The Impact of Microbiome and Microbiota-Derived Sodium Butyrate on Drosophila Transcriptome and Metabolome Revealed by Multi-Omics Analysis
Abstract
1. Introduction
2. Results
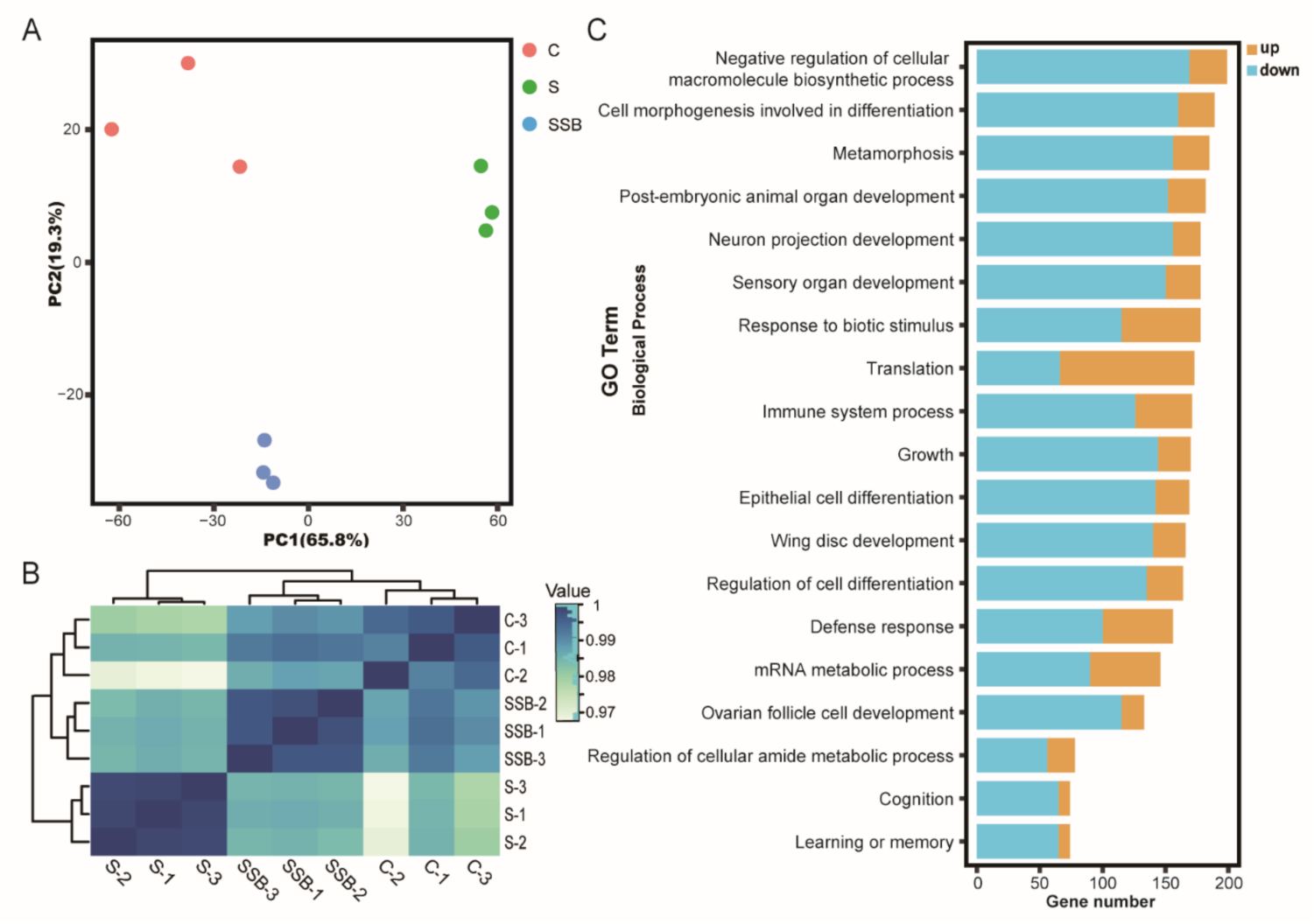
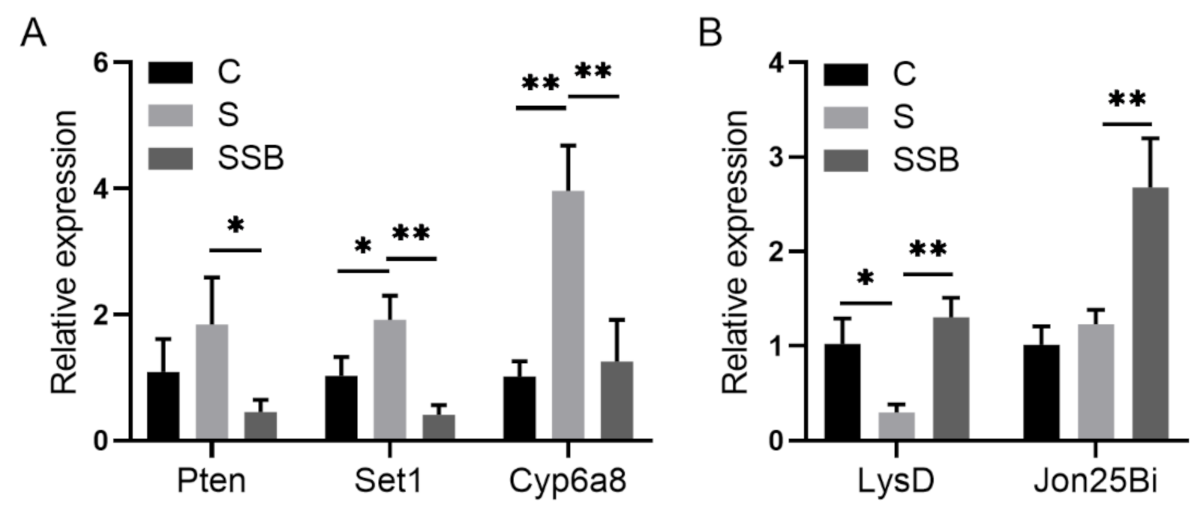
2.1. Drosophila Microbiome and Metabolites Regulate Host Gene Expression under Sterile Condition
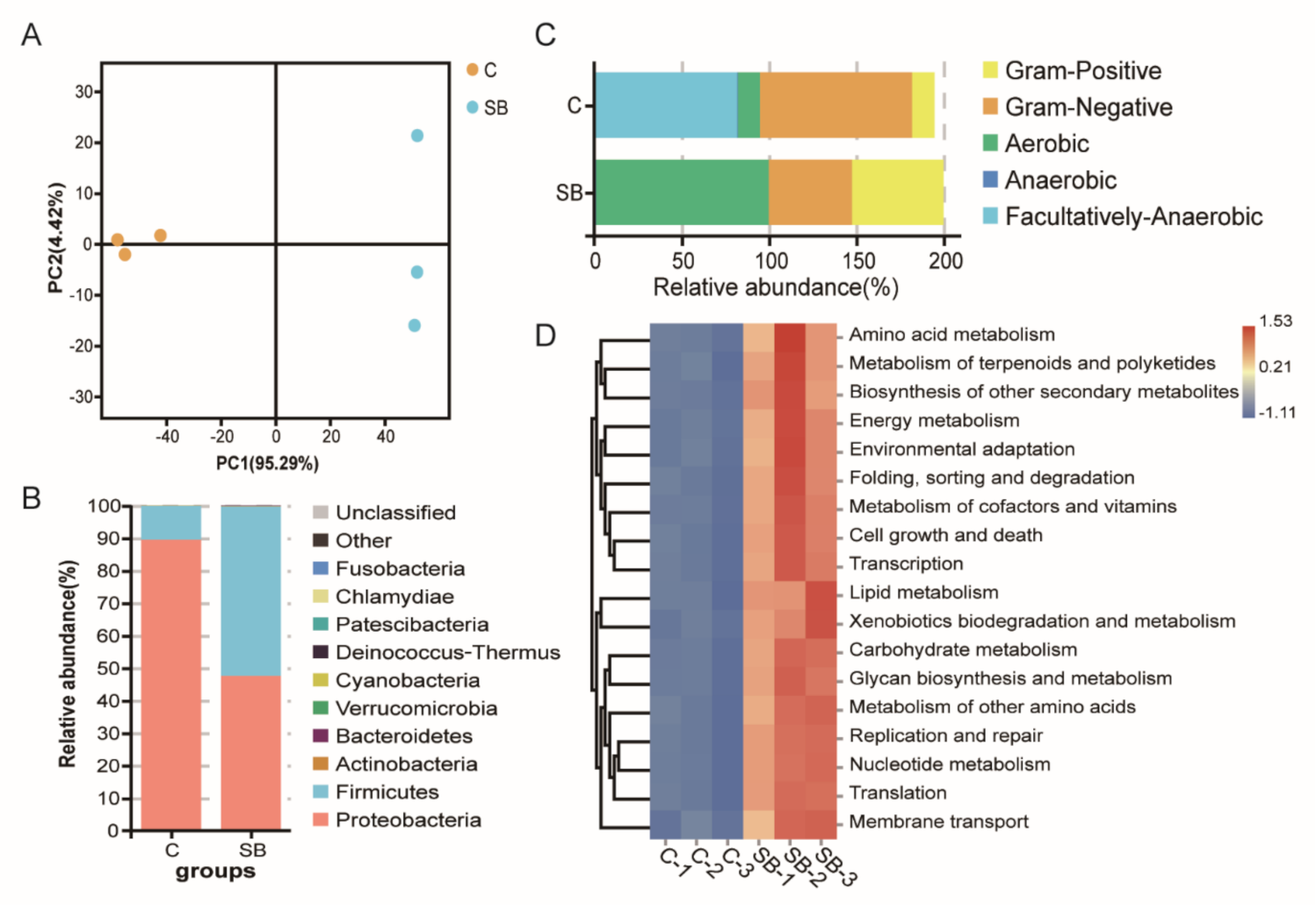
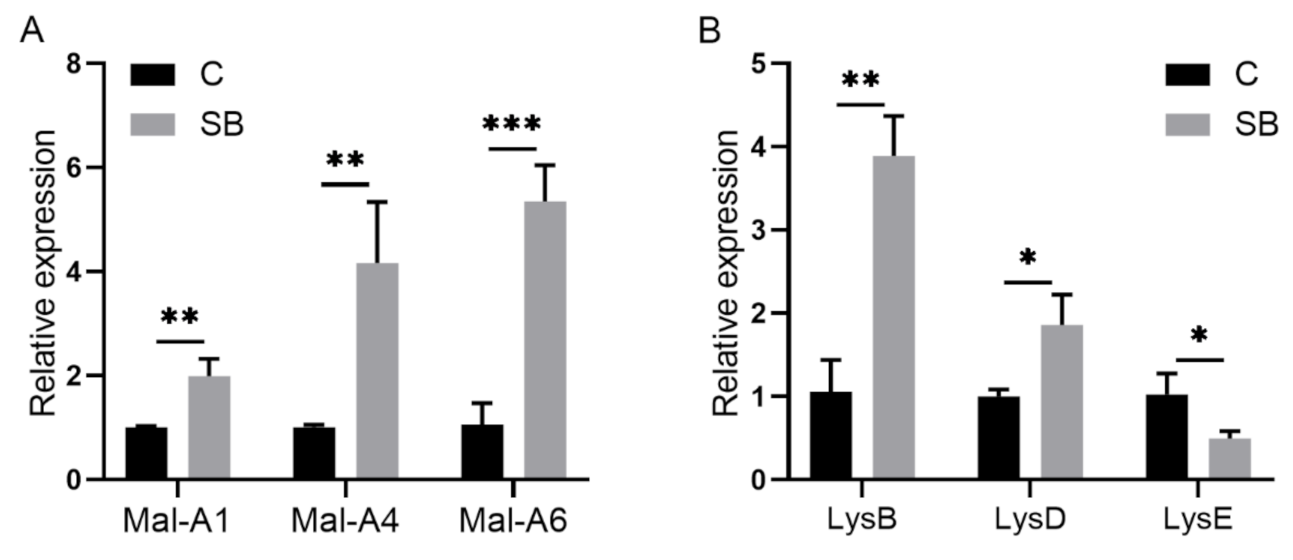
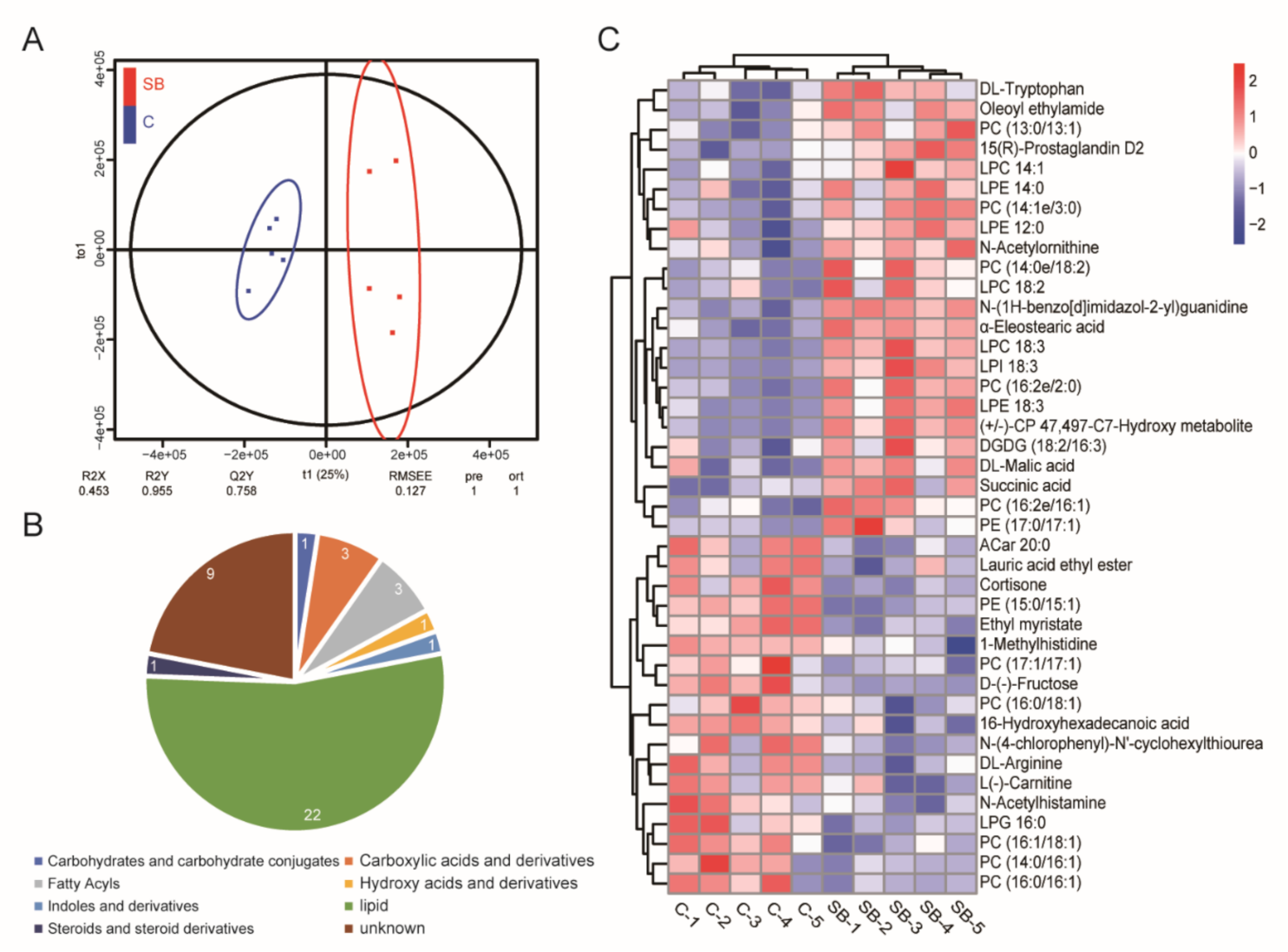
2.2. Drosophila Microbiome and Metabolites Regulate Host Gene Expression under Conventional Condition
3. Discussion
4. Materials and Methods
4.1. Conventional Drosophila and Sterile Drosophila
4.2. Gut Microbiome Analysis
4.3. RNA Sequencing and Data Analysis
4.4. Metabolites Extraction, UHPLC-MS/MS and Metabolome Analysis
4.5. RNA Preparation and RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Fraune, S.; Bosch, T.C. Why bacteria matter in animal development and evolution. Bioessays 2010, 32, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, C.; Jiang, Q.; Yin, Y. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol. Metabol. 2021, 32, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Vaiserman, A.M.; Kolyada, A.K.; Koshel, N.M.; Simonenko, A.V.; Pasyukova, E.G. Effect of histone deacetylase inhibitor sodium butyrate on viability and life span in Drosophila melanogaster. Adv. Gerontol. 2013, 3, 30–34. [Google Scholar] [CrossRef]
