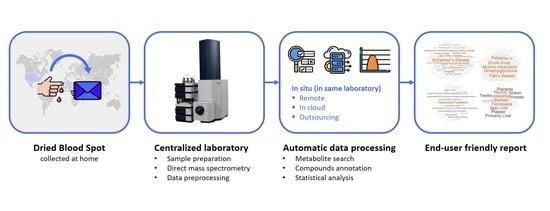
Metabolomic Laboratory-Developed Tests: Current Status and Perspectives
Abstract
1. Introduction
2. History of LDT
3. Metabolomic LDT
4. Metabolomic LDT vs. Metabolomics Study
- There are no government regulations for typical scientific metabolomic studies, and they are conducted solely for research purposes and cannot be commercially scaled up;
- The cost of the scientific metabolomic study is prohibitive for ordinary people, and often such research is funded by public or private grants. Furthermore, the so-called metabolomic case studies are quite rare, which is closest to a personalized metabolomic study, but this is not a scalable research model [70,71];
- The scientific metabolomic study is difficult to reproduce. It is carried out to discover new phenomena and acquire new knowledge. It is based on the experience of a specific research team and reflects the experience and knowledge of only specific people;
- The timing of scientific metabolomic studies can range from several months to several years, which limits their availability for rapid diagnosis;
- The presentation of data in the scientific metabolomic study is available only to scientists in the field. Most people do not have access to data interpretation in scientific metabolomic research due to their specificity and complexity.
- The LDT format simplifies the implementation of metabolomics-based tests, turning protocols and standardization actions into routines of one laboratory, which is regulated to some degree by the local rules;
- Metabolomic LDT can be implemented in the “direct to customer” format since mass spectrometric measurements in metabolomic LDT are compatible with dried blood spot (DBS) samples [72], and there are also methods for simple collection of capillary blood without assistance at home and its subsequent transportation to the laboratory by mail or using a specialized courier service, which makes LDT convenient for customers and available almost everywhere. The LDT results are user-friendly, making them acceptable to a wide range of customers.
- Any educated person can be the end-user of metabolomic LDT. LDT results are available to a wide range of clients: physicians and their patients, researchers, citizen scientists, and any educated person. The process of interpreting complex metabolomic data can be automated, and the data can be presented in an accessible format [22];
- Unlike scientific metabolomic research, LDT can be reproduced an unlimited number of times according to the created workflow (Figure 1) within one laboratory and can be used by people for independent research of the state of health of their body and regular monitoring of their health, which has a pronounced applied value in the modern world [22];
- The method of direct mass spectrometry of blood plasma, based on which it is possible to implement metabolomic LDT, is characterized by a high processing speed and relatively high reproducibility and was also widely used in metabolomics, in particular, in the laboratory where LDT was developed for the study of cancer, diabetes [73], and Parkinson’s disease [74]. The processing of mass spectrometric data was specially developed for high-resolution mass spectra and was successfully used for many years in studies of blood plasma [75], and now it is implemented in the LDT format [22];
- In the LDT format, various options for diagnosing human health can be implemented, for example, as “confirmation of a person’s healthy state”, “score-based diagnostics”, and “disease diagnosis based on metabolite set overrepresentation” [22]. In the future, it is possible to create new and more adaptive methods for diagnosing the state of human health;
- Due to the potentially large number of tests within one laboratory, the cost of metabolomic LDT is expected to be quite low and acceptable for most people.
5. Analytical Limitations
6. Legal Aspects: LDT Regulation and Logistics
6.1. LDT Regulation in the United States
6.2. LDT Regulation in the European Union
6.3. LDT Regulation in Singapore
6.4. International Logistics of DBS Samples
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Marciano, D.P.; Snyder, M.P. Personalized Metabolomics. Methods Mol. Biol. 2019, 1978, 447–456. [Google Scholar] [CrossRef]
- Trifonova, O.; Lokhov, P.; Archakov, A. Postgenomics diagnostics: Metabolomics approaches to human blood profiling. OMICS 2013, 17, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; Carpenter, G.; So, P.-W. Salivary Metabolomics: From Diagnostic Biomarker Discovery to Investigating Biological Function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef]
- Wang, F.-X.; Chen, K.; Huang, F.-Q.; Alolga, R.N.; Ma, J.; Wu, Z.-X.; Fan, Y.; Ma, G.; Guan, M. Cerebrospinal fluid-based metabolomics to characterize different types of brain tumors. J. Neurol. 2020, 267, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Evolution of Translational Omics: Lessons Learned and the Path Forward; Micheel, C.M., Nass, S.J., Omenn, G.S., Eds.; The National Academies Press: Washington, DC, USA, 2012; ISBN 978-0-309-22418-5. [Google Scholar]
- Nass, S.J.; Moses, H.L. Cancer Biomarkers: The Promises and Challenges of Improving Detection and Treatment; National Academies Press: Washington, DC, USA, 2007; ISBN 9780309667111. [Google Scholar]
- FDA. Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs). Draft Guidance. Available online: https://www.fda.gov/media/89841/download (accessed on 5 March 2021).
