Glutamine Homeostasis and Its Role in the Adaptive Strategies of the Blind Mole Rat, Spalax
Abstract
:1. Introduction
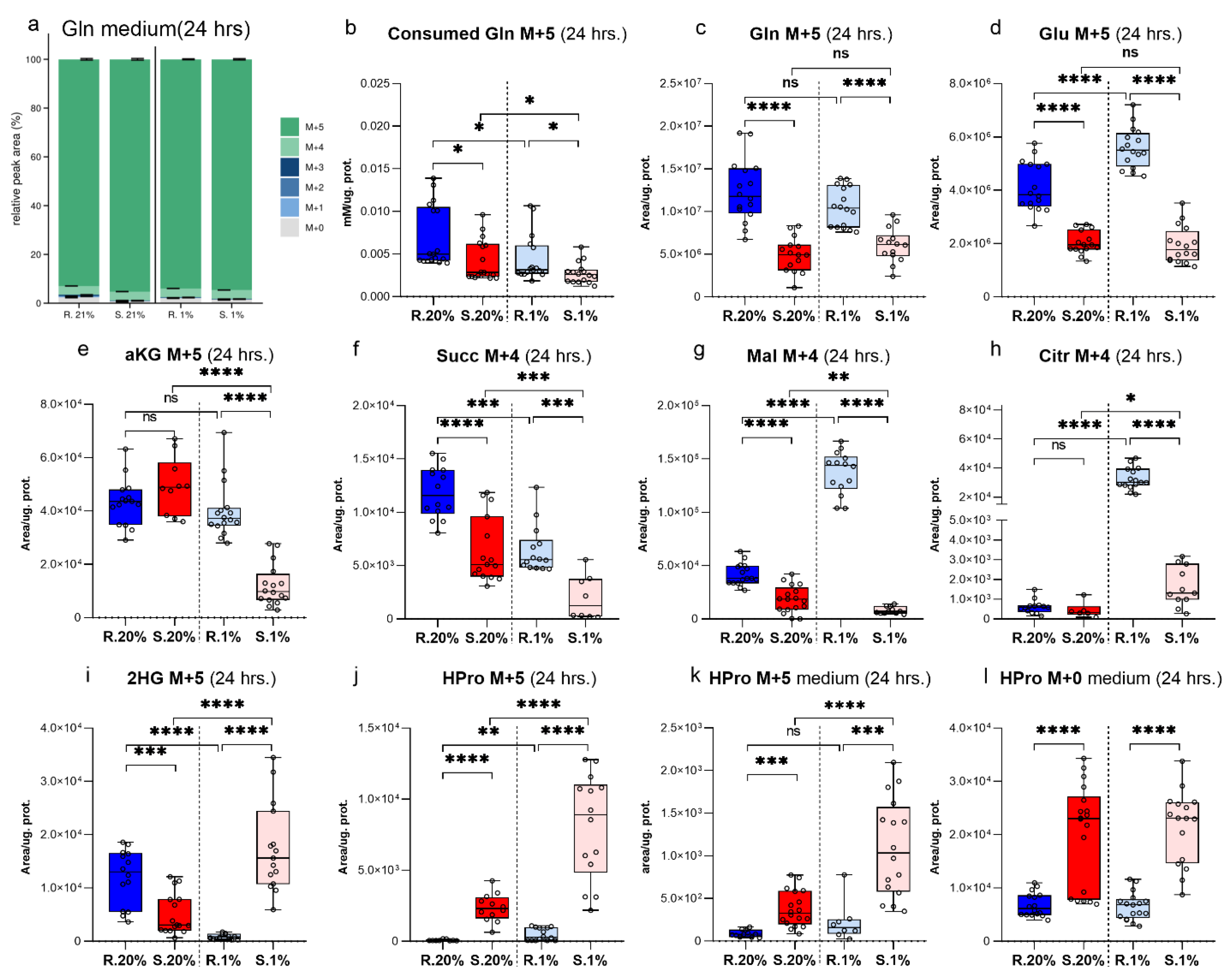
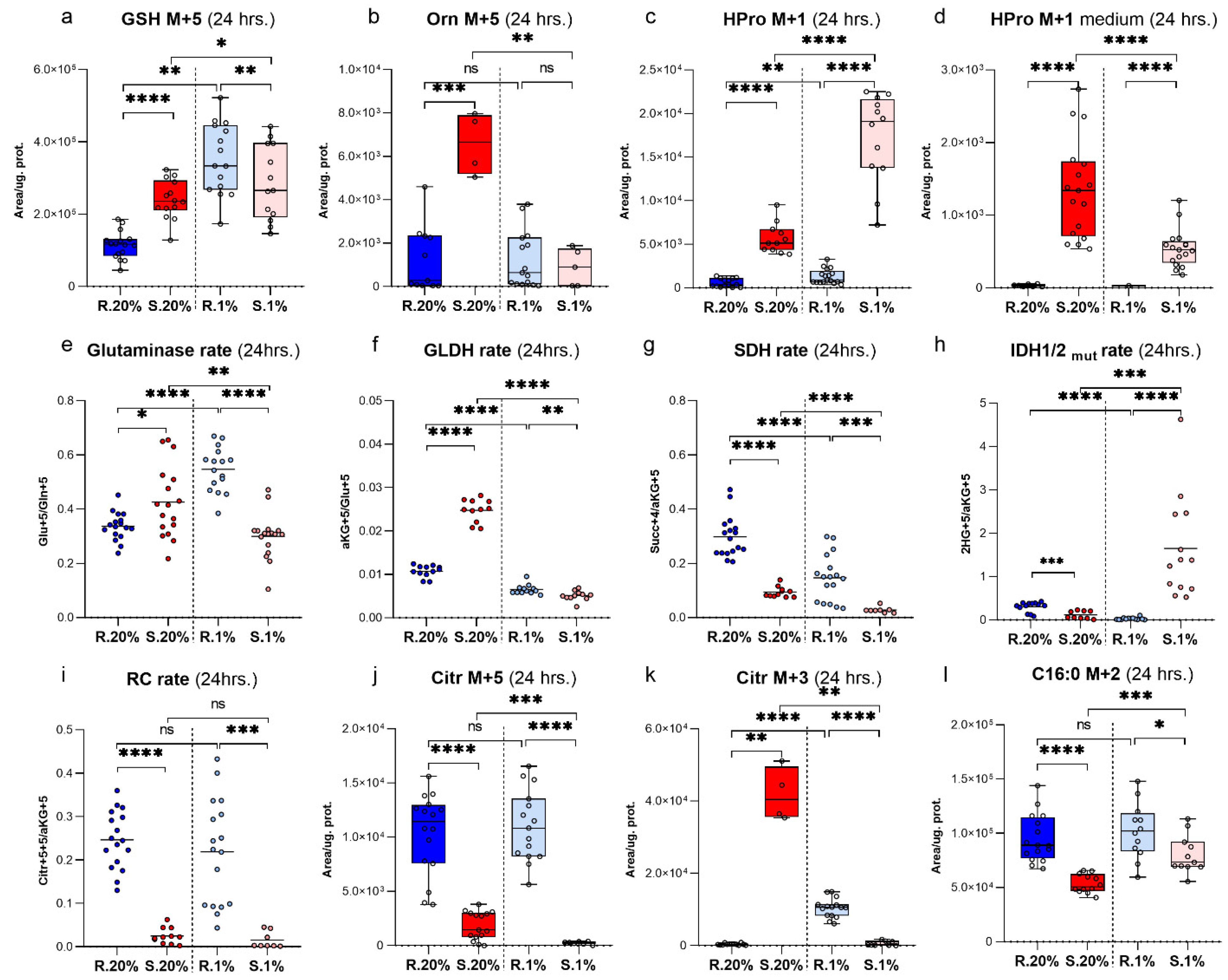
2. Results
Characteristics of αKG Metabolic Domain
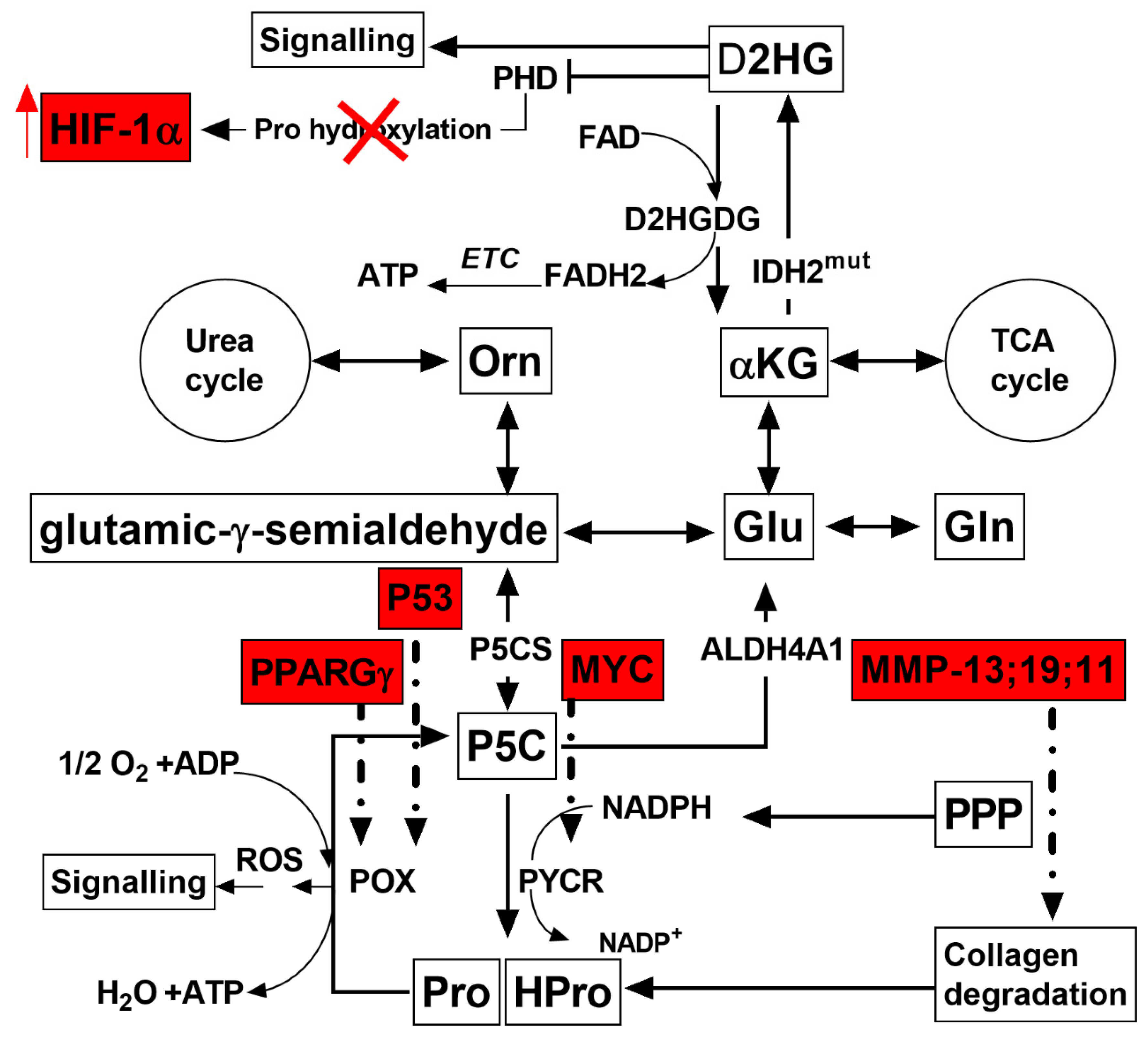
3. Discussion
4. Materials and Methods
4.1. General Experimental Design
4.2. Animals
4.3. Cell Culture
4.4. Metabolomic Analysis
4.5. qPCR Assay
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, E. Nonnutritive effects of glutamine. J. Nutr. 2008, 138, 2025S–2031S. [Google Scholar] [CrossRef]
- Coloff, J.L.; Murphy, J.P.; Braun, C.R.; Harris, I.S.; Shelton, L.M.; Kami, K.; Gygi, S.P.; Selfors, L.M.; Brugge, J.S. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 2016, 23, 867–880. [Google Scholar] [CrossRef]
- Curi, R.; Lagranha, C.J.; Doi, S.Q.; Sellitti, D.F.; Procopio, J.; Pithon-Curi, T.C.; Corless, M.; Newsholme, P. Molecular mechanisms of glutamine action. J. Cell. Physiol. 2005, 204, 392–401. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Ko, B.; Hensley, C.T.; Jiang, L.; Wasti, A.T.; Kim, J.; Sudderth, J.; Calvaruso, M.A.; Lumata, L.; Mitsche, M.; et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 2014, 56, 414–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumvoll, M.; Perriello, G.; Meyer, C.; Gerich, J. Role of glutamine in human carbohydrate metabolism in kidney and other tissues. Kidney Int. 1999, 55, 778–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhakar, N.R.; Semenza, G.L. Oxygen Sensing and Homeostasis. Physiology 2015, 30, 340–348. [Google Scholar] [CrossRef] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakazawa, M.S.; Keith, B.; Simon, M.C. Oxygen availability and metabolic adaptations. Nat. Rev. Cancer 2016, 16, 663–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, N.C.; O’Neill, L.A.J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.C.; Denko, N.C. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014, 19, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watford, M. Glutamine metabolism and function in relation to proline synthesis and the safety of glutamine and proline supplementation. J. Nutr. 2008, 138, 2003S–2007S. [Google Scholar] [CrossRef] [Green Version]