- Brahe, L.K.; Astrup, A.; Larsen, L.H. Is butyrate the link between diet, intestinal microbiota and obesity-related metabolic diseases? Obes. Rev. 2013, 14, 950–959. [Google Scholar] [CrossRef]
- Wang, H.; Ren, E.; Xiang, X.; Su, Y.; Zhu, W. Dynamic Changes in Serum Metabolomic Profiles of Growing Pigs Induced by Intravenous Infusion of Sodium Butyrate. Metabolites 2020, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Kim, S.H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.A.; Yoon, J.H.; Ryu, J.H.; Lee, W.J. Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef]
- Sonnenburg, J.L.; Bäckhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- Lee, W.-J.; Brey, P.T. How Microbiomes Influence Metazoan Development:Insights from History and Drosophila Modeling of Gut-Microbe Interactions. Ann. Rev. Cell Dev. Biol. 2013, 29, 571–592. [Google Scholar] [CrossRef]
- Lee-Sarwar, K.A.; Lasky-Su, J.; Kelly, R.S.; Litonjua, A.A.; Weiss, S.T. Metabolome-Microbiome Crosstalk and Human Disease. Metabolites 2020, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- McMullen, J.G., 2nd; Peters-Schulze, G.; Cai, J.; Patterson, A.D.; Douglas, A.E. How gut microbiome interactions affect nutritional traits of Drosophila melanogaster. J. Exp. Biol. 2020, 223, jeb227843. [Google Scholar] [CrossRef] [PubMed]
- Heys, C.; Lizé, A.; Blow, F.; White, L.; Darby, A.; Lewis, Z.J. The effect of gut microbiota elimination in Drosophila melanogaster: A how-to guide for host-microbiota studies. Ecol. Evol. 2018, 8, 4150–4161. [Google Scholar] [CrossRef]
- Douglas, A.E. The Drosophila model for microbiome research. Lab. Anim. (NY) 2018, 47, 157–164. [Google Scholar] [CrossRef]
- Erkosar, B.; Leulier, F. Transient adult microbiota, gut homeostasis and longevity: Novel insights from the Drosophila model. FEBS Lett. 2014, 588, 4250–4257. [Google Scholar] [CrossRef]
- Broderick, N.A.; Buchon, N.; Lemaitre, B.; McFall-Ngai, M.J. Microbiota-Induced Changes in Drosophila melanogaster Host Gene Expression and Gut Morphology. mBio 2014, 5, e01117-14. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Deng, B.; Tan, C.; Deng, J.; Zhu, G.; Yin, Y.; Ren, W. Implication of G Protein-Coupled Receptor 43 in Intestinal Inflammation: A Mini-Review. Front. Immunol. 2018, 9, 1434. [Google Scholar] [CrossRef]