- FDA. Laboratory Developed Tests. Available online: https://www.fda.gov/medical-devices/vitro-diagnostics/laboratory-developed-tests (accessed on 5 March 2021).
- Genzen, J.R. Regulation of Laboratory-Developed Tests: A Clinical Laboratory Perspective. Am. J. Clin. Pathol. 2019, 152, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Sharfstein, J. FDA Regulation of Laboratory-Developed Diagnostic Tests: Protect the Public, Advance the Science. JAMA 2015, 313, 667–668. [Google Scholar] [CrossRef]
- Schreier, J.; Feeney, R.; Keeling, P. Diagnostics Reform and Harmonization of Clinical Laboratory Testing. J. Mol. Diagn. 2019, 21, 737–745. [Google Scholar] [CrossRef]
- Burchard, P.R.; Abou Tayoun, A.N.; Lefferts, J.A.; Lewis, L.D.; Tsongalis, G.J.; Cervinski, M.A. Development of a rapid clinical TPMT genotyping assay. Clin. Biochem. 2014, 47, 126–129. [Google Scholar] [CrossRef]
- Munari, E.; Zamboni, G.; Lunardi, G.; Marconi, M.; Brunelli, M.; Martignoni, G.; Netto, G.J.; Quatrini, L.; Vacca, P.; Moretta, L.; et al. PD-L1 expression in non-small cell lung cancer: Evaluation of the diagnostic accuracy of a laboratory-developed test using clone E1L3N in comparison with 22C3 and SP263 assays. Hum. Pathol. 2019, 90, 54–59. [Google Scholar] [CrossRef]
- Fiset, P.O.; Labbé, C.; Young, K.; Craddock, K.J.; Smith, A.C.; Tanguay, J.; Pintilie, M.; Wang, R.; Torlakovic, E.; Cheung, C.; et al. Anaplastic lymphoma kinase 5A4 immunohistochemistry as a diagnostic assay in lung cancer: A Canadian reference testing center’s results in population-based reflex testing. Cancer 2019, 125, 4043–4051. [Google Scholar] [CrossRef]
- Tinawi-Aljundi, R.; King, L.; Knuth, S.T.; Gildea, M.; Ng, C.; Kahl, J.; Dion, J.; Young, C.; Schervish, E.W.; Frontera, J.R.; et al. One-year monitoring of an oligonucleotide fluorescence in situ hybridization probe panel laboratory-developed test for bladder cancer detection. Res. Rep. Urol. 2015, 7, 49–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brukner, I.; Eintracht, S.; Forgetta, V.; Papadakis, A.I.; Spatz, A.; Oughton, M. Laboratory-developed test for detection of acute Clostridium difficile infections with the capacity for quantitative sample normalization. Diagn. Microbiol. Infect. Dis. 2019, 95, 113–118. [Google Scholar] [CrossRef]
- Kulis-Horn, R.K.; Tiemann, C. Evaluation of a laboratory-developed test for simultaneous detection of norovirus and rotavirus by real-time RT-PCR on the Panther Fusion® system. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Calvert, J.; Saber, N.; Hoffman, J.; Das, R. Machine-Learning-Based Laboratory Developed Test for the Diagnosis of Sepsis in High-Risk Patients. Diagnostics 2019, 9, 20. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Trifonova, O.P.; Maslov, D.L.; Lichtenberg, S.; Balashova, E.E. Diagnosis of Parkinson’s Disease by A Metabolomics-Based Laboratory-Developed Test (LDT). Diagnostics 2020, 10, 332. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Maslov, D.L.; Lichtenberg, S.; Trifonova, O.P.; Balashova, E.E. Holistic Metabolomic Laboratory-Developed Test (LDT): Development and Use for the Diagnosis of Early-Stage Parkinson’s Disease. Metabolites 2020, 11, 14. [Google Scholar] [CrossRef]
- Centers for Medicare and Medicaid Services. Background Document on CLIA Oversight of LDTs. Available online: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/Downloads/LDT-and-CLIA_FAQs.pdf (accessed on 6 March 2021).
- FDA. The Public Health Evidence for FDA Oversight of Laboratory Developed Tests: 20 Case Studies—The Real and Potential Harms to Patients and to Public Health from Certain Laboratory Developed Tests (LDTs). Available online: http://wayback.archive-it.org/7993/20171115144712/https:/www.fda.gov/downloads/AboutFDA/ReportsManualsForms/Reports/UCM472777.pdf (accessed on 6 March 2021).
