Enhancing Metabolomic Coverage in Positive Ionization Mode Using Dicationic Reagents by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization
Abstract
1. Introduction
2. Results and Discussions
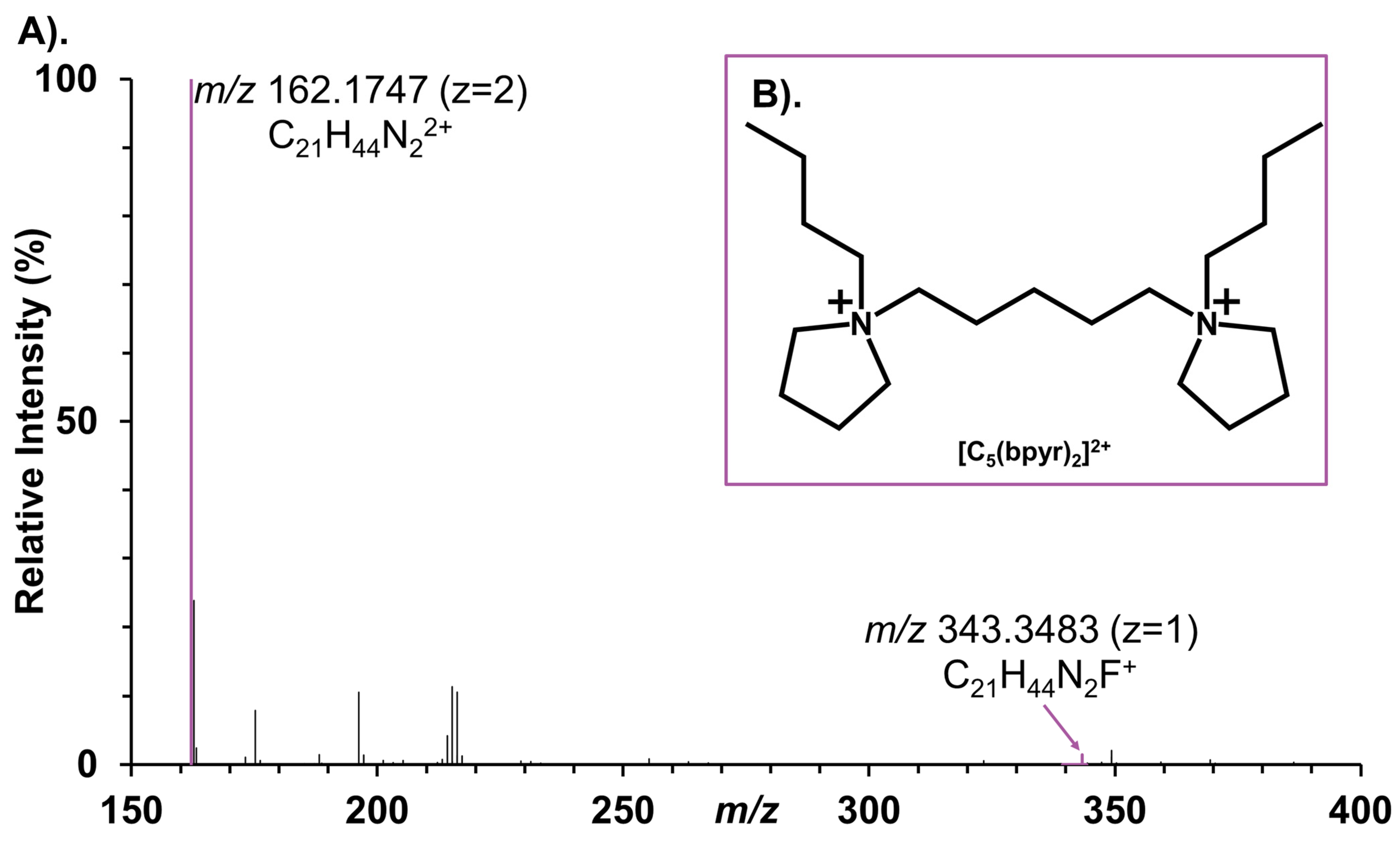
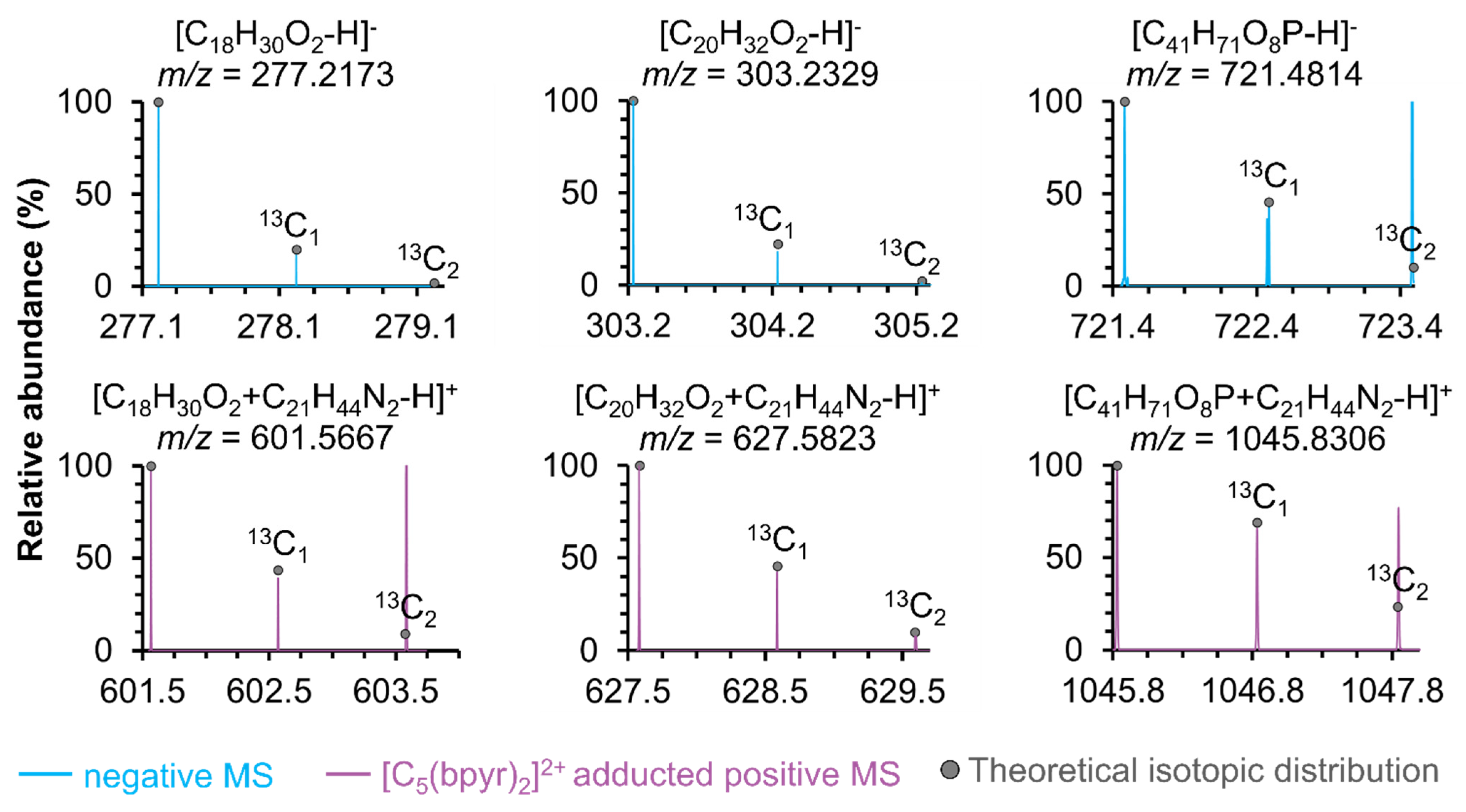
2.1. Detection of Negatively Charged Metabolites in Adducted Positive Ionization Polarity
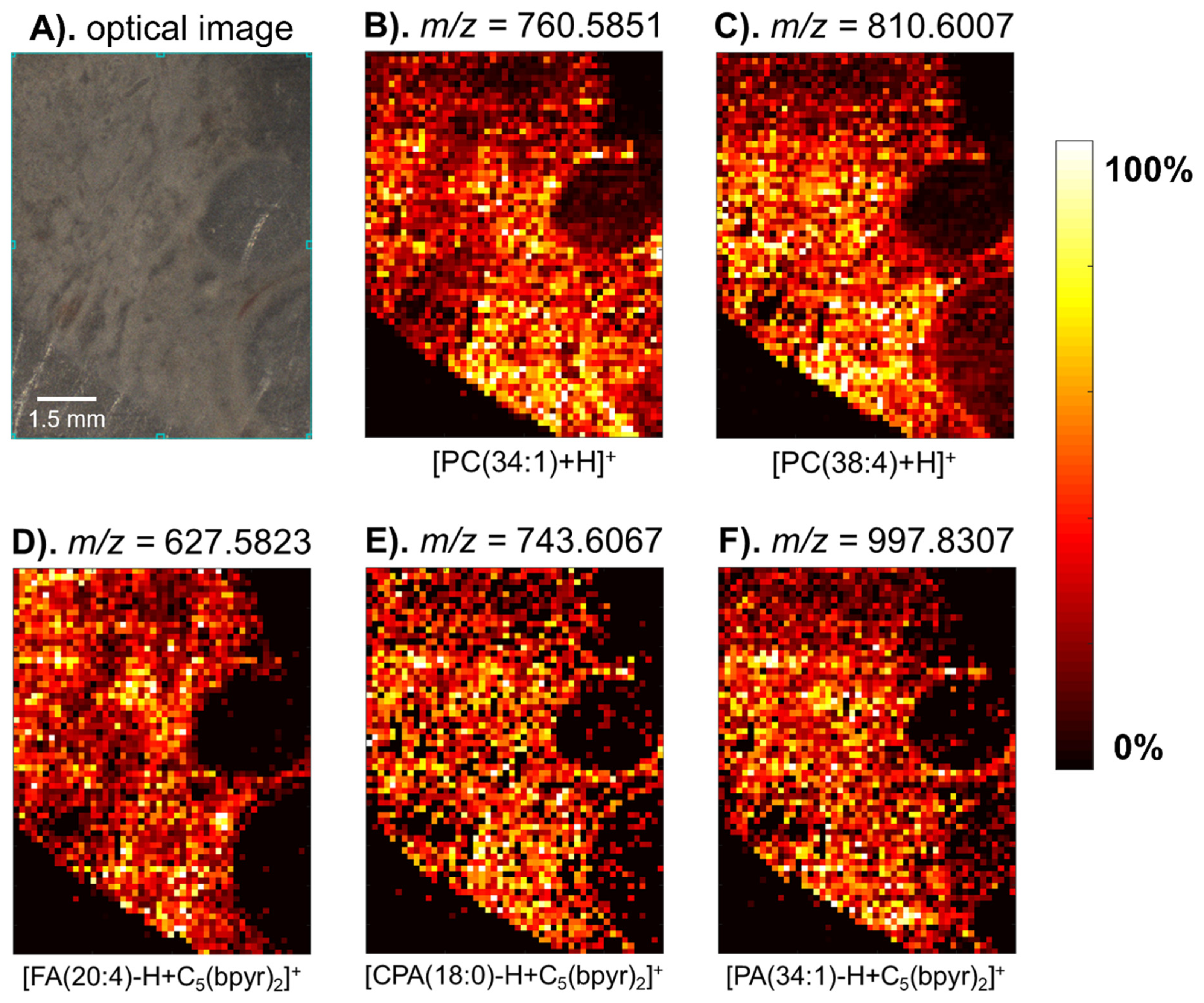
2.2. Mass Spectrometry Imaging of a Hen Ovary Section with the Dicationic Reagent
2.3. MS/MS Analysis of Selected Lipid Adducts with the Dicationic Reagent
3. Materials and Methods
3.1. Materials
3.2. Preparation of Rat Liver and Hen Ovary Sections
3.3. IR-MALDESI Experimental Parameters
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCartney, A.; Vignoli, A.; Biganzoli, L.; Love, R.; Tenori, L.; Luchinat, C.; Di Leo, A. Metabolomics in breast cancer: A decade in review. Cancer Treat. Rev. 2018, 67, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Andrews, W.T.; Donahue, D.; Holmes, A.; Balsara, R.; Castellino, F.J.; Hummon, A.B. In situ metabolite and lipid analysis of GluN2D−/− and wild-type mice after ischemic stroke using MALDI MSI. Anal. Bioanal. Chem. 2020, 412, 6275–6285. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, K.; Zhang, X. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: Mini review. Oncotarget 2017, 8, 115774–115786. [Google Scholar] [CrossRef] [PubMed]
- Ussher, J.R.; Elmariah, S.; Gerszten, R.E.; Dyck, J.R.B. The Emerging Role of Metabolomics in the Diagnosis and Prognosis of Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2850–2870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. BioMed Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- Armiñán, A.; Palomino-Schätzlein, M.; Deladriere, C.; Arroyo-Crespo, J.J.; Vicente-Ruiz, S.; Vicent, M.J.; Pineda-Lucena, A. Metabolomics facilitates the discrimination of the specific anti-cancer effects of free- and polymer-conjugated doxorubicin in breast cancer models. Biomaterials 2018, 162, 144–153. [Google Scholar] [CrossRef]
