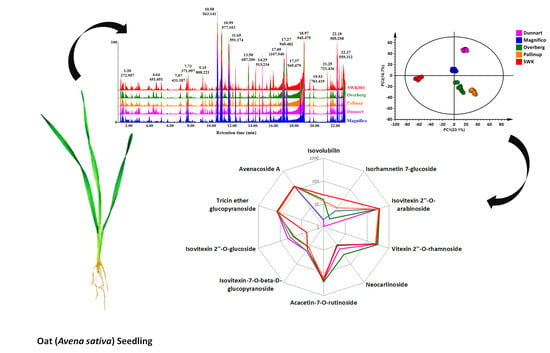
Metabolomics for Biomarker Discovery: Key Signatory Metabolic Profiles for the Identification and Discrimination of Oat Cultivars
Abstract
1. Introduction
2. Results
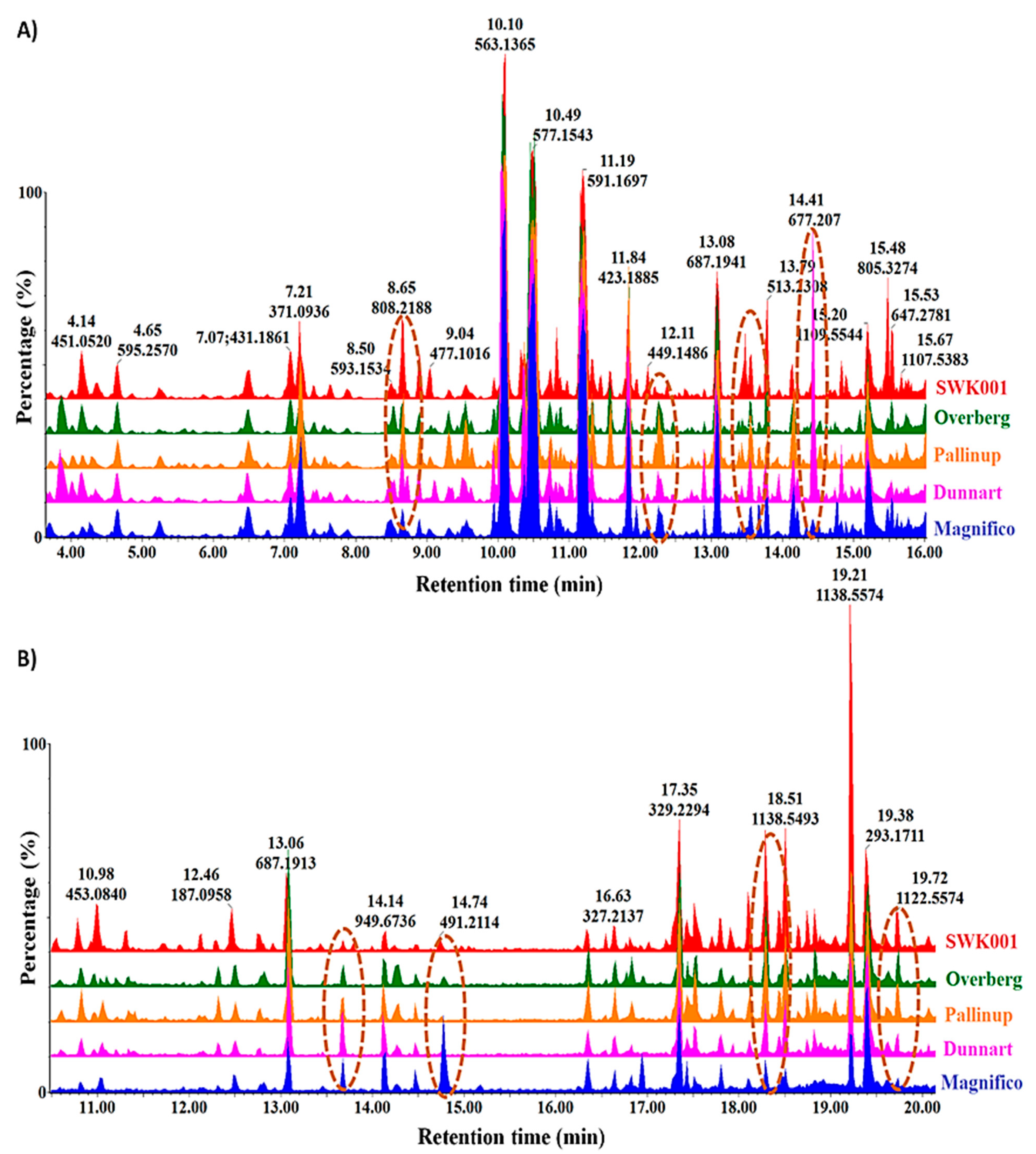
2.1. Differential Chromatographic–Mass Spectrometric Analyses of Respective Oat Cultivars
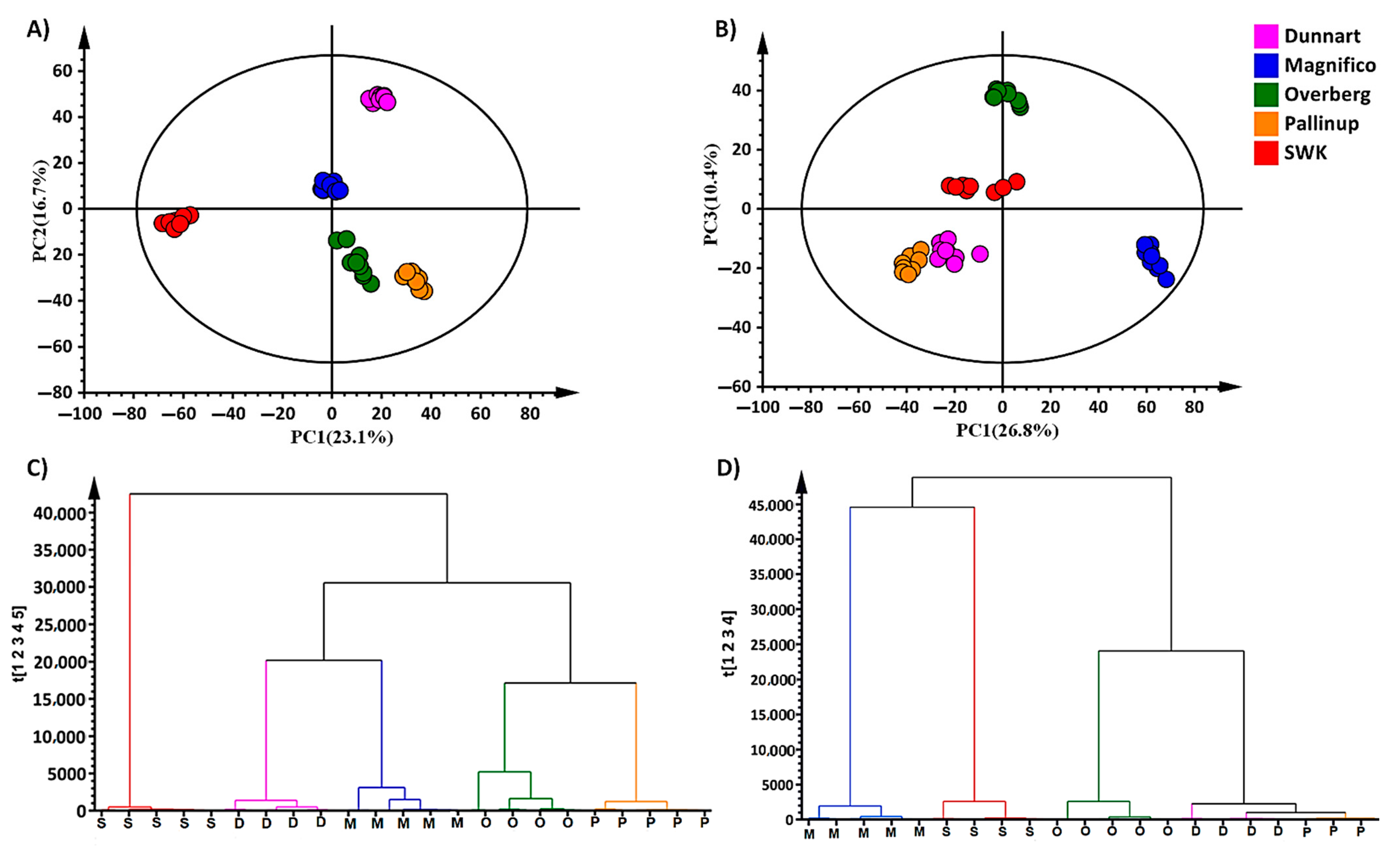
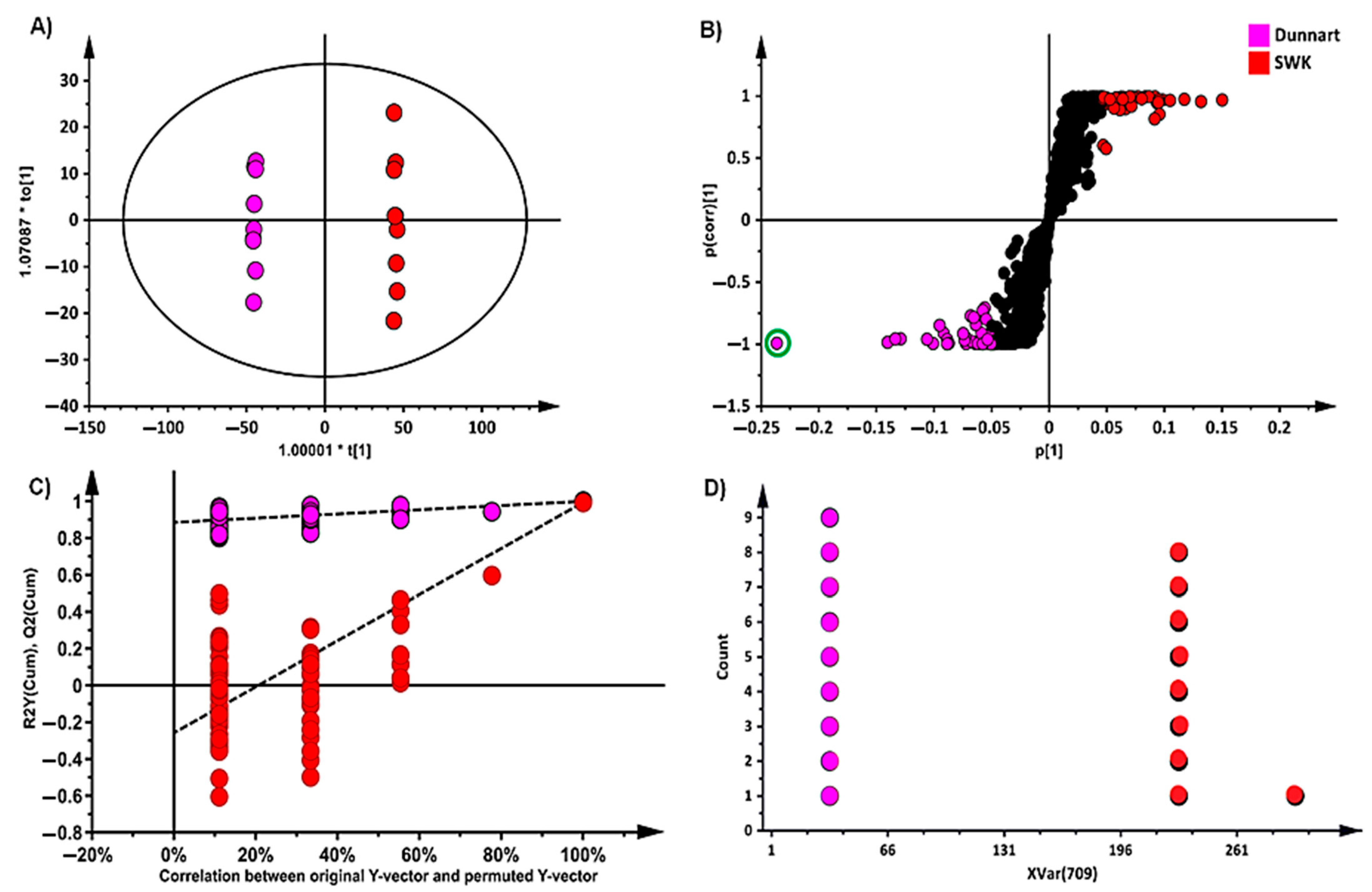
2.2. Chemometric Analyses for Profiling the Oat Cultivar Metabolomes
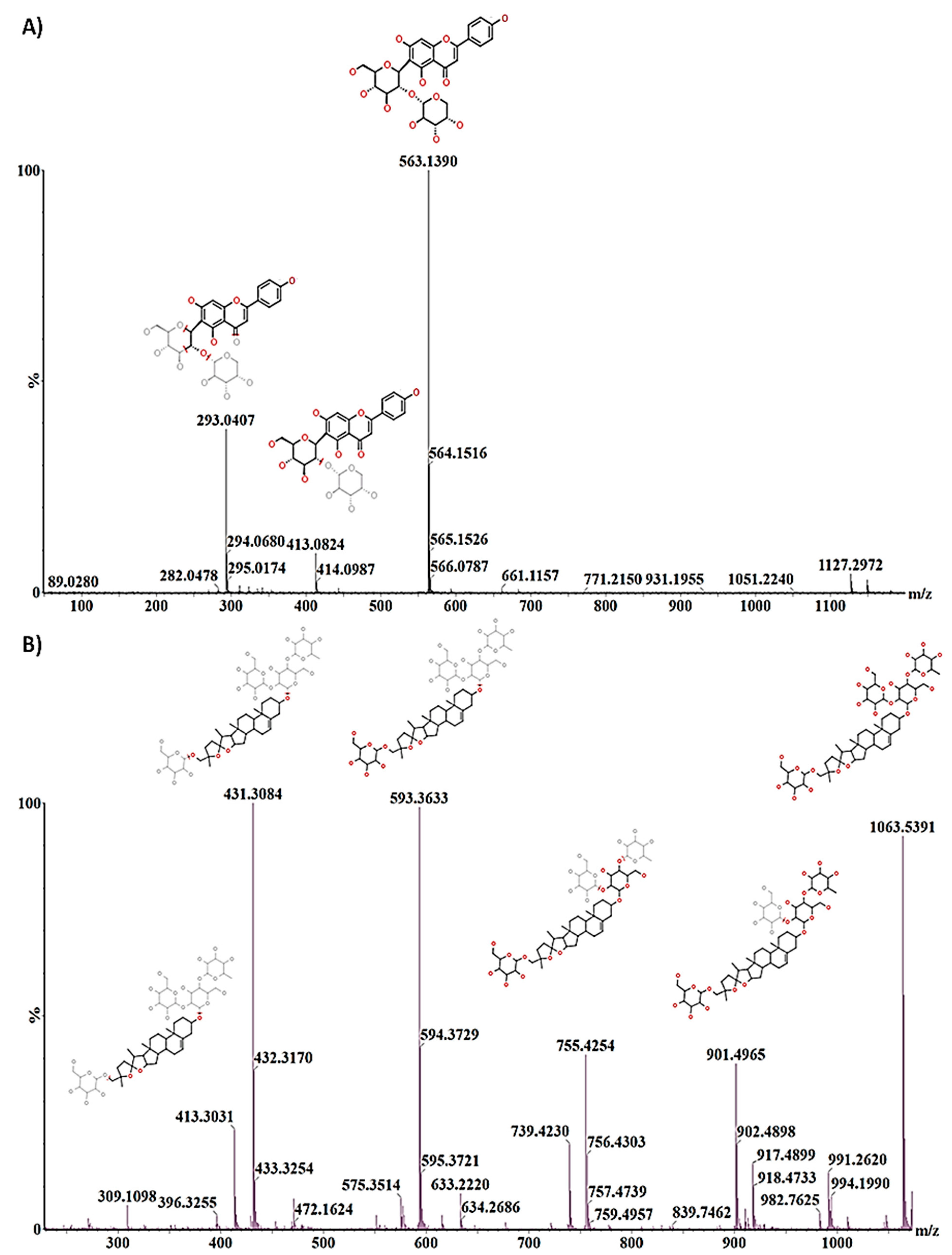
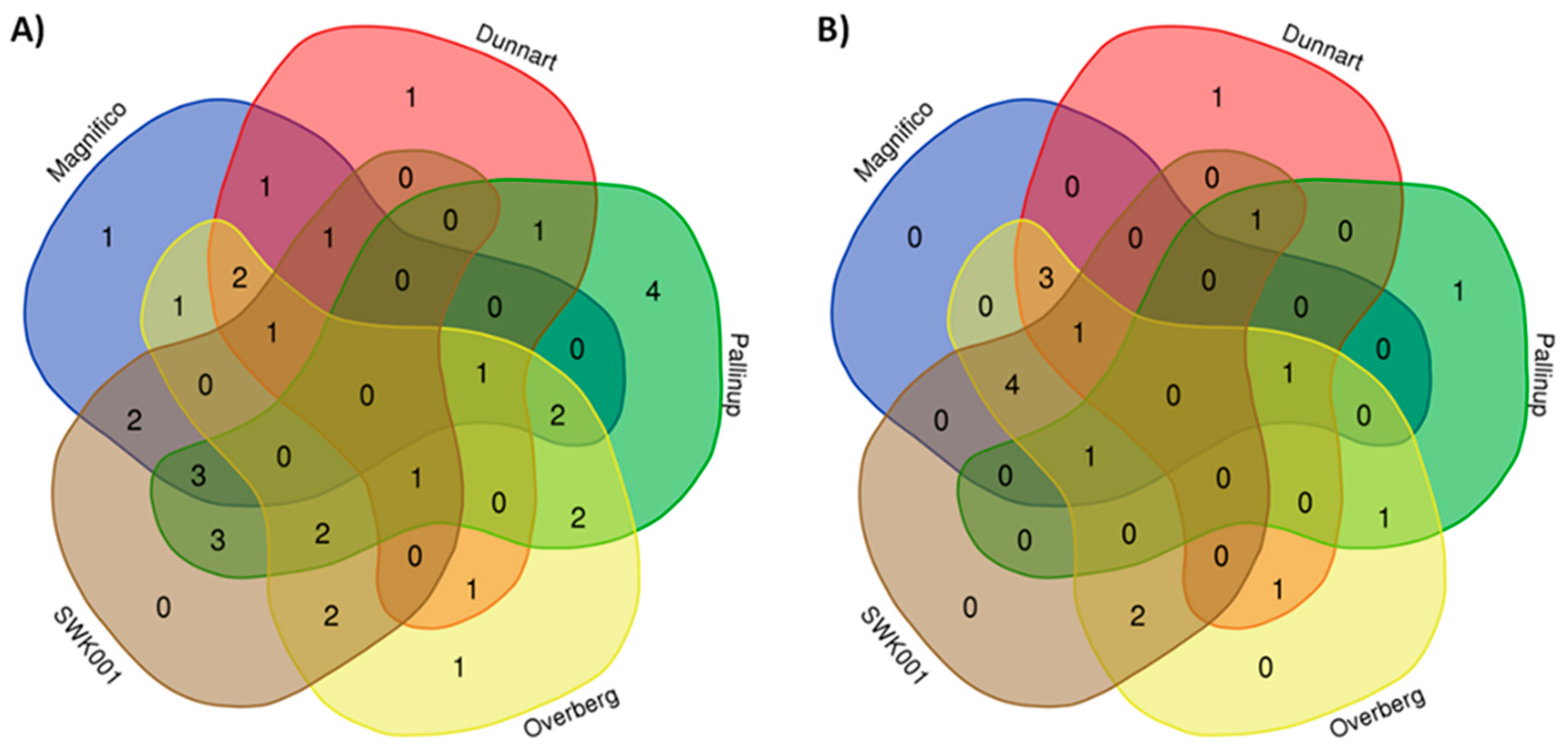
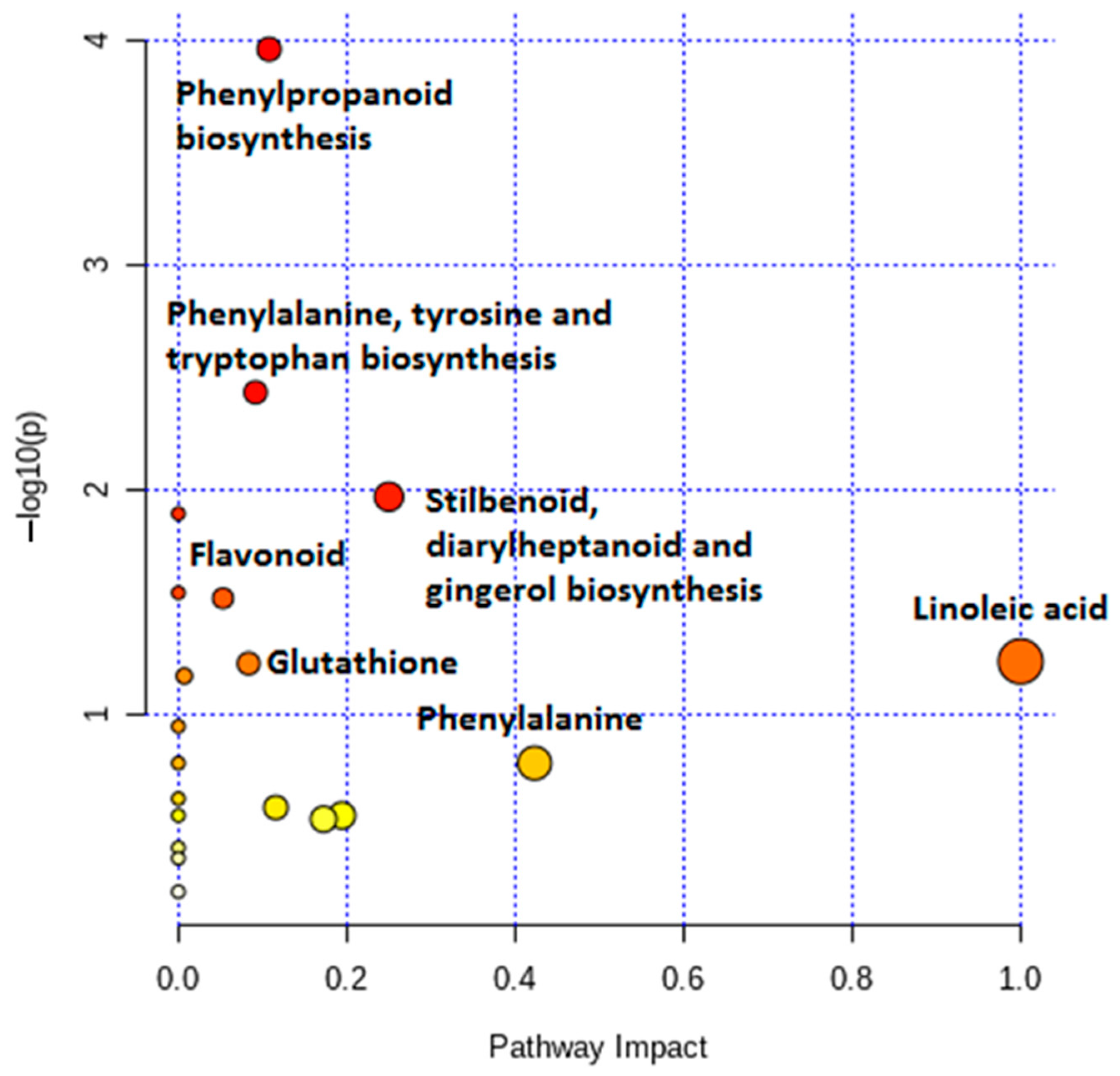
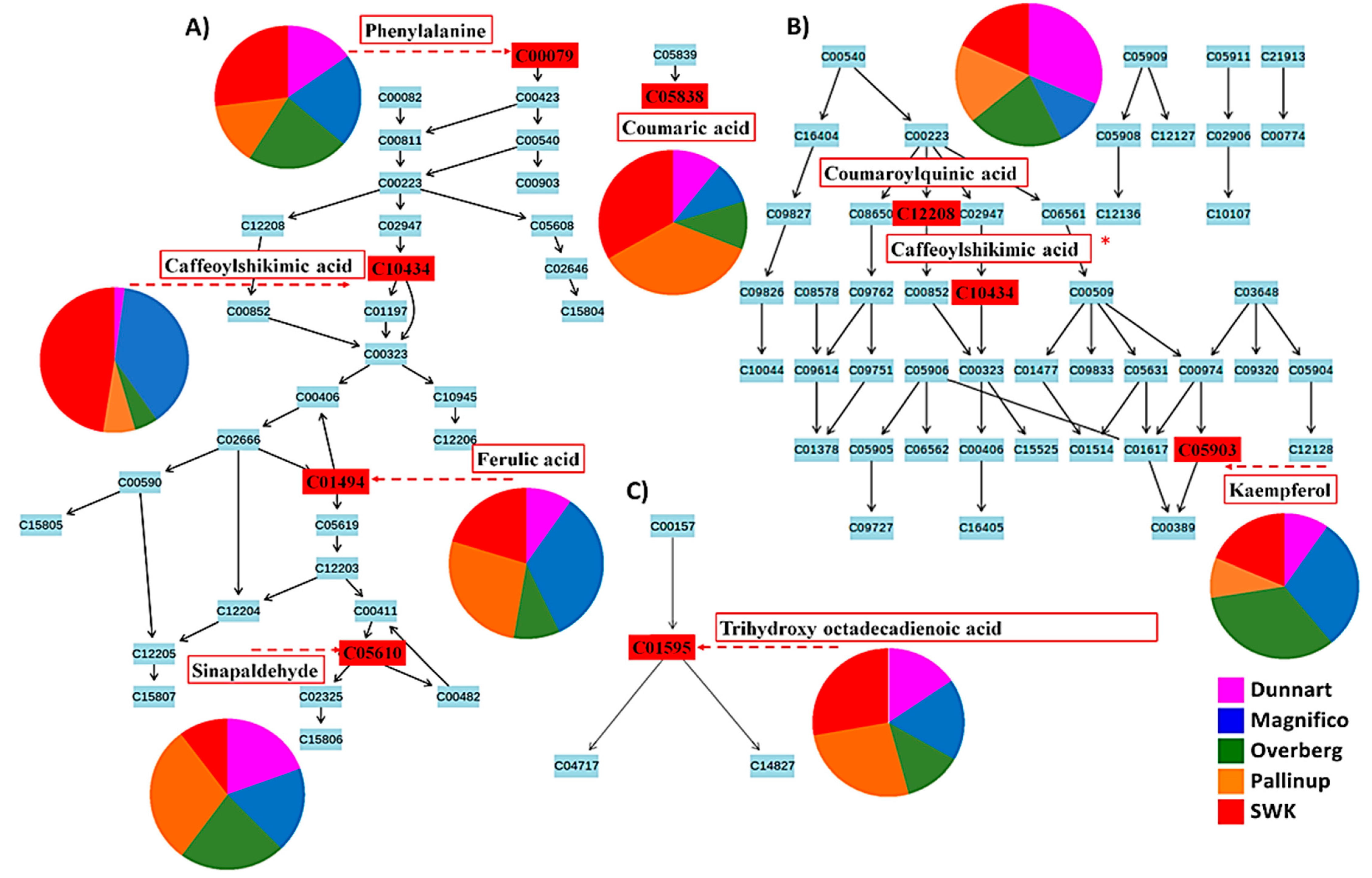
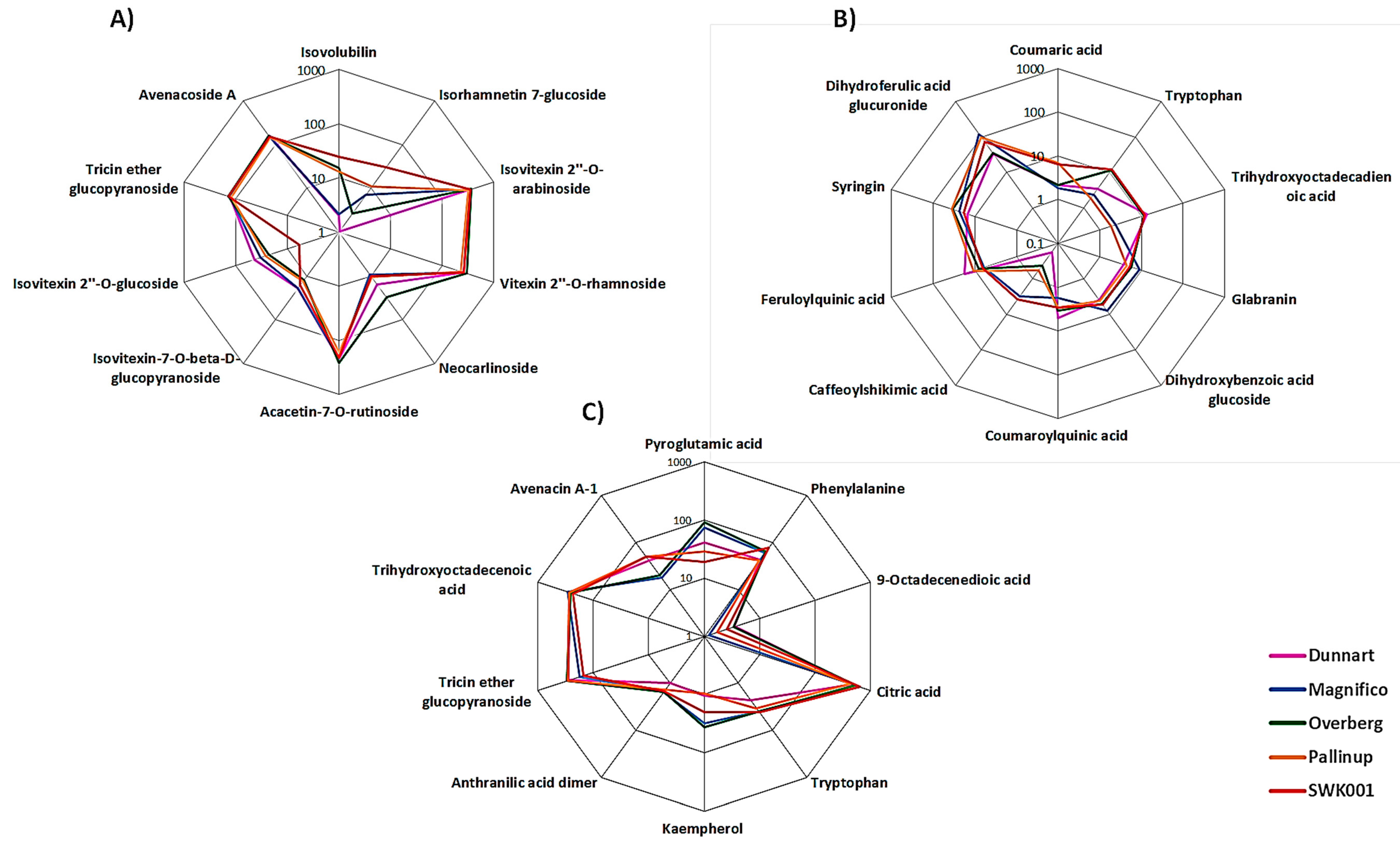
2.3. Differential Metabolic Profiles Based on Discriminatory Ions
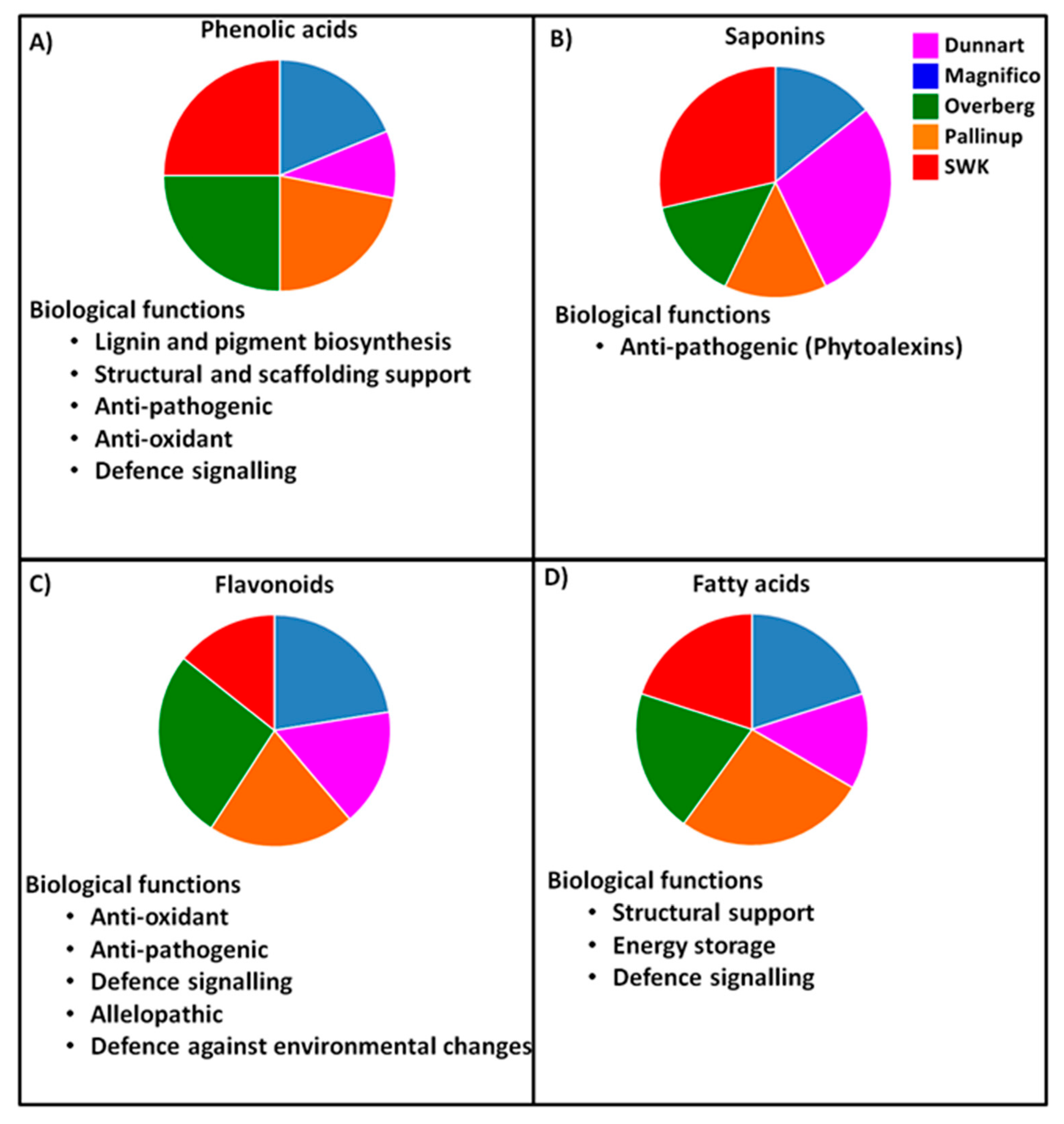
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation
4.2. Metabolite Extraction and Sample Preparation
4.3. Ultra-High Performance Liquid Chromatography (UHPLC) Analyses
4.4. Quadrupole Time-of-Flight Mass Spectrometry (q–TOF–MS)
4.5. Data Analyses
4.6. Metabolite Annotation and Semi-Quantitative Comparisons
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Hundleby, P.A.C.; Harwood, W.A. Impacts of the EU GMO regulatory framework for plant genome editing. Food Energy Secur. 2019, 8, e00161. [Google Scholar] [CrossRef]
- Korir, N.K.; Han, J.; Shangguan, L.; Wang, C.; Kayesh, E.; Zhang, Y.; Fang, J. Plant variety and cultivar identification: Advances and prospects. Crit. Rev. Biotechnol. 2012, 33, 111–125. [Google Scholar] [CrossRef]
- Edwards, D.; Batley, J.; Snowdon, R.J. Accessing complex crop genomes with next-generation sequencing. Theor. Appl. Genet. 2013, 126, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, N.M. Marker-assisted selection (MAS). In Genetic Mapping and Marker Assisted Selection; Springer: Singapore, 2020; pp. 343–388. [Google Scholar]
- Yandeau-Nelson, M.D.; Lauter, N.; Zabotina, O.A. Advances in metabolomic applications in plant genetics and breeding. CAB Rev. 2015, 10, 1–15. [Google Scholar] [CrossRef]