- Gustavsson, I.; Aarnio, R.; Myrnäs, M.; Hedlund-Lindberg, J.; Taku, O.; Meiring, T.; Wikström, I.; Enroth, S.; Williamson, A.-L.; Olovsson, M.; et al. Clinical validation of the HPVIR high-risk HPV test on cervical samples according to the international guidelines for human papillomavirus DNA test requirements for cervical cancer screening. Virol. J. 2019, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, S.; Nayak, S.; Williams, T.; di Santa Maria, F.S.; Guedes, M.S.; Chaves, R.C.; Linder, V.; Marques, A.R.; Horn, E.J.; Wong, S.J.; et al. A Multiplexed Serologic Test for Diagnosis of Lyme Disease for Point-of-Care Use. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Lochman, I.; Pokorná, L.; Mertová, H. Serological diagnosis of whooping cough using immunoblot methods. Epidemiol. Mikrobiol. Imunol. Cas. Spol. pro Epidemiol. a Mikrobiol. Ces. Lek. Spol. J.E. Purkyne 2017, 66, 107–114. [Google Scholar]
- Ingles, J.; Macciocca, I.; Morales, A.; Thomson, K. Genetic Testing in Inherited Heart Diseases. Heart. Lung Circ. 2020, 29, 505–511. [Google Scholar] [CrossRef]
- Lebo, M.S.; Baxter, S.M. New molecular genetic tests in the diagnosis of heart disease. Clin. Lab. Med. 2014, 34, 137–156. [Google Scholar] [CrossRef]
- FDA. Discussion Paper on Laboratory Developed Tests (LDTs). Available online: https://www.fda.gov/media/102367/download (accessed on 6 March 2021).
- FDA. The FDA Warns against the Use of Many Genetic Tests with Unapproved Claims to Predict Patient Response to Specific Medications. Available online: https://www.fda.gov/medical-devices/safety-communications/fda-warns-against-use-many-genetic-tests-unapproved-claims-predict-patient-response-specific (accessed on 6 March 2021).
- FDA. FDA Issues Warning Letter to Genomics Lab for Illegally Marketing Genetic Test That Claims to Predict Patients’ Responses to Specific Medications. Available online: https://www.fda.gov/news-events/press-announcements/fda-issues-warning-letter-genomics-lab-illegally-marketing-genetic-test-claims-predict-patients (accessed on 9 March 2021).
- HHS. Rescission of Guidances and Other Informal Issuances Concerning Premarket Review of Laboratory Developed Tests. Available online: https://www.hhs.gov/coronavirus/testing/recission-guidances-informal-issuances-premarket-review-lab-tests/index.html (accessed on 9 March 2021).
- FDA. In Vitro Diagnostics EUAs. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas (accessed on 13 March 2021).
- Deloitte Insights. 2021 Global Health Care Outlook. Available online: https://www2.deloitte.com/global/en/pages/life-sciences-and-healthcare/articles/global-health-care-sector-outlook.html (accessed on 13 March 2021).
- Huang, W.; Li, J.; Alem, L. Towards Preventative Healthcare: A Review of Wearable and Mobile Applications. Stud. Health Technol. Inform. 2018, 251, 11–14. [Google Scholar]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef]
- Yu, K.-H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Truong, T.; Gilbank, P.; Johnson-Cover, K.; Ieraci, A. A Framework for Applied AI in Healthcare. Stud. Health Technol. Inform. 2019, 264, 1993–1994. [Google Scholar] [CrossRef]
- Noorbakhsh-Sabet, N.; Zand, R.; Zhang, Y.; Abedi, V. Artificial Intelligence Transforms the Future of Health Care. Am. J. Med. 2019, 132, 795–801. [Google Scholar] [CrossRef]
- Chen, M.; Decary, M. Artificial intelligence in healthcare: An essential guide for health leaders. Healthc. Manag. Forum 2020, 33, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Vaishya, R.; Javaid, M.; Khan, I.H.; Haleem, A. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab. Syndr. 2020, 14, 337–339. [Google Scholar] [CrossRef]
- Borsky, A.; Zhan, C.; Miller, T.; Ngo-Metzger, Q.; Bierman, A.S.; Meyers, D. Few Americans Receive All High-Priority, Appropriate Clinical Preventive Services. Health Aff. 2018, 37, 925–928. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. Healthy People 2030. Available online: https://health.gov/healthypeople/objectives-and-data/browse-objectives/preventive-care (accessed on 16 March 2021).