- Nazari, M.; Muddiman, D.C. Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst 2016, 141, 595–605. [Google Scholar] [CrossRef]
- Nazari, M.; Bokhart, M.T.; Loziuk, P.L.; Muddiman, D.C. Quantitative mass spectrometry imaging of glutathione in healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst 2018, 143, 654–661. [Google Scholar] [CrossRef]
- Tu, A.; Said, N.; Muddiman, D.C. Spatially resolved metabolomic characterization of muscle invasive bladder cancer by mass spectrometry imaging. Metabolomics 2021, 17, 70. [Google Scholar] [CrossRef]
- Walker, H.J. Metabolite imaging by mass spectrometry: A new discovery tool. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2021; Volume 98, ISBN 9780128216880. [Google Scholar]
- Schnackenberg, L.K.; Thorn, D.A.; Barnette, D.; Jones, E.E. MALDI imaging mass spectrometry: An emerging tool in neurology. Metab. Brain Dis. 2021. [Google Scholar] [CrossRef]
- Lee, P.Y.; Yeoh, Y.; Omar, N.; Pung, Y.-F.; Lim, L.C.; Low, T.Y. Molecular tissue profiling by MALDI imaging: Recent progress and applications in cancer research. Crit. Rev. Clin. Lab. Sci. 2021, 58, 513–529. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, J.; Chen, Y.; Xiong, S.; Wang, G.; Chen, J.; Yang, G. A Novel Strategy for MALDI-TOF MS Analysis of Small Molecules. J. Am. Soc. Mass Spectrom. 2010, 21, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Monopoli, A.; Cataldi, T.R.I.; Palmisano, F. MALDI matrices for low molecular weight compounds: An endless story? Anal. Bioanal. Chem. 2018, 410, 4015–4038. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Nie, H.; Liang, D.; Bai, Y.; Liu, H. Phospholipid imaging of zebrafish exposed to fipronil using atmospheric pressure matrix-assisted laser desorption ionization mass spectrometry. Talanta 2020, 209, 120357. [Google Scholar] [CrossRef] [PubMed]
- Kompauer, M.; Heiles, S.; Spengler, B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 2016, 14, 90–96. [Google Scholar] [CrossRef]
