The Dysregulation of Eicosanoids and Bile Acids Correlates with Impaired Kidney Function and Renal Fibrosis in Chronic Renal Failure
Abstract
1. Introduction
2. Results
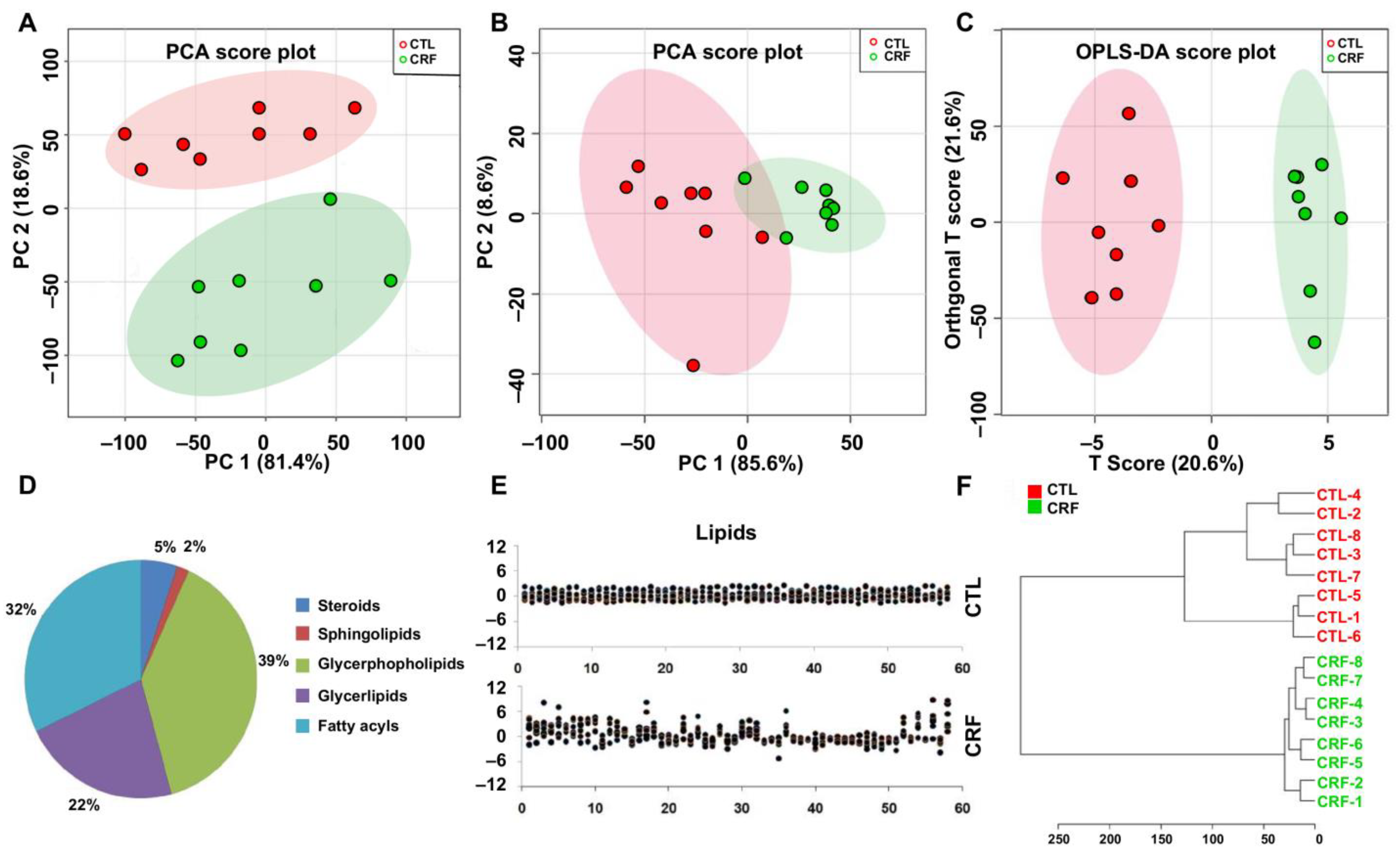
2.1. Multivariate Analysis and Identification of Important Differential Lipid Species
2.2. Correlation Analyses between Important Differential Lipid Species and Serum Creatinine Levels
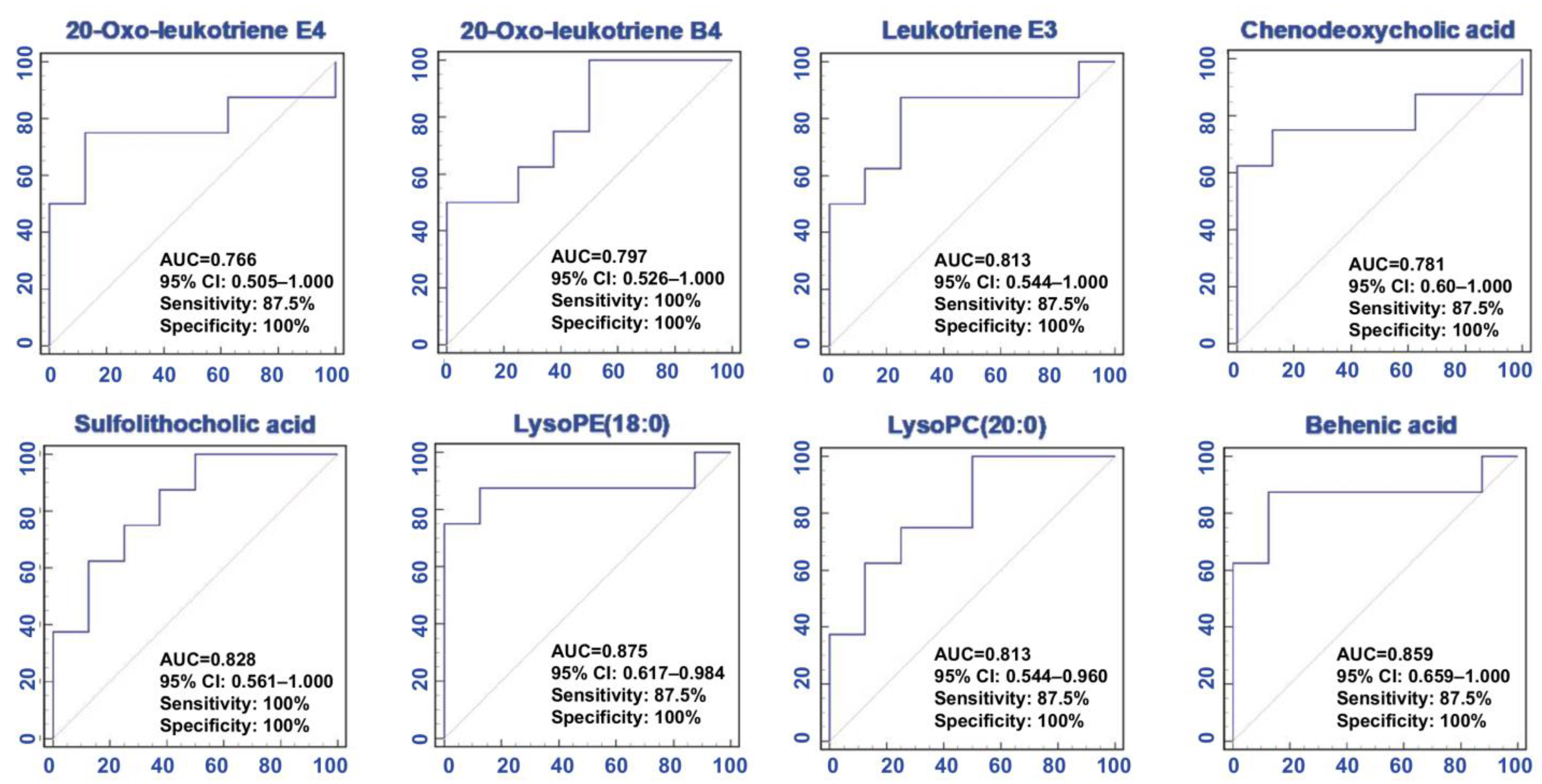
2.3. Predictive Performance Assessment
2.4. Renoprotective Effect of PPU and ERG on Impaired Kidney Function
2.5. Antifibrotic Effect of PPU and ERG
2.6. The Inhibitory Effect of PPU and ERG on the Levels of Eight Lipid Species
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animals Experiment and Sample Collection
4.3. Renal Function Evaluation
4.4. Histological Analysis and Western Blot Analysis
4.5. Metabolomic Analysis
4.6. Linear Correlation Analysis and Receiver Operating Characteristic Curve (ROC) Analysis
4.7. Pattern Recognition Analysis
4.8. Statistics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic kidney disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef]
- Qi, R.; Yang, C. Renal tubular epithelial cells: The neglected mediator of tubulointerstitial fibrosis after injury. Cell Death Dis. 2018, 9, 1126. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.M.; Nikolic-Paterson, D.J.; Lan, H.Y. Macrophages: Versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 2019, 15, 144–158. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Cheng, X.L.; Wei, F.; Xiao, X.Y.; Sun, W.J.; Zhang, Y.; Lin, R.C. Serum metabonomics study of adenine-induced chronic renal failure in rats by ultra performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Biomarkers 2012, 17, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Chen, H.; Tian, T.; Chen, D.Q.; Bai, X.; Wei, F. A pharmaco-metabonomic study on chronic kidney disease and therapeutic effect of ergone by UPLC-QTOF/HDMS. PLoS ONE 2014, 9, e115467. [Google Scholar] [CrossRef]
- Liu, J.R.; Miao, H.; Deng, D.Q.; Vaziri, N.D.; Li, P.; Zhao, Y.Y. Gut microbiota-derived tryptophan metabolism mediates renal fibrosis by aryl hydrocarbon receptor signaling activation. Cell. Mol. Life Sci. 2020, 78, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Dou, F.; Miao, H.; Wang, J.W.; Chen, L.; Wang, M.; Chen, H.; Wen, A.D.; Zhao, Y.Y. An integrated lipidomics and phenotype study reveals protective effect and biochemical mechanism of traditionally used Alisma orientale juzepzuk in chronic kidney disease. Front. Pharmacol. 2018, 9, 53. [Google Scholar] [CrossRef]
- Lydic, T.A.; Goo, Y.H. Lipidomics unveils the complexity of the lipidome in metabolic diseases. Clin. Transl. Med. 2018, 7, 4. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Cheng, X.L.; Lin, R.C. Lipidomics applications for discovering biomarkers of diseases in clinical chemistry. Int. Rev. Cell. Mol. Biol. 2014, 313, 1–26. [Google Scholar]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Ferro, C.J.; Mark, P.B.; Kanbay, M.; Sarafidis, P.; Heine, G.H.; Rossignol, P.; Massy, Z.A.; Mallamaci, F.; Valdivielso, J.M.; Malyszko, J.; et al. Lipid management in patients with chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 727–749. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.L.; Wang, C.H.; Shiao, M.S.; Liu, M.H.; Huang, Y.Y.; Huang, C.Y.; Mao, C.T.; Lin, J.F.; Ho, H.Y.; Yang, N.I. Metabolic disturbances identified in plasma are associated with outcomes in patients with heart failure: Diagnostic and prognostic value of metabolomics. J. Am. Coll. Cardiol. 2015, 65, 1509–1520. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Cheng, X.L.; Lin, R.C.; Wei, F. Lipidomics applications for disease biomarker discovery in mammal models. Biomark. Med. 2015, 9, 153–168. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Vaziri, N.D.; Lin, R.C. Lipidomics: New insight into kidney disease. Adv. Clin. Chem. 2015, 68, 153–175. [Google Scholar]
- Zhang, Z.H.; Chen, H.; Vaziri, N.D.; Mao, J.R.; Zhang, L.; Bai, X.; Zhao, Y.Y. Metabolomic signatures of chronic kidney disease of diverse etiologies in the rats and humans. J. Proteome Res. 2016, 15, 3802–3812. [Google Scholar] [CrossRef]
- Hu, J.R.; Coresh, J.; Inker, L.A.; Levey, A.S.; Zheng, Z.; Rebholz, C.M.; Tin, A.; Appel, L.J.; Chen, J.; Sarnak, M.J.; et al. Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int. 2018, 94, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Cao, G.; Wu, X.Q.; Chen, Y.Y.; Chen, D.Q.; Chen, L.; Vaziri, N.D.; Feng, Y.L.; Su, W.; Gao, Y.; et al. Identification of endogenous 1-aminopyrene as a novel mediator of progressive chronic kidney disease via aryl hydrocarbon receptor activation. Br. J. Pharmacol. 2020, 177, 3415–3435. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.L.; Chen, H.; Chen, D.Q.; Vaziri, N.D.; Su, W.; Ma, S.X.; Shang, Y.Q.; Mao, J.R.; Yu, X.Y.; Zhang, L.; et al. Activated NF-κB/Nrf2 and Wnt/β-catenin pathways are associated with lipid metabolism in CKD patients with microalbuminuria and macroalbuminuria. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2317–2332. [Google Scholar] [CrossRef]
- Hocher, B.; Adamski, J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Afshinnia, F.; Rajendiran, T.M.; Wernisch, S.; Soni, T.; Jadoon, A.; Karnovsky, A.; Michailidis, G.; Pennathur, S. Lipidomics and biomarker discovery in kidney disease. Semin. Nephrol. 2018, 38, 127–141. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Cheng, X.L.; Wei, F.; Bai, X.; Tan, X.J.; Lin, R.C.; Mei, Q. Intrarenal metabolomic investigation of chronic kidney disease and its TGF-β1 mechanism in induced-adenine rats using UPLC Q-TOF/HSMSE. J. Proteome Res. 2013, 12, 692–703. [Google Scholar] [CrossRef]
- Chen, H.; Chen, L.; Liu, D.; Chen, D.Q.; Vaziri, N.D.; Yu, X.Y.; Zhang, L.; Su, W.; Bai, X.; Zhao, Y.Y. Combined clinical phenotype and lipidomic analysis reveals the impact of chronic kidney disease on lipid metabolism. J. Proteome Res. 2017, 16, 1566–1578. [Google Scholar] [CrossRef] [PubMed]