- Trivedi, D.K.; Hollywood, K.A.; Goodacre, R. Metabolomics for the masses: The future of metabolomics in a personalized world. New Horiz. Transl. Med. 2017, 3, 294–305. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites 2019, 9, 108. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Macel, M.; Van Dam, N.M.; Keurentjes, J.J.B. Metabolomics: The chemistry between ecology and genetics. Mol. Ecol. Resour. 2010, 10, 583–593. [Google Scholar] [CrossRef]
- Perez De Souza, L.; Alseekh, S.; Brotman, Y.; Fernie, A.R. Network-based strategies in metabolomics data analysis and interpretation: From molecular networking to biological interpretation. Expert Rev. Proteom. 2020, 17, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.; Gall, W.; Adam, K.-P.; Nakhle, P.; Button, E.; Hathorn, J.; Lawton, K.; Milburn, M.; Perichon, R.; Mitchell, M.; et al. A novel fasting blood test for insulin resistance and prediabetes. J. Diabetes Sci. Technol. 2013, 7, 100–110. [Google Scholar] [CrossRef]
- Jelonek, K.; Widłak, P. Metabolome-based biomarkers: Their potential role in the early detection of lung cancer. Contemp. Oncol. 2018, 22, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.Y.; Wu, H.-J.; Menon, S.S.; Fallah, Y.; Zhong, X.; Rizk, N.; Unger, K.; Mapstone, M.; Fiandaca, M.S.; Federoff, H.J.; et al. Metabolomic biomarkers of pancreatic cancer: A meta-analysis study. Oncotarget 2017, 8, 68899–68915. [Google Scholar] [CrossRef]
- Ellis, D.I.; Dunn, W.B.; Griffin, J.L.; Allwood, J.W.; Goodacre, R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 2007, 8, 1243–1266. [Google Scholar] [CrossRef]
- Suman, S.; Sharma, R.K.; Kumar, V.; Sinha, N.; Shukla, Y. Metabolic fingerprinting in breast cancer stages through (1)H NMR spectroscopy-based metabolomic analysis of plasma. J. Pharm. Biomed. Anal. 2018, 160, 38–45. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Wilson, I.D. Metabolic Profiling: Status, Challenges, and Perspective. Methods Mol. Biol. 2018, 1738, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Mass spectrometry-based metabolomics diagnostics—Myth or reality? Expert Rev. Proteom. 2021, 1–6. [Google Scholar] [CrossRef]
- Haznadar, M.; Cai, Q.; Krausz, K.W.; Bowman, E.D.; Margono, E.; Noro, R.; Thompson, M.D.; Mathé, E.A.; Munro, H.M.; Steinwandel, M.D.; et al. Urinary Metabolite Risk Biomarkers of Lung Cancer: A Prospective Cohort Study. Cancer Epidemiol. Biomark. Prev. 2016, 25, 978–986. [Google Scholar] [CrossRef]
- Mu, Y.; Zhou, Y.; Wang, Y.; Li, W.; Zhou, L.; Lu, X.; Gao, P.; Gao, M.; Zhao, Y.; Wang, Q.; et al. Serum Metabolomics Study of Nonsmoking Female Patients with Non-Small Cell Lung Cancer Using Gas Chromatography-Mass Spectrometry. J. Proteome Res. 2019, 18, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Jin, S.; Zhang, J.; Chen, M.; Xia, Y.; Shu, Y.; Guo, R. Cortisol, cortisone, and 4-methoxyphenylacetic acid as potential plasma biomarkers for early detection of non-small cell lung cancer. Int. J. Biol. Markers 2018, 33, 314–320. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, Z.; Zhong, J.; Li, L.; Min, L.; Xu, L.; Li, H.; Zhang, J.; Wu, W.; Dai, L. Simultaneous quantification of serum monounsaturated and polyunsaturated phosphatidylcholines as potential biomarkers for diagnosing non-small cell lung cancer. Sci. Rep. 2018, 8, 7137. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Ciborowski, M.; Kisluk, J.; Kretowski, A.; Barbas, C. Mass spectrometry based proteomics and metabolomics in personalized oncology. Biochim. Biophys. Acta. Mol. Basis Dis. 2020, 1866, 165690. [Google Scholar] [CrossRef] [PubMed]
- López-López, Á.; López-Gonzálvez, Á.; Barker-Tejeda, T.C.; Barbas, C. A review of validated biomarkers obtained through metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Considine, E.C. The Search for Clinically Useful Biomarkers of Complex Disease: A Data Analysis Perspective. Metabolites 2019, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Metabolon. Metabolon Launches Meta UDxTM Test to Speed Diagnosis of Rare and Undiagnosed Diseases in Children and Adults. Available online: https://www.metabolon.com/metabolon-launches-meta-udx-test-to-speed-diagnosis-of-rare-and-undiagnosed-diseases-in-children-and-adults/ (accessed on 20 March 2021).