- Hayashi, Y.; Zaitsu, K.; Murata, T.; Ohara, T.; Moreau, S.; Kusano, M.; Tanihata, H.; Tsuchihashi, H.; Ishii, A.; Ishikawa, T. Intact metabolite profiling of mouse brain by probe electrospray ionization/triple quadrupole tandem mass spectrometry (PESI/MS/MS) and its potential use for local distribution analysis of the brain. Anal. Chim. Acta 2017, 983, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhao, Y.; Cai, S.; Fu, S.; Yang, C.; Zhang, S.; Zhang, X. Single cell analysis with probe ESI-mass spectrometry: Detection of metabolites at cellular and subcellular levels. Anal. Chem. 2014, 86, 3809–3816. [Google Scholar] [CrossRef]
- Banerjee, S.; Zare, R.N.; Tibshirani, R.J.; Kunder, C.A.; Nolley, R.; Fan, R.; Brooks, J.D.; Sonn, G.A. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. USA 2017, 114, 3334–3339. [Google Scholar] [CrossRef]
- León, M.; Ferreira, C.R.; Eberlin, L.S.; Jarmusch, A.K.; Pirro, V.; Rodrigues, A.C.B.; Favaron, P.O.; Miglino, M.A.; Cooks, R.G. Metabolites and Lipids Associated with Fetal Swine Anatomy via Desorption Electrospray Ionization—Mass Spectrometry Imaging. Sci. Rep. 2019, 9, 7247. [Google Scholar] [CrossRef]
- Bagley, M.C.; Ekelöf, M.; Rock, K.; Patisaul, H.; Muddiman, D.C. IR-MALDESI mass spectrometry imaging of underivatized neurotransmitters in brain tissue of rats exposed to tetrabromobisphenol A. Anal. Bioanal. Chem. 2018, 410, 7979–7986. [Google Scholar] [CrossRef]
- Xi, Y.; Tu, A.; Muddiman, D.C. Lipidomic profiling of single mammalian cells by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Anal. Bioanal. Chem. 2020, 412, 8211–8222. [Google Scholar] [CrossRef] [PubMed]
- Bagley, M.C.; Pace, C.L.; Ekelöf, M.; Muddiman, D.C. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) mass spectrometry imaging analysis of endogenous metabolites in cherry tomatoes. Analyst 2020, 145, 5516–5523. [Google Scholar] [CrossRef] [PubMed]
- Caleb Bagley, M.; Garrard, K.P.; Muddiman, D.C. The development and application of matrix assisted laser desorption electrospray ionization: The teenage years. Mass Spectrom. Rev. 2021, 1–32. [Google Scholar] [CrossRef]