- Gienapp, P.; Laine, V.N.; Mateman, A.C.; van Oers, K.; Visser, M.E. Environment-dependent genotype-phenotype associations in avian breeding time. Front. Genet. 2017, 8, 102. [Google Scholar] [CrossRef] [PubMed]
- Rosato, A.; Tenori, L.; Cascante, M.; Carulla, P.R.D.A.; Dos Santos, V.A.P.M.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Handakumbura, P.P.; Stanfill, B.; Rivas-Ubach, A.; Fortin, D.; Vogel, J.P.; Jansson, C. Metabotyping as a stopover in genome-to-phenome mapping. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. J. Agron. 2020, 10, 831. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Kapoore, R.V.; Vaidyanathan, S. Towards quantitative mass spectrometry-based metabolomics in microbial and mammalian systems. Philos. Trans. R. Soc. A 2016, 374, 20150363. [Google Scholar] [CrossRef]
- Perez, E.R.; Knapp, J.A.; Horn, C.K.; Stillman, S.L.; Evans, J.E.; Arfsten, D.P. Comparison of LC–MS-MS and GC–MS Analysis of Benzodiazepine Compounds Included in the Drug Demand Reduction Urinalysis Program. J. Anal. Toxicol. 2016, 40, 201–207. [Google Scholar] [CrossRef]
- Shimizu, T.; Watanabe, M.; Fernie, A.R.; Tohge, T. Targeted LC-MS analysis for plant secondary metabolites. In Plant Metabolomics; Humana Press: New York, NY, USA, 2018; pp. 171–181. [Google Scholar]
- Gathungu, R.M.; Kautz, R.; Kristal, B.S.; Bird, S.S.; Vouros, P. The integration of LC-MS and NMR for the analysis of low molecular weight trace analytes in complex matrices. Mass Spectrom. Rev. 2020, 39, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Razzaq, A.; Sadia, B.; Raza, A.; Hameed, M.K.; Saleem, F. Metabolomics: A Way Forward for Crop Improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Want, E.J. From Samples to Insights into Metabolism: Uncovering Biologically Relevant Information in LC-HRMS Metabolomics Data. Metabolites 2019, 9, 308. [Google Scholar] [CrossRef]
- Hussein, R.A.; El-Anssary, A.A. Plants secondary metabolites: The Key drivers of the pharmacological actions of medicinal plants. In Herbal Medicine; Builders, P.H., Ed.; IntechOpen: London, UK, 2018; pp. 11–30. [Google Scholar]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 1–25. [Google Scholar] [CrossRef]