- Kell, D.B.; Oliver, S.G. The metabolome 18 years on: A concept comes of age. Metabolomics 2016, 12, 148. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Peccatori, F.A.; Codacci-Pisanelli, G.; Mellgren, G.; Buonomo, B.; Baldassarre, E.; Lien, E.A.; Bifulco, E.; Hustad, S.; Zachariassen, E.; Johansson, H.; et al. First-in-human pharmacokinetics of tamoxifen and its metabolites in the milk of a lactating mother: A case study. ESMO Open 2020, 5, e000859. [Google Scholar] [CrossRef]
- O’Shea, K.; Cameron, S.J.S.; Lewis, K.E.; Lu, C.; Mur, L.A.J. Metabolomic-based biomarker discovery for non-invasive lung cancer screening: A case study. Biochim. Biophys. Acta 2016, 1860, 2682–2687. [Google Scholar] [CrossRef]
- Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Evaluation of Dried Blood Spot Sampling for Clinical Metabolomics: Effects of Different Papers and Sample Storage Stability. Metabolites 2019, 9, 277. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Archakov, A.I.; Shestakova, E.A.; Shestakova, M.V.; Dedov, I.I. Diagnosing impaired glucose tolerance using direct infusion mass spectrometry of blood plasma. PLoS ONE 2014, 9, e105343. [Google Scholar] [CrossRef] [PubMed]
- Balashova, E.E.; Lokhov, P.G.; Maslov, D.L.; Trifonova, O.P.; Khasanova, D.M.; Zalyalova, Z.A.; Nigmatullina, R.R.; Ugrumov, A.I.A. and M. V Plasma Metabolome Signature in Patients with Early-stage Parkinson Disease. Curr. Metab. 2018, 6, 75–82. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Maslov, D.L.; Kharibin, O.N.; Balashova, E.E.; Archakov, A.I. Label-free data standardization for clinical metabolomics. BioData Min. 2017, 10, 10. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Ponomarenko, E.A.; Archakov, A.I. Mass Spectrometry-Based Metabolomics Analysis of Obese Patients’ Blood Plasma. Int. J. Mol. Sci. 2020, 21, 568. [Google Scholar] [CrossRef]
- Naryshkin, S.; Austin, R.M. Limitations of widely used high-risk human papillomavirus laboratory-developed testing in cervical cancer screening. Drug. Healthc. Patient Saf. 2012, 4, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Brennan, L.; Fiehn, O.; Hankemeier, T.; Kristal, B.S.; van Ommen, B.; Pujos-Guillot, E.; Verheij, E.; Wishart, D.; Wopereis, S. Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 2009, 5, 435. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, A.; Miao, J.; Sun, H.; Han, Y.; Yan, G.; Wu, F.; Wang, X. Metabolomics biotechnology, applications, and future trends: A systematic review. RSC Adv. 2019, 9, 37245–37257. [Google Scholar] [CrossRef]
- Johnson, C.H.; Gonzalez, F.J. Challenges and opportunities of metabolomics. J. Cell. Physiol. 2012, 227, 2975–2981. [Google Scholar] [CrossRef]
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.D.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef]
- Gowda, G.A.N.; Djukovic, D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods Mol. Biol. 2014, 1198, 3–12. [Google Scholar] [CrossRef]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef]
- Nassar, A.F.; Wu, T.; Nassar, S.F.; Wisnewski, A. V UPLC-MS for metabolomics: A giant step forward in support of pharmaceutical research. Drug Discov. Today 2017, 22, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Yu, S.; Ronis, M.J.; Badger, T.M. Matrix effects break the LC behavior rule for analytes in LC-MS/MS analysis of biological samples. Exp. Biol. Med. 2015, 240, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Heyman, H.M. Mass Spectrometry-Based Metabolomics. Methods Mol. Biol. 2018, 1775, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.V.; Schmitt-Kopplin, P. Capillary Electrophoresis in Metabolomics. Methods Mol. Biol. 2016, 1483, 437–470. [Google Scholar] [CrossRef]
- Sasaki, K.; Sagawa, H.; Suzuki, M.; Yamamoto, H.; Tomita, M.; Soga, T.; Ohashi, Y. Metabolomics Platform with Capillary Electrophoresis Coupled with High-Resolution Mass Spectrometry for Plasma Analysis. Anal. Chem. 2019, 91, 1295–1301. [Google Scholar] [CrossRef]