- Bokhart, M.T.; Muddiman, D.C. Infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging analysis of biospecimens. Analyst 2016, 141, 5236–5245. [Google Scholar] [CrossRef]
- Dixon, R.B.; Muddiman, D.C. Study of the ionization mechanism in hybrid laser based desorption techniques. Analyst 2010, 135, 880–882. [Google Scholar] [CrossRef]
- Rosen, E.P.; Bokhart, M.T.; Ghashghaei, H.T.; Muddiman, D.C. Influence of desorption conditions on analyte sensitivity and internal energy in discrete tissue or whole body imaging by IR-MALDESI. J. Am. Soc. Mass Spectrom. 2015, 26, 899–910. [Google Scholar] [CrossRef][Green Version]
- Tu, A.; Muddiman, D.C. Internal Energy Deposition in Infrared Matrix-Assisted Laser Desorption Electrospray Ionization With and Without the Use of Ice as a Matrix. J. Am. Soc. Mass Spectrom. 2019, 30, 2380–2391. [Google Scholar] [CrossRef]
- Bai, H.; Linder, K.E.; Muddiman, D.C. Three-dimensional (3D) imaging of lipids in skin tissues with infrared matrix-assisted laser desorption electrospray ionization (MALDESI) mass spectrometry. Anal. Bioanal. Chem. 2021, 413, 2793–2801. [Google Scholar] [CrossRef]
- McClory, P.J.; Håkansson, K. Corona Discharge Suppression in Negative Ion Mode Nanoelectrospray Ionization via Trifluoroethanol Addition. Anal. Chem. 2017, 89, 10188–10193. [Google Scholar] [CrossRef] [PubMed]
- Cech, N.B.; Enke, C.G. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001, 20, 362–387. [Google Scholar] [CrossRef]
- Tsai, S.T.; Chen, C.W.; Huang, L.C.L.; Huang, M.C.; Chen, C.H.; Wang, Y.S. Simultaneous mass analysis of positive and negative ions using a dual-polarity time-of-flight mass spectrometer. Anal. Chem. 2006, 78, 7729–7734. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.L.; He, L.; Tu, Y.P.; Alexandrova, L. Application of polarity switching in the identification of the metabolites of RO9237. Rapid Commun. Mass Spectrom. 2007, 21, 1661–1668. [Google Scholar] [CrossRef]