- Steinfath, M.; Strehmel, N.; Peters, R.; Schauer, N.; Groth, D.; Hummel, J.; Steup, M.; Selbig, J.; Kopka, J.; Geigenberger, P.; et al. Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol. J. 2010, 8, 900–911. [Google Scholar] [CrossRef]
- Peng, B.; Li, H.; Peng, X.-X. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. [Google Scholar] [CrossRef]
- Sarwar, M.H.; Sarwar, M.F.; Sarwar, M.; Qadri, N.A.; Moghal, S. The importance of cereals (Poaceae: Gramineae) nutrition in human health: A review. J. Cereals Oilseeds 2013, 4, 32–35. [Google Scholar] [CrossRef]
- Jing, P.; Hu, X. Nutraceutical properties and health benefits of oats. In Cereals and Pulses: Nutraceutical Properties and Health Benefits; Yu, L., Tsao, R., Shahidi, F., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 21–36. [Google Scholar]
- Rasane, P.; Jha, A.; Sabikhi, L.; Kumar, A.; Unnikrishnan, V.S. Nutritional advantages of oats and opportunities for its processing as value added foods—A review. J. Food Sci. Technol. 2013, 52, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Gani, M.; Saini, P.; Bhat, J.A.; Francies, R.M.; Negi, N.; Chauhan, S.S. Molecular breeding for resistance to economically important diseases of fodder oat. In Disease Resistance in Crop Plants; Wani, S.H., Ed.; Springer: Cham, Switzerland, 2019; pp. 199–239. [Google Scholar]
- Boccard, J.; Rudaz, S. Harnessing the complexity of metabolomic data with chemometrics. J. Chemom. 2014, 28, 1–9. [Google Scholar] [CrossRef]
- Brereton, R.G. Pattern recognition in chemometrics. Chemom. Intell. Lab. Syst. 2015, 149, 90–96. [Google Scholar] [CrossRef]
- Tebani, A.; Afonso, C.; Bekri, S. Advances in metabolome information retrieval: Turning chemistry into biology. Part II: Biological information recovery. J. Inherit. Metab. Dis. 2018, 41, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Handorf, E.A.; Heckman, C.J.; Darlow, S.; Slifker, M.; Ritterband, L. A hierarchical clustering approach to identify repeated enrollments in web survey data. PLoS ONE 2018, 13, e0204394. [Google Scholar] [CrossRef]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef]