- Kalim, S.; Rhee, E.P. An overview of renal metabolomics. Kidney Int. 2017, 91, 61–69. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Wang, H.L.; Cheng, X.L.; Wei, F.; Bai, X.; Lin, R.C.; Vaziri, N.D. Metabolomics analysis reveals the association between lipid abnormalities and oxidative stress, inflammation, fibrosis, and Nrf2 dysfunction in aristolochic acid-induced nephropathy. Sci. Rep. 2015, 5, 12936. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cao, G.; Chen, D.Q.; Wang, M.; Vaziri, N.D.; Zhang, Z.H.; Mao, J.R.; Bai, X.; Zhao, Y.Y. Metabolomics insights into activated redox signaling and lipid metabolism dysfunction in chronic kidney disease progression. Redox Biol. 2016, 10, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Tyurina, Y.Y.; St. Croix, C.M.; Watkins, S.C.; Watson, A.M.; Epperly, M.W.; Anthonymuthu, T.S.; Kisin, E.R.; Vlasova, I.I.; Krysko, O.; Krysko, D.V.; et al. Redox (phospho)lipidomics of signaling in inflammation and programmed cell death. J. Leukoc. Biol. 2019, 106, 57–81. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Ahn, S.H.; Choi, P.; Ko, Y.A.; Han, S.H.; Chinga, F.; Park, A.S.; Tao, J.; Sharma, K.; Pullman, J.; et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015, 21, 37–46. [Google Scholar] [CrossRef]
- Chen, D.Q.; Feng, Y.L.; Cao, G.; Zhao, Y.Y. Natural products as a source for antifibrosis therapy. Trends Pharmacol. Sci. 2018, 39, 937–952. [Google Scholar] [CrossRef]
- Feng, Y.L.; Chen, D.Q.; Vaziri, N.D.; Guo, Y.; Zhao, Y.Y. Small molecule inhibitors of epithelial-mesenchymal transition for the treatment of cancer and fibrosis. Med. Res. Rev. 2020, 40, 54–78. [Google Scholar] [CrossRef]
- Wang, M.; Chen, D.Q.; Chen, L.; Cao, G.; Zhao, H.; Liu, D.; Vaziri, N.D.; Guo, Y.; Zhao, Y.Y. Novel inhibitors of the cellular renin-angiotensin system components, poricoic acids, target smad3 phosphorylation and Wnt/β-catenin pathway against renal fibrosis. Br. J. Pharmacol. 2018, 175, 2689–2708. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.H.; Cao, G.; Wu, X.Q.; Vaziri, N.D.; Zhao, Y.Y. Wnt signaling pathway in aging-related tissue fibrosis and therapies. Ageing Res. Rev. 2020, 60, 101063. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Li, M.H.; Liu, D.; Chen, H.; Chen, D.Q.; Tan, N.H.; Ma, S.C.; Zhao, Y.Y. Rhubarb protect against tubulointerstitial fibrosis by inhibiting TGF-β/smad pathway and improving abnormal metabolome in chronic kidney disease. Front. Pharmacol. 2018, 9, 1029. [Google Scholar] [CrossRef]
- Zhao, Y.Y. Traditional uses, phytochemistry, pharmacology, pharmacokinetics and quality control of Polyporus umbellatus (pers.) fries: A review. J. Ethnopharmacol. 2013, 149, 35–48. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Chao, X.; Zhang, Y.; Lin, R.C.; Sun, W.J. Cytotoxic steroids from Polyporus umbellatus. Planta Med. 2010, 76, 1755–1758. [Google Scholar] [CrossRef]
- Li, H.; Yan, Z.; Xiong, Q.; Chen, X.; Lin, Y.; Xu, Y.; Bai, L.; Jiang, W.; Zheng, D.; Xing, C. Renoprotective effect and mechanism of polysaccharide from Polyporus umbellatus sclerotia on renal fibrosis. Carbohydr. Polym. 2019, 212, 1–10. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Shen, X.; Chao, X.; Ho, C.C.; Cheng, X.L.; Zhang, Y.; Lin, R.C.; Du, K.J.; Luo, W.J.; Chen, J.Y.; et al. Ergosta-4,6,8(14),22-tetraen-3-one induces G2/M cell cycle arrest and apoptosis in human hepatocellular carcinoma HepG2 cells. Biochim. Biophys. Acta Gen. Subj. 2011, 1810, 384–390. [Google Scholar] [CrossRef]