- Breitbach, Z.S.; Berthod, A.; Huang, K.; Armstrong, D.W. Mass spectrometric detection of trace anions: The evolution of paired-ion electrospray ionization (PIESI). Mass Spectrom. Rev. 2016, 35, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Dodbiba, E.; Xu, C.; Payagala, T.; Wanigasekara, E.; Moon, M.H.; Armstrong, D.W. Use of ion pairing reagents for sensitive detection and separation of phospholipids in the positive ion mode LC-ESI-MS. Analyst 2011, 136, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Santos, I.C.; Guo, H.; Mesquita, R.B.R.; Rangel, A.O.S.S.; Armstrong, D.W.; Schug, K.A. Paired-ion electrospray ionization—Triple quadrupole tandem mass spectrometry for quantification of anionic surfactants in waters. Talanta 2015, 143, 320–327. [Google Scholar] [CrossRef]
- Remsburg, J.W.; Soukup-Hein, R.J.; Crank, J.A.; Breitbach, Z.S.; Payagala, T.; Armstrong, D.W. Evaluation of Dicationic Reagents for Their Use in Detection of Anions Using Positive Ion Mode ESI-MS Via Gas Phase Ion Association. J. Am. Soc. Mass Spectrom. 2008, 19, 261–269. [Google Scholar] [CrossRef]
- Rao, W.; Pan, N.; Tian, X.; Yang, Z. High-Resolution Ambient MS Imaging of Negative Ions in Positive Ion Mode: Using Dicationic Reagents with the Single-Probe. J. Am. Soc. Mass Spectrom. 2016, 27, 124–134. [Google Scholar] [CrossRef]
- Pan, N.; Rao, W.; Standke, S.J.; Yang, Z. Using dicationic ion-pairing compounds to enhance the single cell mass spectrometry analysis using the single-probe: A microscale sampling and ionization device. Anal. Chem. 2016, 88, 6812–6819. [Google Scholar] [CrossRef]
- Cotte-Rodríguez, I.; Takáts, Z.; Talaty, N.; Chen, H.; Cooks, R.G. Desorption electrospray ionization of explosives on surfaces: Sensitivity and selectivity enhancement by reactive desorption electrospray ionization. Anal. Chem. 2005, 77, 6755–6764. [Google Scholar] [CrossRef]
- Lostun, D.; Perez, C.J.; Licence, P.; Barrett, D.A.; Ifa, D.R. Reactive DESI-MS Imaging of Biological Tissues with Dicationic Ion-Pairing Compounds. Anal. Chem. 2015, 87, 3286–3293. [Google Scholar] [CrossRef]