- White, A.C.; Rogers, A.; Rees, M.; Osborne, C.P. How can we make plants grow faster? A source-sink perspective on growth rate. J. Exp. Bot. 2015, 67, 31–45. [Google Scholar] [CrossRef]
- Mugford, S.T.; Osbourn, A. Saponin Synthesis and Function. In Isoprenoid Synthesis in Plants and Microorganisms; Bach, T., Rohmer, M., Eds.; Springer Sciences and Business Media: New York, NY, USA, 2012; pp. 405–424. [Google Scholar]
- Worley, B.; Powers, R. PCA as a practical indicator of OPLS-DA model reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Fraser, C.M.; Chapple, C. The Phenylpropanoid Pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.M.; Niksiar, P. Multidimensional mechanics: Performance mapping of natural biological systems using permutated radar charts. PLoS ONE 2018, 13, e0204309. [Google Scholar] [CrossRef]
- Stewart, D.; McDougall, G. Oat agriculture, cultivation and breeding targets: Implications for human nutrition and health. Br. J. Nutr. 2014, 112, S50–S57. [Google Scholar] [CrossRef]
- Belobrajdic, D.P.; Bird, A.R. The potential role of phytochemicals in wholegrain cereals for the prevention of type-2 diabetes. Nutr. J. 2013, 12, 1–62. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Chu, Y. Whole grain oats, more than just a fiber: Role of unique phytochemicals. Mol. Nutr. Food Res. 2017, 61, 1600715. [Google Scholar] [CrossRef]
- Badea, G.I.; Radu, G.L. Introductory chapter: Carboxylic acids—Key role in life sciences. In Carboxylic Acid: Key Role in Life Sciences; IntechOpen: London, UK, 2018; p. 1. [Google Scholar] [CrossRef]
- Zhang, Y.; Fernie, A.R. On the role of the tricarboxylic acid cycle in plant productivity. J. Integr. Plant Biol. 2018, 60, 1199–1216. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. New Insights into the Shikimate and Aromatic Amino Acids Biosynthesis Pathways in Plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef]