- Chen, L.; Chen, D.Q.; Liu, J.R.; Zhang, J.; Vaziri, N.D.; Zhuang, S.; Chen, H.; Feng, Y.L.; Guo, Y.; Zhao, Y.Y. Unilateral ureteral obstruction causes gut microbial dysbiosis and metabolome disorders contributing to tubulointerstitial fibrosis. Exp. Mol. Med. 2019, 51, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Zhang, L.; Mao, J.R.; Cheng, X.H.; Lin, R.C.; Zhang, Y.; Sun, W.J. Ergosta-4,6,8(14),22-tetraen-3-one isolated from Polyporus umbellatus prevents early renal injury in aristolochic acid-induced nephropathy rats. J. Pharm. Pharmacol. 2011, 63, 1581–1586. [Google Scholar] [CrossRef]
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef]
- Madsen, T.; Christensen, J.H.; Toft, E.; Aardestrup, I.; Lundbye-Christensen, S.; Schmidt, E.B. Effect of intravenous ω-3 fatty acid infusion and hemodialysis on fatty acid composition of free fatty acids and phospholipids in patients with end-stage renal disease. J. Parenter. Enteral. Nutr. 2011, 35, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.N.; Yu, Z.; Denski, C.; Tamez, H.; Wenger, J.; Thadhani, R.; Li, Y.; Watkins, B. Fatty acids and other risk factors for sudden cardiac death in patients starting hemodialysis. Am. J. Nephrol. 2013, 38, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Kárpáti, I.; Paragh, G.; Buris, L.; Kakuk, G. Relative abundance of some free fatty acids in plasma of uremic patients: Relationship between fatty acids, lipid parameters, and diseases. Nephron 1997, 77, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, C.; Liu, S.; Chang, M.; Gao, P.; Wang, L.; Pan, Z.; Xu, G. Plasma lipidomics investigation of hemodialysis effects by using liquid chromatography-mass spectrometry. J. Proteome Res. 2016, 15, 1986–1994. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Su, Y.; Ju, Y.; Ma, K.; Li, W.; Li, W. Astragalosides iv protected the renal tubular epithelial cells from free fatty acids-induced injury by reducing oxidative stress and apoptosis. Biomed. Pharmacother. 2018, 108, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, Z.O.; Sattar, M.A.; Abdullah, N.A.; Rathore, H.A.; Johns, E.J. Effect of high saturated free fatty acids feeding on progression of renal failure in rat model of experimental nephrotoxicity. Bosn. J. Basic Med. Sci. 2012, 12, 26–32. [Google Scholar] [CrossRef]
- Liu, Z.X.; Hong, Q.; Peng, D.H.; Yang, Y.; Yu, W.L.; Shui, H.; Zhou, X.; Liu, S.M. Evaluation of serum free fatty acids in chronic renal failure: Evidence from a rare case with undetectable serum free fatty acids and population data. Lipids Health Dis. 2019, 18, 151. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Palygin, O.; Guijas, C.; Palermo, A.; Palacio-Escat, N.; Domingo-Almenara, X.; Montenegro-Burke, R.; Saez-Rodriguez, J.; Staruschenko, A.; Siuzdak, G. Metabolic rewiring of the hypertensive kidney. Sci. Signal. 2019, 12, eaax9760. [Google Scholar] [CrossRef]
- Shimanaka, Y.; Kono, N.; Taketomi, Y.; Arita, M.; Okayama, Y.; Tanaka, Y.; Nishito, Y.; Mochizuki, T.; Kusuhara, H.; Adibekian, A.; et al. ω-3 fatty acid epoxides are autocrine mediators that control the magnitude of ige-mediated mast cell activation. Nat. Med. 2017, 23, 1287–1297. [Google Scholar] [CrossRef]