- Phang, J.M.; Liu, W.; Hancock, C.; Christian, K.J. The proline regulatory axis and cancer. Front. Oncol. 2012, 2, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Glunde, K.; Bhujwalla, Z.M.; Raman, V.; Sharma, A.; Phang, J.M. Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer Res. 2012, 72, 3677–3686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phang, J.M.; Downing, S.J.; Yeh, G.C. Linkage of the HMP pathway to ATP generation by the proline cycle. Biochem. Biophys. Res. Commun. 1980, 93, 462–470. [Google Scholar] [CrossRef]
- Hagedorn, C.H.; Phang, J.M. Transfer of reducing equivalents into mitochondria by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch. Biochem. Biophys. 1983, 225, 95–101. [Google Scholar] [CrossRef]
- Krishnan, N.; Dickman, M.B.; Becker, D.F. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol. Med. 2008, 44, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Nagano, T.; Nakashima, A.; Onishi, K.; Kawai, K.; Awai, Y.; Kinugasa, M.; Iwasaki, T.; Kikkawa, U.; Kamada, S. Proline dehydrogenase promotes senescence through the generation of reactive oxygen species. J. Cell Sci. 2017, 130, 1413–1420. [Google Scholar] [CrossRef] [Green Version]
- Wadhawan, S.; Gautam, S.; Sharma, A. Involvement of proline oxidase (PutA) in programmed cell death of Xanthomonas. PLoS ONE 2014, 9, e96423. [Google Scholar] [CrossRef] [Green Version]
- Rivera, A.; Maxwell, S.A. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J. Biol. Chem. 2005, 280, 29346–29354. [Google Scholar] [CrossRef] [Green Version]
- Goncalves, R.L.; Rothschild, D.E.; Quinlan, C.L.; Scott, G.K.; Benz, C.C.; Brand, M.D. Sources of superoxide/H2O2 during mitochondrial proline oxidation. Redox Biol. 2014, 2, 901–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaraffia, P.Y.; Wells, M.A. Proline can be utilized as an energy substrate during flight of Aedes aegypti females. J. Insect Physiol. 2003, 49, 591–601. [Google Scholar] [CrossRef]
- Pandhare, J.; Donald, S.P.; Cooper, S.K.; Phang, J.M. Regulation and function of proline oxidase under nutrient stress. J. Cell Biochem. 2009, 107, 759–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shams, I.; Avivi, A.; Nevo, E. Hypoxic stress tolerance of the blind subterranean mole rat: Expression of erythropoietin and hypoxia-inducible factor 1 alpha. Proc. Natl. Acad. Sci. USA 2004, 101, 9698–9703. [Google Scholar] [CrossRef] [Green Version]