- Ishihara, A.; Matsuda, F.; Miyagawa, H.; Wakasa, K. Metabolomics for metabolically manipulated plants: Effects of tryptophan overproduction. Metabolomics 2007, 3, 319–334. [Google Scholar] [CrossRef][Green Version]
- Kokubo, Y.; Nishizaka, M.; Ube, N.; Yabuta, Y.; Tebayashi, S.-I.; Ueno, K.; Taketa, S.; Ishihara, A. Distribution of the tryptophan pathway-derived defensive secondary metabolites gramine and benzoxazinones in Poaceae. Biosci. Biotechnol. Biochem. 2017, 81, 431–440. [Google Scholar] [CrossRef]
- Okazaki, Y.; Isobe, T.; Iwata, Y.; Matsukawa, T.; Matsuda, F.; Miyagawa, H.; Ishihara, A.; Nishioka, T.; Iwamura, H. Metabolism of avenanthramide phytoalexins in oats. Plant J. 2004, 39, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Y.; Meesapyodsuk, D.; Qiu, X. The Biosynthetic Pathway of Major Avenanthramides in Oat. Metabolites 2019, 9, 163. [Google Scholar] [CrossRef]
- Ferreyra, M.L.E.; Rius, S.P.; Ecasati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Peterson, D.M. Oat Antioxidants. J. Cereal Sci. 2001, 33, 115–129. [Google Scholar] [CrossRef]
- Krošlák, E.; Maliar, T.; Maliarová, M.; Nemeček, P.; Hozlár, P.; Ondrejovič, M.; Havrlentová, M.; Kraic, J. Antioxidant and protease-inhibitory potential of extracts from grains of oat. Open Chem. 2016, 14, 324–334. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Lafay, S.; Gil-Izquierdo, A. Bioavailability of phenolic acids. Phytochem. Rev. 2007, 7, 301–311. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Sood, P.; Citovsky, V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010, 11, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Aid, F. Plant lipid metabolism. In Advances in Lipid Metabolism; IntechOpen: London, UK, 2019; pp. 1–16. [Google Scholar]
- Walley, J.W.; Kliebenstein, D.J.; Bostock, R.M.; Dehesh, K. Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 2013, 16, 520–526. [Google Scholar] [CrossRef]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Pecio, Ł.; Wawrzyniak-Szołkowska, A.; Oleszek, W.; Stochmal, A. Rapid analysis of avenacosides in grain and husks of oats by UPLC–qTOF–MS. Food Chem. 2013, 141, 2300–2304. [Google Scholar] [CrossRef]