- Huang, X.; Stenvinkel, P.; Qureshi, A.R.; Risérus, U.; Cederholm, T.; Bárány, P.; Heimbürger, O.; Lindholm, B.; Carrero, J.J. Essential polyunsaturated fatty acids, inflammation and mortality in dialysis patients. Nephrol. Dial. Transplant. 2012, 27, 3615–3620. [Google Scholar] [CrossRef]
- Maaløe, T.; Schmidt, E.B.; Svensson, M.; Aardestrup, I.V.; Christensen, J.H. The effect of n-3 polyunsaturated fatty acids on leukotriene B₄ and leukotriene B₅ production from stimulated neutrophil granulocytes in patients with chronic kidney disease. Prostaglandins Leukot. Essent. Fatty. Acids 2011, 85, 37–41. [Google Scholar] [CrossRef]
- Tran, M.; Tam, D.; Bardia, A.; Bhasin, M.; Rowe, G.C.; Kher, A.; Zsengeller, Z.K.; Akhavan-Sharif, M.R.; Khankin, E.V.; Saintgeniez, M.; et al. PGC-1α promotes recovery after acute kidney injury during systemic inflammation in mice. J. Clin. Investig. 2011, 121, 4003–4014. [Google Scholar] [CrossRef] [PubMed]
- Chau, B.N.; Xin, C.; Hartner, J.; Ren, S.; Castano, A.P.; Linn, G.; Li, J.; Tran, P.T.; Kaimal, V.; Huang, X.; et al. Microrna-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 2012, 4, 121ra118. [Google Scholar] [CrossRef] [PubMed]
- Afshinnia, F.; Zeng, L.; Byun, J.; Wernisch, S.; Deo, R.; Chen, J.; Hamm, L.; Miller, E.R.; Rhee, E.P.; Fischer, M.J.; et al. Elevated lipoxygenase and cytochrome P450 products predict progression of chronic kidney disease. Nephrol. Dial. Transpl. 2020, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.S.; Cuzzocrea, S.; Chatterjee, P.K.; Di Paola, R.; Sautebin, L.; Britti, D.; Thiemermann, C. Reduction of renal ischemia-reperfusion injury in 5-lipoxygenase knockout mice and by the 5-lipoxygenase inhibitor zileuton. Mol. Pharmacol. 2004, 66, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.H.; Bresnahan, B.A.; Lianos, E.A. Hemodynamic role of arachidonate 12- and 5-lipoxygenases in nephrotoxic serum nephritis. Kidney Int. 1993, 43, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Xu, Z.G.; Reddy, M.A.; Li, S.L.; Lanting, L.; Sharma, K.; Adler, S.G.; Natarajan, R. Novel interactions between TGF-β1 actions and the 12/15-lipoxygenase pathway in mesangial cells. J. Am. Soc. Nephrol. 2005, 16, 352–362. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile acids activated receptors regulate innate immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Zhang, K.; Zhang, Y.; Jin, X.; Jiang, H. Mechanism underlying an elevated serum bile acid level in chronic renal failure patients. Int. Urol. Nephrol. 2015, 47, 345–351. [Google Scholar] [CrossRef]
- Li, R.; Zeng, L.; Xie, S.; Chen, J.; Yu, Y.; Zhong, L. Targeted metabolomics study of serum bile acid profile in patients with end-stage renal disease undergoing hemodialysis. PeerJ 2019, 7, e7145. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, S.; Li, S.; Zhao, L.; Hao, Y.; Qin, J.; Zhang, L.; Zhang, C.; Bian, W.; Zuo, L.; et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 2020, 69, 2131–2142. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ren, L.; Wang, C.; Liu, B.; Song, G. Effect of chenodeoxycholic acid on fibrosis, inflammation and oxidative stress in kidney in high-fructose-fed wistar rats. Kidney Blood Press Res. 2012, 36, 85–97. [Google Scholar] [CrossRef]