- Zhang, W.; Ramautar, R. CE-MS for metabolomics: Developments and applications in the period 2018–2020. Electrophoresis 2021, 42, 381–401. [Google Scholar] [CrossRef]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Qiu, S.; Wang, X. Metabolomics insights into pathophysiological mechanisms of nephrology. Int. Urol. Nephrol. 2014, 46, 1025–1030. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, A.; Song, Q.; Fang, H.; Liu, X.; Su, J.; Yang, L.; Yu, M.; Wang, X. Functional metabolomics discover pentose and glucuronate interconversion pathways as promising targets for Yang Huang syndrome treatment with Yinchenhao Tang. RSC Adv. 2018, 8, 36831–36839. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Xu, H.; Qiu, S.; Wang, X. Cell metabolomics. OMICS 2013, 17, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, J.A.; Brennan, L.; Broadhurst, D.; Fiehn, O.; Cascante, M.; Dunn, W.B.; Schmidt, M.A.; Velagapudi, V. Preanalytical Processing and Biobanking Procedures of Biological Samples for Metabolomics Research: A White Paper, Community Perspective (for “Precision Medicine and Pharmacometabolomics Task Group”—The Metabolomics Society Initiative). Clin. Chem. 2018, 64, 1158–1182. [Google Scholar] [CrossRef]
- Gong, Z.-G.; Hu, J.; Wu, X.; Xu, Y.-J. The Recent Developments in Sample Preparation for Mass Spectrometry-Based Metabolomics. Crit. Rev. Anal. Chem. 2017, 47, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Villas-Boas, S.G.; Aggio, R. Analysis of Intracellular Metabolites from Microorganisms: Quenching and Extraction Protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lv, H.; Qiu, S.; Gao, L.; Ai, H. Plasma metabolic profiling and novel metabolite biomarkers for diagnosing prostate cancer. RSC Adv. 2017, 7, 30060–30069. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Han, Y.; Wang, X. Metabolomics in diagnosis and biomarker discovery of colorectal cancer. Cancer Lett. 2014, 345, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Lynn, K.-S.; Cheng, M.-L.; Chen, Y.-R.; Hsu, C.; Chen, A.; Lih, T.M.; Chang, H.-Y.; Huang, C.; Shiao, M.-S.; Pan, W.-H.; et al. Metabolite identification for mass spectrometry-based metabolomics using multiple types of correlated ion information. Anal. Chem. 2015, 87, 2143–2151. [Google Scholar] [CrossRef]
- Kwak, M.; Kang, K.; Wang, Y. Methods of Metabolite Identification Using MS/MS Data. J. Comput. Inf. Syst. 2019, 1–7. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Ulmer, C.Z.; Jones, C.M.; Yost, R.A.; Bowden, J.A. Common cases of improper lipid annotation using high-resolution tandem mass spectrometry data and corresponding limitations in biological interpretation. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2017, 1862, 766–770. [Google Scholar] [CrossRef]
- Vaysse, P.-M.; Heeren, R.M.A.; Porta, T.; Balluff, B. Mass spectrometry imaging for clinical research—latest developments, applications, and current limitations. Analyst 2017, 142, 2690–2712. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, K.; Misra, B.B. Software tools, databases and resources in metabolomics: Updates from 2018 to 2019. Metabolomics 2020, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Department of Health (Wadsworth Center). Test Approval. Available online: https://www.wadsworth.org/regulatory/clep/clinical-labs/obtain-permit/test-approval (accessed on 5 April 2021).
- Congressional Research Service. FDA Regulation of Laboratory-Developed Tests (LDTs). Available online: https://crsreports.congress.gov/product/pdf/IF/IF11389 (accessed on 5 April 2021).
- 116th CONGRESS 2d Session. Verifying Accurate Leading-Edge IVCT Development Act. Available online: https://www.congress.gov/bill/116th-congress/senate-bill/3404/text (accessed on 5 March 2020).
- Directive 98/79/EC of the European Parliament and of the Council on In Vitro Diagnostic Medical Devices (IVDMD). Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:01998L0079-20120111 (accessed on 5 April 2021).
- Regulation (EU) 2017/746 of the European Parliament and of the Council of 5 April 2017 on In Vitro Diagnostic Medical Devices. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02017R0746-20170505 (accessed on 5 April 2021).
- Health Products Act (HPA). Singapore. Available online: https://sso.agc.gov.sg/Act/HPA2007 (accessed on 5 April 2021).
- Health Products (Medical Devices) Regulations 2010. Singapore. Available online: https://sso.agc.gov.sg/SL/HPA2007-S436-2010 (accessed on 5 April 2021).