- Rao, W.; Mitchell, D.; Licence, P.; Barrett, D.A. The use of dicationic ion-pairing compounds to enhance the ambient detection of surface lipids in positive ionization mode using desorption electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 616–624. [Google Scholar] [CrossRef]
- Xu, C.; Guo, H.; Breitbach, Z.S.; Armstrong, D.W. Mechanism and sensitivity of anion detection using rationally designed unsymmetrical dications in paired ion electrospray ionization mass spectrometry. Anal. Chem. 2014, 86, 2665–2672. [Google Scholar] [CrossRef]
- Breitbach, Z.S.; Wanigasekara, E.; Dodbiba, E.; Schug, K.A.; Armstrong, D.W. Mechanisms of ESI-MS selectivity and sensitivity enhancements when detecting anions in the positive mode using cationic pairing agents. Anal. Chem. 2010, 82, 9066–9073. [Google Scholar] [CrossRef]
- Guo, H.; Breitbach, Z.S.; Armstrong, D.W. Reduced matrix effects for anionic compounds with paired ion electrospray ionization mass spectrometry. Anal. Chim. Acta 2016, 912, 74–84. [Google Scholar] [CrossRef]
- Khodjaniyazova, S.; Nazari, M.; Garrard, K.P.; Matos, M.P.V.; Jackson, G.P.; Muddiman, D.C. Characterization of the Spectral Accuracy of an Orbitrap Mass Analyzer Using Isotope Ratio Mass Spectrometry. Anal. Chem. 2018, 90, 1897–1906. [Google Scholar] [CrossRef]
- Robichaud, G.; Barry, J.A.; Garrard, K.P.; Muddiman, D.C. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J. Am. Soc. Mass Spectrom. 2013, 24, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, G.; Barry, J.A.; Muddiman, D.C. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J. Am. Soc. Mass Spectrom. 2014, 25, 319–328. [Google Scholar] [CrossRef]
- Ekelöf, M.; Manni, J.; Nazari, M.; Bokhart, M.; Muddiman, D.C. Characterization of a novel miniaturized burst-mode infrared laser system for IR-MALDESI mass spectrometry imaging. Anal. Bioanal. Chem. 2018, 410, 2395–2402. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Schramm, T.; Hester, A.; Klinkert, I.; Both, J.P.; Heeren, R.M.A.; Brunelle, A.; Laprévote, O.; Desbenoit, N.; Robbe, M.F.; Stoeckli, M.; et al. ImzML—A common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteomics 2012, 75, 5106–5110. [Google Scholar] [CrossRef] [PubMed]
