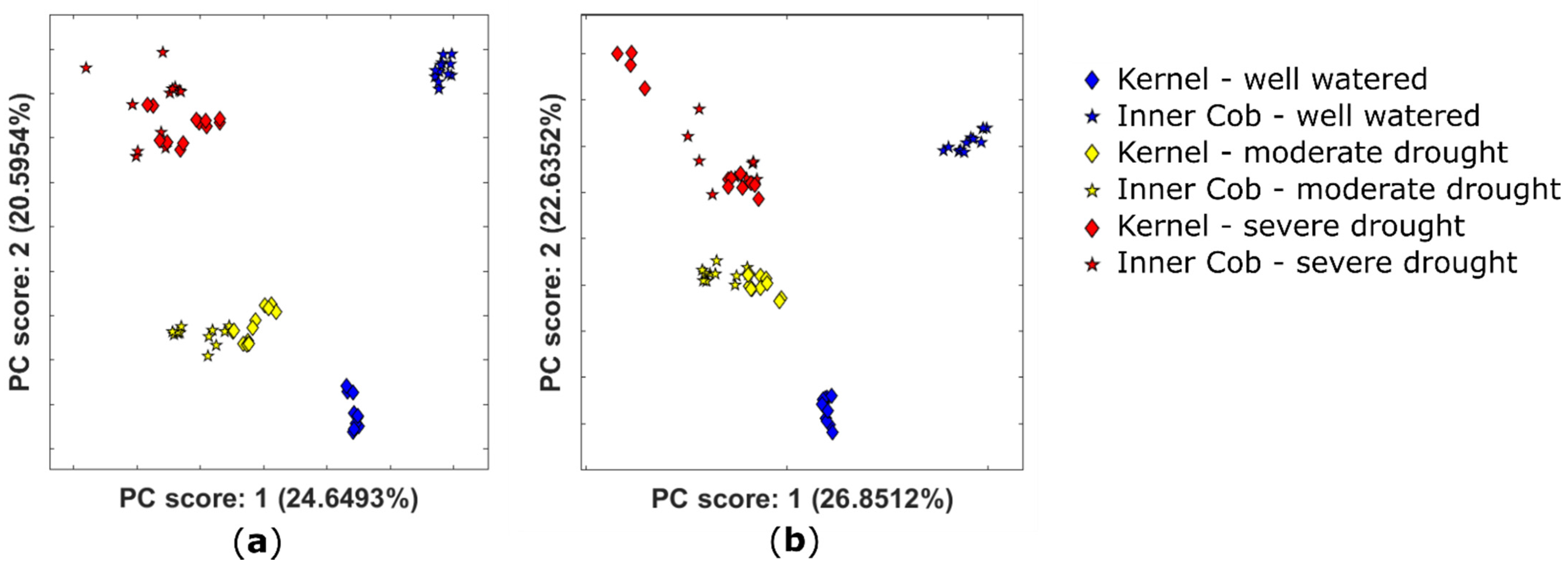
| Tissue Type | Scan Mode | m/z | tR | Molecular Species | MF | ppm | p-val | Possible Identification | Fragmentation Score |
|---|---|---|---|---|---|---|---|---|---|
| K | N * | 279.1238 | 9.52 | [M − H]− | C15H20O5 | −0.04 | 9.60 × 10−15 | Hydroxyabscisic acid or Neophaseic acid (neoPA) | 50.1 * |
| K | N * | 143.0349 | 3.88 and 3.89 | [M − H]− | C6H8O4 | 0.30 | 1.78 × 10−14 | Methyl itaconate | 58.5 |
| K | P * | 480.3075 | 18.87 | [M + H]+ | C23H46NO7P | −0.21 | 1.19 × 10−22 | PE(18:1/0:0) | 55.1 |
| K | P | 520.3398 | 17.75 | [M + H]+ | C26H50NO7P | −0.10 | 1.28 × 10−18 | PC(18:2/0:0) | 88.8 |
| K | P | 522.3556 | 18.99 | [M + H]+ | C26H52NO7P | −0.46 | 2.42 × 10−16 | PC(18:1/0:0) | 78.4 |
| IC | N | 540.3305 | 18.51 | [M + FA − H]− | C24H50NO7P | 0.37 | 1.81 × 10−8 | Lysolecithin | 63.5 |
| Condition Group | Details |
|---|---|
| Well watered | Samples were well watered for the duration of the experiment |
| Moderate drought | Drought stressed for two one-week periods. The first week starting at the three-leaf stage (approximately 4 weeks after seeding). The plants were watered for a further week and then droughted for another one-week period |
| Severe drought | Drought stressed as described for moderate drought conditions and then for a further two weeks during grain-fill (approximately 14 weeks after seeding) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaffney, I.; Sallach, J.B.; Wilson, J.; Bergström, E.; Thomas-Oates, J. Metabolomic Approaches to Studying the Response to Drought Stress in Corn (Zea mays) Cobs. Metabolites 2021, 11, 438. https://doi.org/10.3390/metabo11070438
Gaffney I, Sallach JB, Wilson J, Bergström E, Thomas-Oates J. Metabolomic Approaches to Studying the Response to Drought Stress in Corn (Zea mays) Cobs. Metabolites. 2021; 11(7):438. https://doi.org/10.3390/metabo11070438
Chicago/Turabian StyleGaffney, Isabella, Jonathan Brett Sallach, Julie Wilson, Edmund Bergström, and Jane Thomas-Oates. 2021. "Metabolomic Approaches to Studying the Response to Drought Stress in Corn (Zea mays) Cobs" Metabolites 11, no. 7: 438. https://doi.org/10.3390/metabo11070438
APA StyleGaffney, I., Sallach, J. B., Wilson, J., Bergström, E., & Thomas-Oates, J. (2021). Metabolomic Approaches to Studying the Response to Drought Stress in Corn (Zea mays) Cobs. Metabolites, 11(7), 438. https://doi.org/10.3390/metabo11070438