- Rhee, E.P. Metabolomics and renal disease. Curr. Opin. Nephrol. Hypertens. 2015, 24, 371–379. [Google Scholar] [CrossRef]
- Ho, Y.C.; Wu, M.L.; Su, C.H.; Chen, C.H.; Ho, H.H.; Lee, G.L.; Lin, W.S.; Lin, W.Y.; Hsu, Y.J.; Kuo, C.C.; et al. A novel protective function of 5-methoxytryptophan in vascular injury. Sci. Rep. 2016, 5, 25374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Xie, R.M.; Chao, X.; Zhang, Y.; Lin, R.C.; Sun, W.J. Bioactivity-directed isolation, identification of diuretic compounds from Polyporus umbellatus. J. Ethnopharmacol. 2009, 126, 184–187. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Wei, F.; Vaziri, N.D.; Cheng, X.L.; Bai, X.; Lin, R.C.; Zhao, Y.Y. Metabolomics insights into chronic kidney disease and modulatory effect of rhubarb against tubulointerstitial fibrosis. Sci. Rep. 2015, 5, 14472. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Vaziri, N.D.; Wei, F.; Cheng, X.L.; Bai, X.; Zhao, Y.Y. An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci. Rep. 2016, 6, 22151. [Google Scholar] [CrossRef]
- Chen, D.Q.; Cao, G.; Chen, H.; Liu, D.; Su, W.; Yu, X.Y.; Vaziri, N.D.; Liu, X.H.; Bai, X.; Zhang, L.; et al. Gene and protein expressions and metabolomics exhibit activated redox signaling and Wnt/β-catenin pathway are associated with metabolite dysfunction in patients with chronic kidney disease. Redox Biol. 2017, 12, 505–521. [Google Scholar] [CrossRef]
- Wang, M.; Chen, D.Q.; Wang, M.C.; Chen, H.; Chen, L.; Liu, D.; Zhao, H.; Zhao, Y.Y. Poricoic acid za, a novel RAS inhibitor, attenuates tubulo-interstitial fibrosis and podocyte injury by inhibiting TGF-β/smad signaling pathway. Phytomedicine 2017, 36, 243–253. [Google Scholar] [CrossRef]
- Chen, D.Q.; Cao, G.; Chen, H.; Argyopoulos, C.P.; Yu, H.; Su, W.; Chen, L.; Samuels, D.C.; Zhuang, S.; Bayliss, G.P.; et al. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat. Commun. 2019, 10, 1476. [Google Scholar] [CrossRef]




| Lipids | FC a | pb | pc | FDR d | AUC | R | Formula | Class |
|---|---|---|---|---|---|---|---|---|
| 20-Oxo-leukotriene E4 | 1.642 | 4.23 × 10−2 | 7.40 × 10−2 | 4.72 × 10−2 | 0.77 | 0.942 | C23H35NO6S | Fatty acids |
| Behenic acid | 1.510 | 2.42 × 10−2 | 8.59 × 10−1 | 4.69 × 10−2 | 0.01 | 0.920 | C22H44O2 | Fatty acids |
| LysoPC (20:0) | 2.517 | 3.82 × 10−2 | 3.50 × 10−2 | 4.27 × 10−2 | 0.81 | 0.897 | C28H58NO7P | GPs |
| Leukotriene E3 | 1.396 | 4.06 × 10−2 | 3.60 × 10−2 | 5.02 × 10−2 | 0.81 | 0.885 | C23H39NO5S | Fatty acids |
| 20-Oxo- leukotriene B4 | 2.031 | 4.22 × 10−2 | 4.60 × 10−2 | 4.80 × 10−2 | 0.80 | 0.871 | C20H30O5 | Fatty acids |
| LysoPE (18:0) | 1.405 | 1.67 × 10−2 | 1.20 × 10−2 | 5.39 × 10−2 | 0.88 | 0.870 | C23H48NO7P | GPs |
| Sulfolithocholic acid | 1.626 | 3.38 × 10−2 | 2.70 × 10−2 | 5.29 × 10−2 | 0.83 | 0.862 | C24H40O6S | Steroids |
| CDCA | 1.858 | 2.55 × 10−2 | 4.60 × 10−2 | 4.77 × 10−2 | 0.80 | 0.851 | C24H40O4 | Steroids |
| TG(60:6) | 2.930 | 5.51 × 10−3 | 3.50 × 10−2 | 4.99 × 10−2 | 0.81 | 0.800 | C63H110O6 | Glycerolipids |