- Miyamoto, J.; Hasegawa, S.; Kasubuchi, M.; Ichimura, A.; Nakajima, A.; Kimura, I. Nutritional Signaling via Free Fatty Acid Receptors. Int. J. Mol. Sci. 2016, 17, 450. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Wu, S.E.; Hashimoto-Hill, S.; Woo, V.; Eshleman, E.M.; Whitt, J.; Engleman, L.; Karns, R.; Denson, L.A.; Haslam, D.B.; Alenghat, T. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature 2020, 586, 108–112. [Google Scholar] [CrossRef]
- Reolon, G.K.; Maurmann, N.; Werenicz, A.; Garcia, V.A.; Schröder, N.; Wood, M.A.; Roesler, R. Posttraining systemic administration of the histone deacetylase inhibitor sodium butyrate ameliorates aging-related memory decline in rats. Behav. Brain Res. 2011, 221, 329–332. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gao, S.; Chen, J.; Zhao, R.; Yang, X. Maternal sodium butyrate supplement elevates the lipolysis in adipose tissue and leads to lipid accumulation in offspring liver of weaning-age rats. Lipids Health Dis. 2016, 15, 119. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Liu, S.; Chen, L.; Shen, J.; Niu, Y.; Wang, T.; Zhang, W.; Fu, L. Effect of exercise and butyrate supplementation on microbiota composition and lipid metabolism. J. Endocrinol. 2019, 243, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Simple animal models for microbiome research. Nat. Rev. Microbiol. 2019, 17, 764–775. [Google Scholar] [CrossRef]