- Shams, I.; Avivi, A.; Nevo, E. Oxygen and carbon dioxide fluctuations in burrows of subterranean blind mole rats indicate tolerance to hypoxic-hypercapnic stresses. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005, 142, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Tacutu, R.; Craig, T.; Budovsky, A.; Wuttke, D.; Lehmann, G.; Taranukha, D.; Costa, J.; Fraifeld, V.E.; de Magalhaes, J.P. Human Ageing Genomic Resources: Integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013, 41, D1027–D1033. [Google Scholar] [CrossRef]
- Caballero, B.; Tomas-Zapico, C.; Vega-Naredo, I.; Sierra, V.; Tolivia, D.; Hardeland, R.; Rodriguez-Colunga, M.J.; Joel, A.; Nevo, E.; Avivi, A.; et al. Antioxidant activity in Spalax ehrenbergi: A possible adaptation to underground stress. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2006, 192, 753–759. [Google Scholar] [CrossRef]
- Manov, I.; Hirsh, M.; Iancu, T.C.; Malik, A.; Sotnichenko, N.; Band, M.; Avivi, A.; Shams, I. Pronounced cancer resistance in a subterranean rodent, the blind mole-rat, Spalax: In vivo and in vitro evidence. BMC Biol. 2013, 11, 91. [Google Scholar] [CrossRef] [Green Version]
- Viltard, M.; Durand, S.; Perez-Lanzon, M.; Aprahamian, F.; Lefevre, D.; Leroy, C.; Madeo, F.; Kroemer, G.; Friedlander, G. The metabolomic signature of extreme longevity: Naked mole rats versus mice. Aging 2019, 11, 4783–4800. [Google Scholar] [CrossRef]
- Park, T.J.; Reznick, J.; Peterson, B.L.; Blass, G.; Omerbasic, D.; Bennett, N.C.; Kuich, P.; Zasada, C.; Browe, B.M.; Hamann, W.; et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 2017, 356, 307–311. [Google Scholar] [CrossRef] [Green Version]
- Faulkes, C.G.; Eykyn, T.R.; Aksentijevic, D. Cardiac metabolomic profile of the naked mole-rat-glycogen to the rescue. Biol. Lett. 2019, 15, 20190710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ahn, W.S.; Gameiro, P.A.; Keibler, M.A.; Zhang, Z.; Stephanopoulos, G. 13C isotope-assisted methods for quantifying glutamine metabolism in cancer cells. Methods Enzymol. 2014, 542, 369–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.; Stephanopoulos, G.; Kelleher, J.K. Quantifying carbon sources for de novo lipogenesis in wild-type and IRS-1 knockout brown adipocytes. J. Lipid Res. 2004, 45, 1324–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Le, A.; Hancock, C.; Lane, A.N.; Dang, C.V.; Fan, T.W.; Phang, J.M. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. USA 2012, 109, 8983–8988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Fendt, S.M.; Bell, E.L.; Keibler, M.A.; Olenchock, B.A.; Mayers, J.R.; Wasylenko, T.M.; Vokes, N.I.; Guarente, L.; Vander Heiden, M.G.; Stephanopoulos, G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat. Commun. 2013, 4, 2236. [Google Scholar] [CrossRef] [Green Version]