- Race, A.M.; Styles, I.B.; Bunch, J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics 2012, 75, 5111–5112. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, G.; Garrard, K.P.; Barry, J.A.; Muddiman, D.C. MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on matlab platform. J. Am. Soc. Mass Spectrom. 2013, 24, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Bokhart, M.T.; Nazari, M.; Garrard, K.P.; Muddiman, D.C. MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. J. Am. Soc. Mass Spectrom. 2018, 29, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.; Phapale, P.; Chernyavsky, I.; Lavigne, R.; Fay, D.; Tarasov, A.; Kovalev, V.; Fuchser, J.; Nikolenko, S.; Pineau, C.; et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods 2016, 14, 57–60. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, Y.; Muddiman, D.C. Enhancing Metabolomic Coverage in Positive Ionization Mode Using Dicationic Reagents by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization. Metabolites 2021, 11, 810. https://doi.org/10.3390/metabo11120810
Xi Y, Muddiman DC. Enhancing Metabolomic Coverage in Positive Ionization Mode Using Dicationic Reagents by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization. Metabolites. 2021; 11(12):810. https://doi.org/10.3390/metabo11120810
Chicago/Turabian StyleXi, Ying, and David C. Muddiman. 2021. "Enhancing Metabolomic Coverage in Positive Ionization Mode Using Dicationic Reagents by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization" Metabolites 11, no. 12: 810. https://doi.org/10.3390/metabo11120810
APA StyleXi, Y., & Muddiman, D. C. (2021). Enhancing Metabolomic Coverage in Positive Ionization Mode Using Dicationic Reagents by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization. Metabolites, 11(12), 810. https://doi.org/10.3390/metabo11120810