- Broderick, N.A.; Lemaitre, B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012, 3, 307–321. [Google Scholar] [CrossRef]
- Koyle, M.L.; Veloz, A.M.; Judd, A.M.; Wong, A.C.N.; Newell, P.D.; Douglas, A.E.; Chaston, J.M. Rearing the Fruit Fly Drosophila melanogaster Under Axenic and Gnotobiotic Conditions. JoVE 2016. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Dobson, A.J.; Chaston, J.M.; Newell, P.D.; Donahue, L.; Hermann, S.L.; Sannino, D.R.; Westmiller, S.; Wong, A.C.N.; Clark, A.G.; Lazzaro, B.P.; et al. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat. Commun. 2015, 6, 6312. [Google Scholar] [CrossRef]
- Bergman, P.; Seyedoleslami Esfahani, S.; Engström, Y. Drosophila as a Model for Human Diseases-Focus on Innate Immunity in Barrier Epithelia. Curr. Top. Dev. Biol. 2017, 121, 29–81. [Google Scholar]
- Wong, A.C.; Dobson, A.J.; Douglas, A.E. Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 2014, 217, 1894–1901. [Google Scholar] [CrossRef]
- Walters, A.W.; Hughes, R.C.; Call, T.B.; Walker, C.J.; Wilcox, H.; Petersen, S.C.; Rudman, S.M.; Newell, P.D.; Douglas, A.E.; Schmidt, P.S.; et al. The microbiota influences the Drosophila melanogaster life history strategy. Mol. Ecol. 2020, 29, 639–653. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, F.; Liu, B.; Liu, X.; Li, Y.; Wang, L.; Huang, J.; Luo, G.; Wang, X. The Impact of Microbiome and Microbiota-Derived Sodium Butyrate on Drosophila Transcriptome and Metabolome Revealed by Multi-Omics Analysis. Metabolites 2021, 11, 298. https://doi.org/10.3390/metabo11050298
Zhou F, Liu B, Liu X, Li Y, Wang L, Huang J, Luo G, Wang X. The Impact of Microbiome and Microbiota-Derived Sodium Butyrate on Drosophila Transcriptome and Metabolome Revealed by Multi-Omics Analysis. Metabolites. 2021; 11(5):298. https://doi.org/10.3390/metabo11050298
Chicago/Turabian StyleZhou, Fan, Biaodi Liu, Xin Liu, Yan Li, Luoluo Wang, Jia Huang, Guanzheng Luo, and Xiaoyun Wang. 2021. "The Impact of Microbiome and Microbiota-Derived Sodium Butyrate on Drosophila Transcriptome and Metabolome Revealed by Multi-Omics Analysis" Metabolites 11, no. 5: 298. https://doi.org/10.3390/metabo11050298
APA StyleZhou, F., Liu, B., Liu, X., Li, Y., Wang, L., Huang, J., Luo, G., & Wang, X. (2021). The Impact of Microbiome and Microbiota-Derived Sodium Butyrate on Drosophila Transcriptome and Metabolome Revealed by Multi-Omics Analysis. Metabolites, 11(5), 298. https://doi.org/10.3390/metabo11050298