- Taylor, L.; Curthoys, N.P. Glutamine metabolism: Role in acid-base balance. Biochem. Mol. Biol. Educ. 2004, 32, 291–304. [Google Scholar] [CrossRef]
- Yelamanchi, S.D.; Jayaram, S.; Thomas, J.K.; Gundimeda, S.; Khan, A.A.; Singhal, A.; Keshava Prasad, T.S.; Pandey, A.; Somani, B.L.; Gowda, H. A pathway map of glutamate metabolism. J. Cell Commun. Signal. 2016, 10, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Häussinger, D.; Sies, H. Glutamine Metabolism in Mammalian Tissues; Springer: Berlin/Heidelberg, Germany, 1984. [Google Scholar]
- McKeehan, W.L. Glycolysis, glutaminolysis and cell proliferation. Cell Biol. Int. Rep. 1982, 6, 635–650. [Google Scholar] [CrossRef]
- Reitzer, L.J.; Wice, B.M.; Kennell, D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979, 254, 2669–2676. [Google Scholar] [CrossRef]
- Ros, S.; Schulze, A. Balancing glycolytic flux: The role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 2013, 1, 8. [Google Scholar] [CrossRef] [Green Version]
- Chandel, N.S.; Maltepe, E.; Goldwasser, E.; Mathieu, C.E.; Simon, M.C.; Schumacker, P.T. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 11715–11720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Groot, H.; Littauer, A. Hypoxia, reactive oxygen, and cell injury. Free Radic Biol. Med. 1989, 6, 541–551. [Google Scholar] [CrossRef]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Ballester, M.; Sentandreu, E.; Luongo, G.; Santamaria, R.; Bolonio, M.; Alcoriza-Balaguer, M.I.; Palomino-Schatzlein, M.; Pineda-Lucena, A.; Castell, J.; Lahoz, A.; et al. Glutamine/glutamate metabolism rewiring in reprogrammed human hepatocyte-like cells. Sci. Rep. 2019, 9, 17978. [Google Scholar] [CrossRef]
- Bertolo, R.F.; Burrin, D.G. Comparative aspects of tissue glutamine and proline metabolism. J. Nutr. 2008, 138, 2032S–2039S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pegg, A.E.; Casero, R.A., Jr. Current status of the polyamine research field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar] [CrossRef] [Green Version]