- Health Sciences Authority. Risk Classification of Medical Devices. Singapore. Available online: https://www.hsa.gov.sg/medical-devices/registration/risk-classification-rule (accessed on 5 April 2021).
- Health Sciences Authority. Class B Medical Device Registration via Full Route. Singapore. Available online: https://www.hsa.gov.sg/medical-devices/registration/guides/class-b-full-registration (accessed on 8 April 2021).
- Health Sciences Authority. Priority Review Scheme. Singapore. Available online: https://www.hsa.gov.sg/medical-devices/registration/priority-review-scheme (accessed on 8 April 2021).
- Health Sciences Authority. SINGAPORE MEDICAL DEVICE REGISTER (SMDR). Singapore. Available online: https://eservice.hsa.gov.sg/medics/md/mdEnquiry.do?action=getAllDevices (accessed on 8 April 2021).
- Health Sciences Authority. Dealer’s Licence. Singapore. Available online: https://www.hsa.gov.sg/medical-devices/dealers-licence/apply (accessed on 8 April 2021).
- Centers for Disease Control and Prevention. Shipping Guidelines for Dried-Blood Spot Specimens. Available online: https://www.cdc.gov/labstandards/pdf/nsqap/Bloodspot_Transportation_Guidelines.pdf (accessed on 21 April 2021).
- USPS. Publication 52, Hazardous, Restricted, and Perishable Mail. Toxic Substances and Infectious Substances (Hazard Class 6). Available online: https://pe.usps.com/text/pub52/pub52c3_024.htm (accessed on 21 April 2021).
- FedEx. International Countries/Territories: Dangerous Goods Acceptance. Available online: https://www.fedex.com/en-us/service-guide/dangerous-goods/international-locations.html (accessed on 21 April 2021).
- UPS. Approved Countries or Territories List. International Dangerous Goods (IDG) Authorized Origins and Destinations. Available online: https://www.ups.com/us/en/help-center/packaging-and-supplies/special-care-shipments/international-dangerous-goods/approved-countries.page (accessed on 21 April 2021).

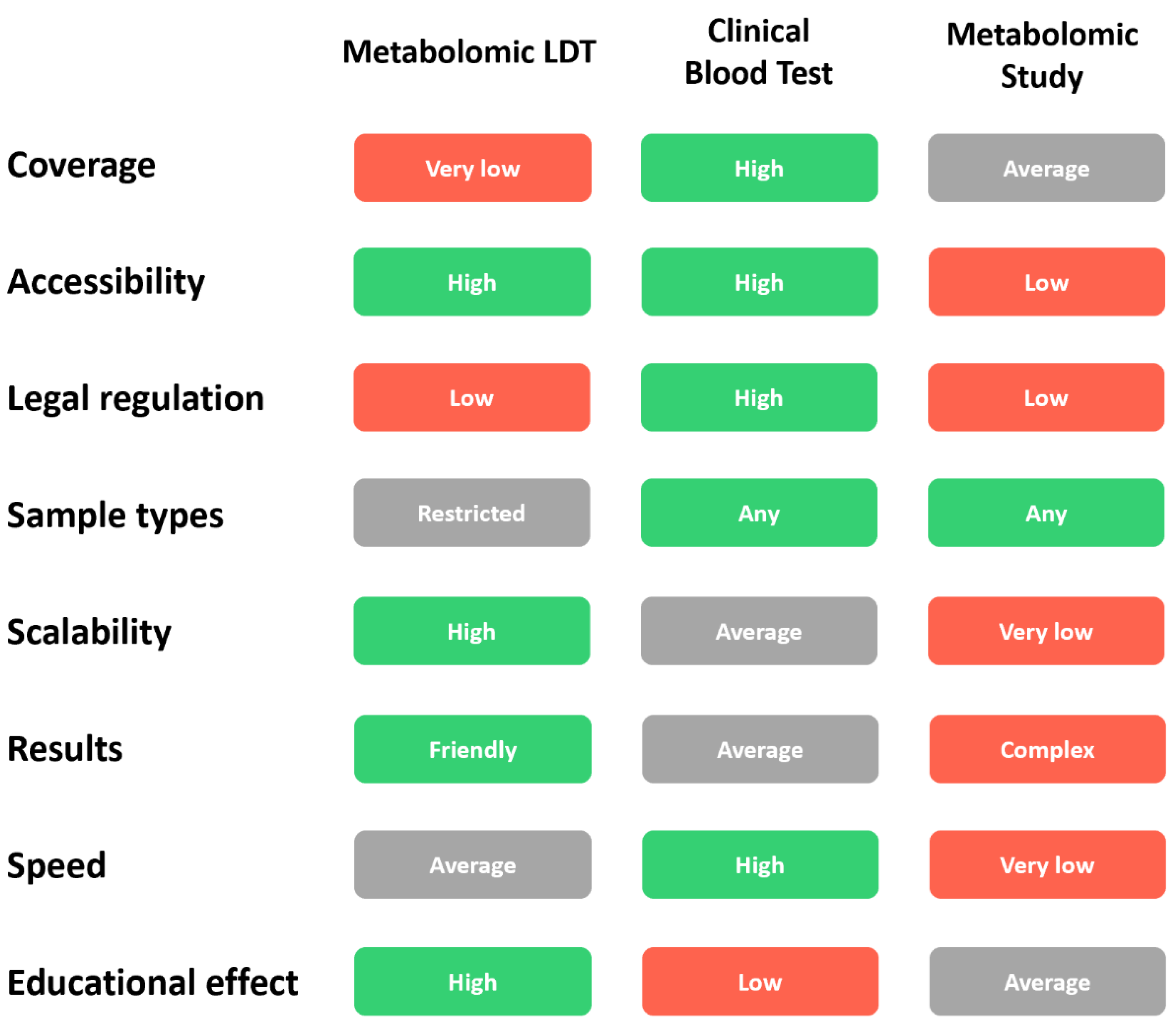
| Parameter | Metabolomic LDT | Clinical Blood Test | Metabolomics Study |
|---|---|---|---|
| Accessibility | Convenient logistics, availability of test kits, understandable test results. | Many clinical laboratories, understandable results for doctors, and relatively understandable results for people. | Available to scientists. Not publicly available to people. |
| Legal regulation | Moderate regulation. | Strict regulation. | Generally not regulated. |
| Samples | Dried Blood Spot (DBS). Small volume, easy collection, the ability to collect capillary blood from a finger at home. Convenience for children. | Venous blood. Large volume. Blood sampling only in the laboratory with the help of a physician. Inconvenience for children. | Generally, blood plasma with a particular anticoagulant (EDTA) is suitable. |