| LPC(16:1) | 1.741 | 4.12 × 10−2 | 5.90 × 10−2 | 4.98 × 10−2 | 0.78 | 0.773 | C24H48NO7P | GPs |
| MG(18:2) | 0.530 | 7.83 × 10−3 | 6.00 × 10−3 | 3.79 × 10−2 | 0.91 | 0.772 | C21H38O4 | Glycerolipids |
| LysoPE(20:0) | 1.713 | 2.83 × 10−2 | 2.70 × 10−2 | 4.82 × 10−2 | 0.83 | 0.771 | C25H52NO7P | GPs |
| LysoPC(14:1) | 1.584 | 4.47 × 10−2 | 2.70 × 10−2 | 4.80 × 10−2 | 0.83 | 0.770 | C22H44NO7P | GPs |
| Cibaric acid | 2.121 | 9.41 × 10−3 | 1.10 × 10−2 | 4.20 × 10−2 | 0.88 | 0.744 | C18H28O5 | Fatty acids |
| MG(20:3) | 0.453 | 5.24 × 10−3 | 2.00 × 10−3 | 1.01 × 10−2 | 0.95 | 0.717 | C23H40O4 | Glycerolipids |
| Palmitelaidic acid | 2.135 | 3.86 × 10−2 | 3.00 × 10−2 | 5.09 × 10−2 | 0.82 | 0.694 | C16H30O2 | Fatty acids |
| LysoPC (20:4) | 2.866 | 3.94 × 10−2 | 2.10 × 10−2 | 5.08 × 10−2 | 0.84 | 0.691 | C28H50NO7P | GPs |
| 2,3-DOPS | 1.465 | 3.63 × 10−2 | 1.60 × 10−2 | 5.40 × 10−2 | 0.86 | 0.691 | C25H46O6 | Glycerolipids |
| PC(42:9) | 0.499 | 4.70 × 10−2 | 9.20 × 10−2 | 4.79 × 10−2 | 0.75 | 0.670 | C50H82NO8P | GPs |
| PA(33:4) | 0.736 | 4.30 × 10−2 | 4.60 × 10−2 | 4.70 × 10−2 | 0.80 | 0.667 | C36H63O8P | GPs |
| LysoPC(22:6) | 0.518 | 1.21 × 10−2 | 1.20 × 10−2 | 4.69 × 10−2 | 0.88 | 0.666 | C30H50NO7P | GPs |
| CPCA | 2.237 | 4.13 × 10−2 | 5.90 × 10−2 | 4.88 × 10−2 | 0.78 | 0.663 | C27H46O5 | Steroids |
| PA(21:0) | 0.510 | 3.40 × 10−2 | 2.10 × 10−2 | 5.20 × 10−2 | 0.84 | 0.622 | C24H47O8P | GPs |
| LysoPC(18:0) | 2.161 | 1.90 × 10−2 | 9.00 × 10−3 | 5.01 × 10−2 | 0.89 | 0.614 | C26H54NO7P | GPs |
| MG(19:0) | 0.513 | 2.19 × 10−2 | 1.60 × 10−2 | 4.89 × 10−2 | 0.86 | 0.601 | C22H44O4 | Glycerolipids |
| PC(20:1) | 0.416 | 6.58 × 10−3 | 6.00 × 10−3 | 4.63 × 10−2 | 0.91 | 0.598 | C28H56NO7P | GPs |
| Homophytanic acid | 1.296 | 2.27 × 10−2 | 9.00 × 10−3 | 4.55 × 10−2 | 0.89 | 0.597 | C21H42O2 | Fatty acids |
| PA(22:0) | 0.662 | 2.22 × 10−2 | 2.70 × 10−2 | 4.77 × 10−2 | 0.83 | 0.574 | C25H49O8P | GPs |
| MG(22:4) | 0.543 | 2.87 × 10−2 | 9.00 × 10−3 | 4.75 × 10−2 | 0.89 | 0.571 | C25H42O4 | Glycerolipids |
| TG(58:13) | 3.66 | 2.26 × 10−2 | 1.10 × 10−2 | 4.69 × 10−2 | 0.88 | 0.514 | C61H92O6 | Glycerolipids |
| MG(12:0) | 1.510 | 3.96 × 10−2 | 2.70 × 10−2 | 4.99 × 10−2 | 0.83 | 0.509 | C15H30O4 | Glycerolipids |
| 5-TCA | 2.883 | 3.85 × 10−2 | 4.30 × 10−2 | 4.19 × 10−2 | 0.80 | 0.449 | C14H26O2 | Fatty acids |
| PA(36:6) | 0.156 | 2.27 × 10−3 | 2.00 × 10−3 | 1.32 × 10−2 | 0.95 | 0.402 | C39H65O8P | GPs |
| PS(42:0) | 0.269 | 3.17 × 10−2 | 5.00 × 10−3 | 5.01 × 10−2 | 0.92 | 0.359 | C48H94O10P | GPs |
| Docosadienoate | 0.601 | 2.09 × 10−2 | 3.60 × 10−2 | 4.85 × 10−2 | 0.81 | 0.322 | C22H40O2 | Fatty acids |
| LysoPC(22:5) | 2.268 | 1.78 × 10−2 | 6.00 × 10−3 | 5.07 × 10−2 | 0.91 | 0.317 | C30H52NO7P | GPs |
| Arachidonic acid | 0.257 | 2.57 × 10−2 | 5.50 × 10−2 | 4.65 × 10−2 | 0.78 | 0.314 | C20H32O2 | Fatty acids |
| Nonadecanoic acid | 0.267 | 7.02 × 10−3 | 9.00 × 10−3 | 5.09 × 10−2 | 0.89 | 0.311 | C19H38O2 | Fatty acids |
| PGP(36:3) | 0.208 | 3.80 × 10−2 | 6.00 × 10−3 | 4.38 × 10−2 | 0.91 | 0.308 | C42H78O13P2 | GPs |
| PA(23:0) | 0.676 | 1.92 × 10−2 | 2.10 × 10−2 | 4.84 × 10−2 | 0.84 | 0.304 | C26H51O8P | GPs |