- Morrissey, J.P.; Wubben, J.P.; Osbourn, A.E. Stagonospora avenae secretes multiple enzymes that hydrolyze oat leaf saponins. Mol. Plant Microbe Interact. 2000, 13, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Du Fall, L.A.; Solomon, P.S. Role of Cereal Secondary Metabolites Involved in Mediating the Outcome of Plant-Pathogen Interactions. Metabolites 2011, 1, 64–78. [Google Scholar] [CrossRef]
- Moses, T.; Papadopoulou, K.K.; Osbourn, A. Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 439–462. [Google Scholar] [CrossRef] [PubMed]
- Armah, C.; Mackie, A.; Roy, C.; Price, K.; Osbourn, A.; Bowyer, P.; Ladha, S. The Membrane-Permeabilizing Effect of Avenacin A-1 Involves the Reorganization of Bilayer Cholesterol. Biophys. J. 1999, 76, 281–290. [Google Scholar] [CrossRef]
- Owatworakit, A.; Townsend, B.; Louveau, T.; Jenner, H.; Rejzek, M.; Hughes, R.K.; Saalbach, G.; Qi, X.; Bakht, S.; Roy, A.D.; et al. Glycosyltransferases from Oat (Avena) Implicated in the Acylation of Avenacins. J. Biol. Chem. 2013, 288, 3696–3704. [Google Scholar] [CrossRef]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Multi-Platform Metabolomic Analyses of Ergosterol-Induced Dynamic Changes in Nicotiana tabacum Cells. PLoS ONE 2014, 9, e87846. [Google Scholar] [CrossRef] [PubMed]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Comparative Metabolic Phenotyping of Tomato (Solanum lycopersicum) for the Identification of Metabolic Signatures in Cultivars Differing in Resistance to Ralstonia solanacearum. Int. J. Mol. Sci. 2018, 19, 2558. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, A.E.; Lundstedt, T. Chemometrics in Metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2012, 1, 92–107. [Google Scholar] [CrossRef][Green Version]
- MetaCyc. Available online: https://metacyc.org/ (accessed on 21 September 2020).
- PlantCyc. Available online: https://plantcyc.org/ (accessed on 21 September 2020).
- MassBank. Available online: https://massbank.eu/MassBank/Search (accessed on 4 September 2020).
- Dictionary of Natural Products. Available online: www.dnp.chemnetbase.com (accessed on 4 September 2020).