| Scalability | Good and simple scalability due to the sale of test kits and the possibility of further transportation of DBS samples through the postal service even to another country. | Moderate scalability. Expansion requires opening new laboratories, purchasing expensive equipment, and going through certification and regulatory procedures each time. | Not scalable. Each scientific laboratory is unique. |
| Test output (result, report) | Understandable to most people. In one analysis, it is possible to analyze many metabolomic pathways, confirm general health status, or identify potential abnormalities at the deep molecular. | Understandable to doctors. To analyze many analytes, a large volume of blood is required, and many separate tests are needed. It is expensive for comprehensive health diagnosis. | The results of scientific research are very complex and are intended for scientists with experience in the same field of science. |
| Speed (time expenditure) | Fast testing speed due to the automation of all processes. Testing speed may change in the case of sending DBS samples to another country. | High testing speed due to the presence of laboratories in the city. The speed may increase due to the amount of analyzed biomaterial. | Very low. Scientific study can take from several months to several years. |
| Cost | Moderate. The sample processing rate is relatively low due to their sequential processing by mass spectrometers. | Low for one or more measured parameters. Moderate when measuring a large number of parameters. | Very high. Individual design of each study, highly qualified staff, a large volume of work, long-term implementation. |
| Educational effect | Any interested citizen can engage in in-depth research of his body using Metabolomic LDT, thereby receiving personalized data and being able to track their change over time. | The purpose and parameters of a clinical blood test are usually prescribed by a doctor and, therefore, as a rule, are not used independently by citizens for self-monitoring of their state of health. | Restricted educational effect for the general public. The results of scientific study are mainly intended for scientists. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lichtenberg, S.; Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Metabolomic Laboratory-Developed Tests: Current Status and Perspectives. Metabolites 2021, 11, 423. https://doi.org/10.3390/metabo11070423
Lichtenberg S, Trifonova OP, Maslov DL, Balashova EE, Lokhov PG. Metabolomic Laboratory-Developed Tests: Current Status and Perspectives. Metabolites. 2021; 11(7):423. https://doi.org/10.3390/metabo11070423
Chicago/Turabian StyleLichtenberg, Steven, Oxana P. Trifonova, Dmitry L. Maslov, Elena E. Balashova, and Petr G. Lokhov. 2021. "Metabolomic Laboratory-Developed Tests: Current Status and Perspectives" Metabolites 11, no. 7: 423. https://doi.org/10.3390/metabo11070423
APA StyleLichtenberg, S., Trifonova, O. P., Maslov, D. L., Balashova, E. E., & Lokhov, P. G. (2021). Metabolomic Laboratory-Developed Tests: Current Status and Perspectives. Metabolites, 11(7), 423. https://doi.org/10.3390/metabo11070423