| DG(37:6) | 0.379 | 1.72 × 10−2 | 4.40 × 10−2 | 5.00 × 10−2 | 0.80 | 0.301 | C40H66O5 | Glycerolipids |
| PA(32:3) | 0.470 | 4.15 × 10−2 | 1.60 × 10−2 | 4.82 × 10−2 | 0.86 | 0.294 | C35H63O8P | GPs |
| MG(18:0) | 0.240 | 1.01 × 10−2 | 1.20 × 10−2 | 4.17 × 10−2 | 0.88 | 0.291 | C21H42O4 | Glycerolipids |
| Cetoleic acid | 0.271 | 7.16 × 10−3 | 9.00 × 10−3 | 4.62 × 10−2 | 0.89 | 0.278 | C22H42O2 | Fatty acids |
| 5-HETE | 0.261 | 6.89 × 10−3 | 9.00 × 10−3 | 4.71 × 10−2 | 0.89 | 0.271 | C20H32O3 | Fatty acids |
| DG(39:1) | 0.284 | 7.51 × 10−3 | 2.00 × 10−2 | 3.96 × 10−2 | 0.84 | 0.265 | C42H80O5 | Glycerolipids |
| MG(10:0) | 0.571 | 2.82 × 10−2 | 2.10 × 10−2 | 4.95 × 10−2 | 0.84 | 0.253 | C13H26O4 | Glycerolipids |
| MG(18:4) | 0.559 | 4.56 × 10−2 | 2.70 × 10−2 | 4.72 × 10−2 | 0.83 | 0.232 | C21H34O4 | Glycerolipids |
| Sphinganine 1-phosphate | 0.447 | 2.64 × 10−3 | 5.00 × 10−2 | 4.65 × 10−2 | 0.92 | 0.214 | C18H40NO5P | Sphingolipids |
| Eicosapentaenoic acid | 0.264 | 6.76 × 10−3 | 9.00 × 10−2 | 4.54 × 10−2 | 0.89 | 0.174 | C22H42O2 | Fatty acids |
| Hydroxymyristic acid | 0.609 | 1.50 × 10−2 | 2.10 × 10−2 | 4.32 × 10−2 | 0.84 | 0.129 | C14H28O3 | Fatty acids |
| ProstaglandinE2 | 1.345 | 3.71 × 10−2 | 4.60 × 10−2 | 4.38 × 10−2 | 0.80 | 0.125 | C22H37NO5 | Fatty acids |
| Colnelenate | 0.205 | 1.84 × 10−2 | 9.00 × 10−3 | 5.09 × 10−2 | 0.89 | 0.114 | C18H28O3 | Fatty acids |
| LysoPE(15:0) | 4.687 | 4.96 × 10−2 | 2.00 × 10−2 | 4.96 × 10−2 | 0.84 | 0.107 | C20H42NO7P | GPs |
| PS(42:11) | 0.192 | 4.48 × 10−2 | 6.00 × 10−3 | 4.72 × 10−2 | 0.91 | 0.090 | C48H72O10P | GPs |
| LysoPE(18:1) | 6.434 | 2.07 × 10−2 | 4.30 × 10−2 | 5.01 × 10−2 | 0.80 | 0.075 | C23H46NO7P | GPs |
| HOCA | 0.713 | 1.45 × 10−2 | 1.20 × 10−2 | 4.25 × 10−2 | 0.88 | 0.042 | C16H32O3 | Fatty acids |
| Eicosadienoic acid | 11.09 | 7.44 × 10−3 | 1.00 × 10−3 | 4.31 × 10−2 | 0.97 | 0.012 | C20H36O2 | Fatty acids |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-N.; Hu, H.-H.; Zhang, D.-D.; Wu, X.-Q.; Liu, J.-L.; Guo, Y.; Miao, H.; Zhao, Y.-Y. The Dysregulation of Eicosanoids and Bile Acids Correlates with Impaired Kidney Function and Renal Fibrosis in Chronic Renal Failure. Metabolites 2021, 11, 127. https://doi.org/10.3390/metabo11020127
Wang Y-N, Hu H-H, Zhang D-D, Wu X-Q, Liu J-L, Guo Y, Miao H, Zhao Y-Y. The Dysregulation of Eicosanoids and Bile Acids Correlates with Impaired Kidney Function and Renal Fibrosis in Chronic Renal Failure. Metabolites. 2021; 11(2):127. https://doi.org/10.3390/metabo11020127
Chicago/Turabian StyleWang, Yan-Ni, He-He Hu, Dan-Dan Zhang, Xia-Qing Wu, Jian-Ling Liu, Yan Guo, Hua Miao, and Ying-Yong Zhao. 2021. "The Dysregulation of Eicosanoids and Bile Acids Correlates with Impaired Kidney Function and Renal Fibrosis in Chronic Renal Failure" Metabolites 11, no. 2: 127. https://doi.org/10.3390/metabo11020127
APA StyleWang, Y.-N., Hu, H.-H., Zhang, D.-D., Wu, X.-Q., Liu, J.-L., Guo, Y., Miao, H., & Zhao, Y.-Y. (2021). The Dysregulation of Eicosanoids and Bile Acids Correlates with Impaired Kidney Function and Renal Fibrosis in Chronic Renal Failure. Metabolites, 11(2), 127. https://doi.org/10.3390/metabo11020127