- Pegg, A.E. Regulation of ornithine decarboxylase. J. Biol. Chem. 2006, 281, 14529–14532. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, H.; Malik, A.; Bicker, A.; Poetzsch, G.; Avivi, A.; Shams, I.; Hankeln, T. Hypoxia tolerance, longevity and cancer-resistance in the mole rat Spalax—A liver transcriptomics approach. Sci. Rep. 2017, 7, 14348. [Google Scholar] [CrossRef]
- Imberman, M.; Oppenheim, F.; Franzblau, C. The appearance of free hydroxyproline as the major product of degradation of newly synthesized collagen in cell culture. Biochim. Biophys. Acta 1982, 719, 480–487. [Google Scholar] [CrossRef]
- Hu, S.; He, W.; Wu, G. Hydroxyproline in animal metabolism, nutrition, and cell signaling. Amino Acids 2021. [Google Scholar] [CrossRef] [PubMed]
- Heinz, A. Elastases and elastokines: Elastin degradation and its significance in health and disease. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 252–273. [Google Scholar] [CrossRef]
- Wise, D.R.; Ward, P.S.; Shay, J.E.; Cross, J.R.; Gruber, J.J.; Sachdeva, U.M.; Platt, J.M.; DeMatteo, R.G.; Simon, M.C.; Thompson, C.B. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA 2011, 108, 19611–19616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Shestov, A.A.; Swain, P.; Yang, C.; Parker, S.J.; Wang, Q.A.; Terada, L.S.; Adams, N.D.; McCabe, M.T.; Pietrak, B.; et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016, 532, 255–258. [Google Scholar] [CrossRef]
- Du, J.; Yanagida, A.; Knight, K.; Engel, A.L.; Vo, A.H.; Jankowski, C.; Sadilek, M.; Tran, V.T.; Manson, M.A.; Ramakrishnan, A.; et al. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2016, 113, 14710–14715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaude, E.; Schmidt, C.; Gammage, P.A.; Dugourd, A.; Blacker, T.; Chew, S.P.; Saez-Rodriguez, J.; O’Neill, J.S.; Szabadkai, G.; Minczuk, M.; et al. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol. Cell 2018, 69, 581–593.e587. [Google Scholar] [CrossRef] [Green Version]
- Lemons, J.M.; Feng, X.J.; Bennett, B.D.; Legesse-Miller, A.; Johnson, E.L.; Raitman, I.; Pollina, E.A.; Rabitz, H.A.; Rabinowitz, J.D.; Coller, H.A. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010, 8, e1000514. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.; Antoniewicz, M.R.; Stephanopoulos, G.; Kelleher, J.K. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J. Biol. Chem. 2008, 283, 20621–20627. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.L.; Semenza, G.L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995, 270, 1230–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014, 9, 47–71. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.; Leung, I.K.; Tian, Y.M.; Abboud, M.I.; Ge, W.; Domene, C.; Cantrelle, F.X.; Landrieu, I.; Hardy, A.P.; Pugh, C.W.; et al. Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases. Nat. Commun. 2016, 7, 12673. [Google Scholar] [CrossRef] [PubMed]
- Struys, E.A.; Verhoeven, N.M.; Brunengraber, H.; Jakobs, C. Investigations by mass isotopomer analysis of the formation of D-2-hydroxyglutarate by cultured lymphoblasts from two patients with D-2-hydroxyglutaric aciduria. FEBS Lett. 2004, 557, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Lin, Y.; Xu, W.; Jiang, W.; Zha, Z.; Wang, P.; Yu, W.; Li, Z.; Gong, L.; Peng, Y.; et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 2009, 324, 261–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achouri, Y.; Noel, G.; Vertommen, D.; Rider, M.H.; Veiga-Da-Cunha, M.; Van Schaftingen, E. Identification of a dehydrogenase acting on D-2-hydroxyglutarate. Biochem. J. 2004, 381, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struys, E.A.; Verhoeven, N.M.; Jansen, E.E.; Ten Brink, H.J.; Gupta, M.; Burlingame, T.G.; Quang, L.S.; Maher, T.; Rinaldo, P.; Snead, O.C.; et al. Metabolism of gamma-hydroxybutyrate to d-2-hydroxyglutarate in mammals: Further evidence for d-2-hydroxyglutarate transhydrogenase. Metabolism 2006, 55, 353–358. [Google Scholar] [CrossRef]
- Parker, A.; Engel, P.C. Preliminary evidence for the existence of specific functional assemblies between enzymes of the beta-oxidation pathway and the respiratory chain. Biochem. J. 2000, 345 Pt 3, 429–435. [Google Scholar] [CrossRef]
- Toplak, M.; Brunner, J.; Schmidt, J.; Macheroux, P. Biochemical characterization of human D-2-hydroxyglutarate dehydrogenase and two disease related variants reveals the molecular cause of D-2-hydroxyglutaric aciduria. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140255. [Google Scholar] [CrossRef]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef]
- Karna, E.; Szoka, L.; Huynh, T.Y.L.; Palka, J.A. Proline-dependent regulation of collagen metabolism. Cell. Mol. Life Sci. 2020, 77, 1911–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priest, R.E.; Davies, L.M. Cellular proliferation and synthesis of collagen. Lab. Investig. 1969, 21, 138–142. [Google Scholar] [PubMed]