- KEGG. Available online: http://www.genome.jp/kegg/ (accessed on 10 January 2009).
- Brown, M.; Wedge, D.C.; Goodacre, R.; Kell, D.B.; Baker, P.N.; Kenny, L.C.; Mamas, M.A.; Neyses, L.; Dunn, W.B. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics 2011, 27, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, 652–660. [Google Scholar] [CrossRef] [PubMed]
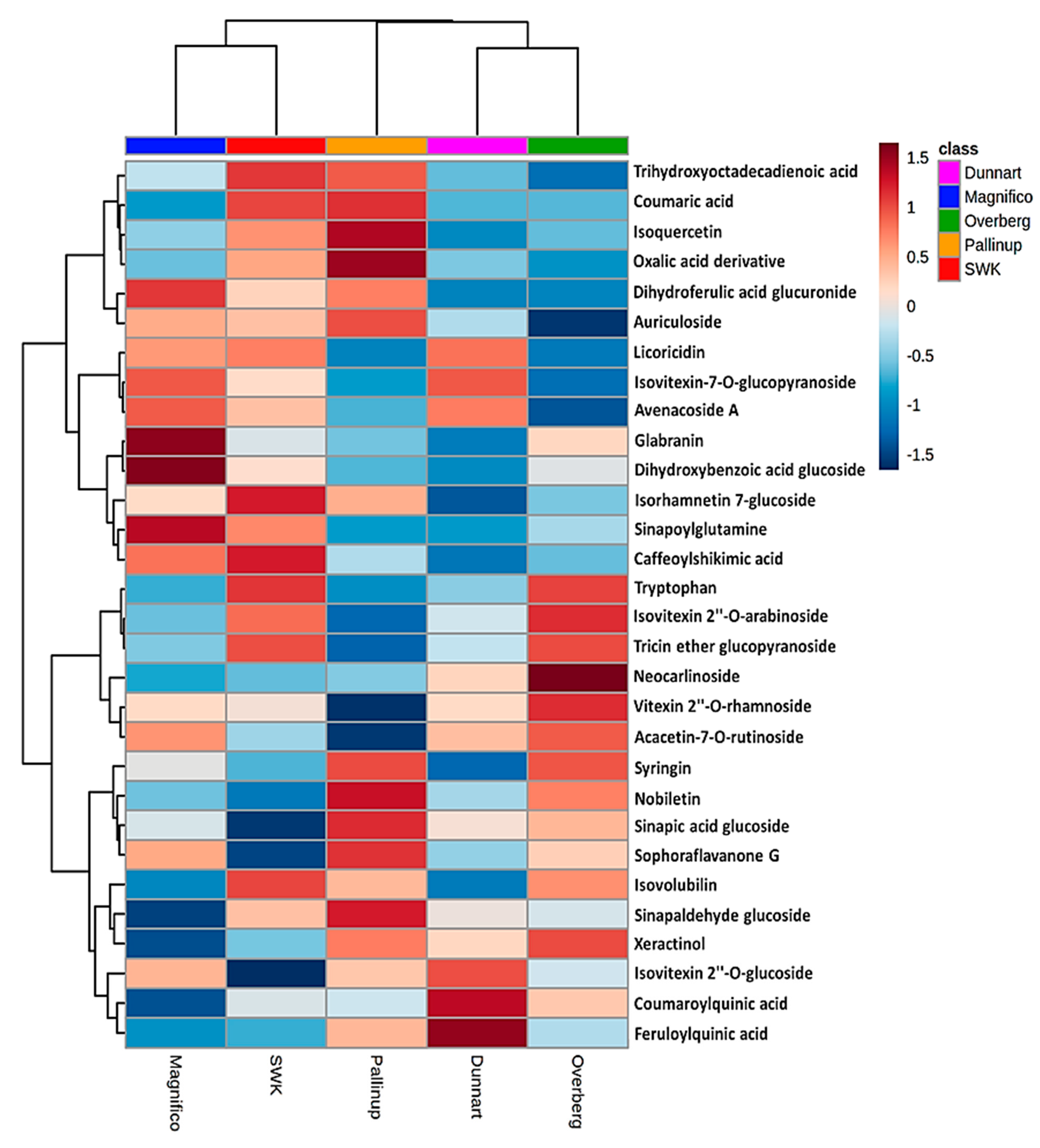
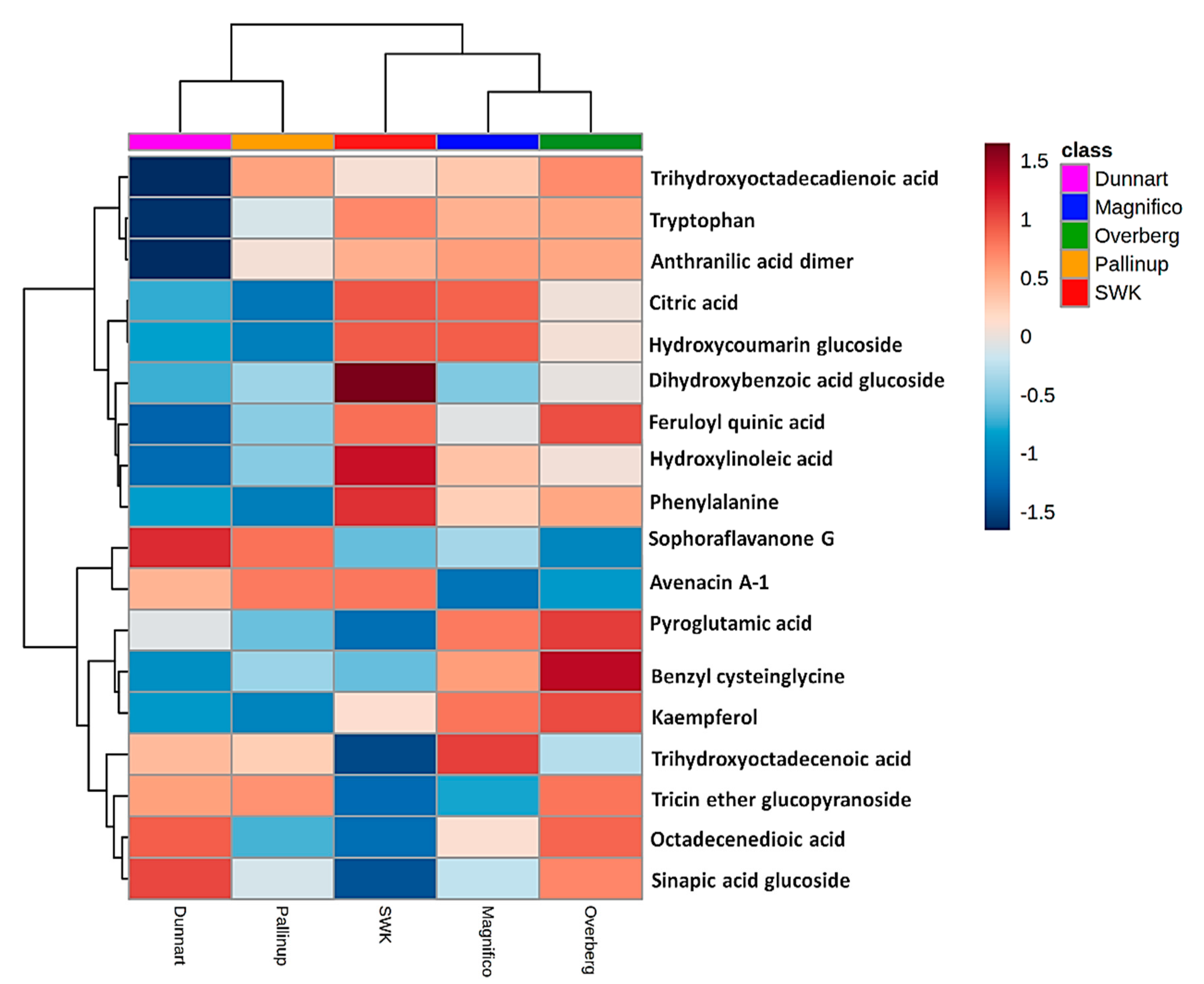
| Annotated Metabolites | Molecular Formula | ESI Mode | m/z | Rt (min) | Metabolite Class | Cultivars | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ‘Mag’ | ‘Dun’ | ‘Pal’ | ‘Over’ | ‘SWK’ | ||||||
| Leaves | ||||||||||
| Coumaric acid | C9H8O3 | Neg | 163.0379 | 3.35 | Phenolic acid | * | * | |||
| Tryptophan | C11H12N2O2 | Neg | 203.081 | 2.48 | Amino acid | * | * | |||
| Hydroxyoctadecatrienoic acid | C18H30O3 | Neg | 293.208 | 21.49 | Fatty acid | * | ||||
| Dihydroxybenzoic acid glucoside | C13H16O9 | Neg | 315.0742 | 2.52 | Phenolic acid | * | * | |||
| Glabranin | C20H20O4 | Neg | 323.1326 | 2.94 | Flavonoid | * | * | |||
| Trihydroxyoctadecadienoic acid | C18H32O5 | Neg | 327.2153 | 16.63 | Fatty acid | * | * | |||
| Trihydroxyoctadecenoic acid | C18H34O5 | Neg | 329.23 | 17.34 | Fatty acid | * | * | |||
| Caffeoylshikimic acid | C16H16O8 | Neg | 335.0422 | 2.24 | Phenolic acid | * | * | |||
| Coumaroylquinic acid | C16H18O8 | Neg | 337.092 | 3.19 | Phenolic acid | * | * | |||
| Sinapoylglutamine | C16H20N2O7 | Neg | 351.1257 | 6.48 | Phenolic acid | * | ||||
| Feruloylquinic acid | C17H20O9 | Neg | 367.1008 | 4.01 | Phenolic acid | * | ||||
| Sinapaldehyde glucoside | C17H22O9 | Neg | 369.1184 | 13.54 | Phenolic acid | * | * | * | * | |
| Dihydroferulic acid glucuronide | C16H20O10 | Neg | 371.0958 | 7.21 | Phenolic acid | * | * | * | ||
| Syringin | C17H24O9 | Neg | 371.1346 | 16.0 | Phenolic acid | * | * | * | ||
| Sinapic acid glucoside | C17H22O10 | Neg | 385.1146 | 3.40 | Phenolic acid | * | ||||
| Auriculoside | C22H26O10 | Neg | 393.1752 | 12.10 | Flavonoid | * | * | * | ||
| Nobiletin | C21H22O8 | Pos | 403.1454 | 9.53 | Flavonoid | * | * | |||
| Sophoraflavanone G | C25H28O6 | Neg | 423.1856 | 11.83 | Flavonoid | * | * | * | ||
| Licoricidin | C26H32O5 | Neg | 423.2204 | 14.81 | Flavonoid | * | * | * | ||
| Isovolubilin | C23H24O9 | Neg | 443.1328 | 16.81 | Flavonoid | * | * | * | ||
| Isoquercetin | C21H20O12 | Neg | 463.0895 | 6.59 | Flavonoid | * | ||||
| Xeractinol | C21H22O12 | Neg | 465.1028 | 12.97 | Flavonoid | * | ||||
| Isorhamnetin 7-glucoside | C22H22O12 | Neg | 477.1038 | 9.03 | Flavonoid | * | * | * | ||
| Oxalate derivative | C25H24O10 | Neg | 483.1281 | 12.20 | Phenolic acid | * | ||||
| Isovitexin 2″-O-arabinoside | C26H28O14 | Pos | 563.1393 | 10.08 | Flavonoid | * | * | |||
| Vitexin 2″-O-rhamnoside | C27H30O14 | Neg | 577.1545 | 10.49 | Flavonoid | * | * | * | ||
| Neocarlinoside | C26H28O15 | Neg | 579.1349 | 8.53 | Flavonoid | * | ||||
| Acacetin-7-O-rutinoside | C28H32O14 | Pos | 593.186 | 11.20 | Flavonoid | * | * | * | ||
| Isovitexin 2″-O-glucoside | C27H30O15 | Neg | 593.1488 | 9.94 | Flavonoid | * | * | * | * | |
| Isovitexin-7-O-glucopyranoside | C27H30O15 | Pos | 595.1499 | 8.50 | Flavonoid | * | * | |||
| Prenylkaempferol diglucoside | C32H38O16 | Neg | 677.207 | 14.41 | Flavonoid | * | * | |||
| Tricin ether glucopyranoside | C33H36O16 | Pos | 689.194 | 13.51 | Flavonoid | * | * | * | ||
| Avenacoside A | C51H82O23 | Pos | 1063.539 | 16.58 | Triterpene | * | * | * | * | |
| Roots | ||||||||||
| Pyroglutamic acid | C5H7NO3 | Neg | 128.033 | 1.16 | Amino acid | * | * | * | ||
| Phenylalanine | C9H11NO2 | Neg | 164.068 | 1.67 | Amino acid | * | * | * | ||
| Citric acid | C6H8O7 | Neg | 191.0163 | 1.16 | Carboxylic acid | * | * | * | * | |
| Tryptophan | C11H12N2O2 | Neg | 203.081 | 2.49 | Amino acid | * | * | * | * | |
| Anthranilic acid dimer | C14H12N2O4 | Neg | 271.07 | 2.49 | Phenolic acid | * | * | |||
| Kaempferol | C15H10O6 | Neg | 285.039 | 12.80 | Flavonoid | * | * | * | ||
| Hydroxylinoleic acid | C18H32O3 | Neg | 295.15 | 23.87 | Fatty acid | * | * | * | ||
| Octadecenedioic acid | C18H32O4 | Neg | 311.165 | 22.78 | Fatty acid | * | * | * | ||
| Dihydroxybenzoic acid glucoside | C13H16O9 | Neg | 315.069 | 1.67 | Phenolic acid | * | * | |||
| Hydroxycoumarin glucoside | C15H16O8 | Neg | 323.097 | 1.68 | Phenolic acid | * | * | * | ||
| Trihydroxyoctadecadienoic acid | C18H32O5 | Neg | 327.214 | 16.63 | Fatty acid | * | ||||
| Trihydroxyoctadecenoic acid | C18H34O5 | Neg | 329.23 | 17.36 | Fatty acid | * | * | * | ||
| Feruloylquinic acid | C17H19O9 | Neg | 367.101 | 4.02 | Phenolic acid | * | * | |||
| Sophoraflavanone G | C25H28O6 | Neg | 423.186 | 7.65 | Flavonoid | * | ||||
| di-Sinapoylglucoside | C28H32O14 | Neg | 591.1693 | 11.20 | Phenolic acid | * | * | |||
| Tricin ether glucopyranoside | C33H36O16 | Neg | 687.192 | 13.06 | Flavonoid | * | * | * | * | |
| Avenacin A-1 | C55H83NO21 | Neg | 1092.55 | 18.49 | Triterpene | * | * | * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pretorius, C.J.; Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomics for Biomarker Discovery: Key Signatory Metabolic Profiles for the Identification and Discrimination of Oat Cultivars. Metabolites 2021, 11, 165. https://doi.org/10.3390/metabo11030165
Pretorius CJ, Tugizimana F, Steenkamp PA, Piater LA, Dubery IA. Metabolomics for Biomarker Discovery: Key Signatory Metabolic Profiles for the Identification and Discrimination of Oat Cultivars. Metabolites. 2021; 11(3):165. https://doi.org/10.3390/metabo11030165
Chicago/Turabian StylePretorius, Chanel J., Fidele Tugizimana, Paul A. Steenkamp, Lizelle A. Piater, and Ian A. Dubery. 2021. "Metabolomics for Biomarker Discovery: Key Signatory Metabolic Profiles for the Identification and Discrimination of Oat Cultivars" Metabolites 11, no. 3: 165. https://doi.org/10.3390/metabo11030165
APA StylePretorius, C. J., Tugizimana, F., Steenkamp, P. A., Piater, L. A., & Dubery, I. A. (2021). Metabolomics for Biomarker Discovery: Key Signatory Metabolic Profiles for the Identification and Discrimination of Oat Cultivars. Metabolites, 11(3), 165. https://doi.org/10.3390/metabo11030165