- Klionsky, D.J.; Abdalla, F.C.; Abeliovich, H.; Abraham, R.T.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [CrossRef] [PubMed]
- Palka, J.A.; Phang, J.M. Prolidase in human breast cancer MCF-7 cells. Cancer Lett. 1998, 127, 63–70. [Google Scholar] [CrossRef]
- Wu, Z.; Hou, Y.; Dai, Z.; Hu, C.A.; Wu, G. Metabolism, Nutrition, and Redox Signaling of Hydroxyproline. Antioxid. Redox Signal. 2019, 30, 674–682. [Google Scholar] [CrossRef]
- Phang, J.M.; Harrop, S.J.; Duff, A.P.; Sokolova, A.V.; Crossett, B.; Walsh, J.C.; Beckham, S.A.; Nguyen, C.D.; Davies, R.B.; Glockner, C.; et al. Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin. Biochem. J. 2016, 473, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Yeh, G.C.; Phang, J.M. Stimulation of phosphoribosyl pyrophosphate and purine nucleotide production by pyrroline 5-carboxylate in human erythrocytes. J. Biol. Chem. 1988, 263, 13083–13089. [Google Scholar] [CrossRef]
- Balboni, E. A proline shuttle in insect flight muscle. Biochem. Biophys. Res. Commun. 1978, 85, 1090–1096. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W.; Zabirnyk, O. Proline metabolism and microenvironmental stress. Annu. Rev. Nutr. 2010, 30, 441–463. [Google Scholar] [CrossRef] [Green Version]
- Boyle, J. Lehninger Principles of Biochemistry, 4th ed.; Biochemistry and Molecular Biology, Education; Nelson, D., Cox, M., Eds.; Freeman and Company: New York, NY, USA, 2005; pp. 74–75. [Google Scholar] [CrossRef]
- Tang, L.; Yang, G.; Ma, M.; Liu, X.; Li, B.; Xie, J.; Fu, Y.; Chen, T.; Yu, Y.; Chen, W.; et al. An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Gao, X.; Yang, J.; Guo, W.; Wu, Z.; Tang, L.; Cao, S.; Cai, X.; Liu, T.; Jia, Q.; et al. Treatment strategies and outcomes for spinal rhabdomyosarcoma: A series of 11 cases in a single center and review of the literature. Clin. Neurol. Neurosurg. 2020, 192, 105729. [Google Scholar] [CrossRef]
- Polyak, K.; Xia, Y.; Zweier, J.L.; Kinzler, K.W.; Vogelstein, B. A model for p53-induced apoptosis. Nature 1997, 389, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Stoner, G.D.; Merchant, D.J. Amino acid utilization by L-M strain mouse cells in a chemically defined medium. In Vitro 1972, 7, 330–343. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Matrix metalloproteinases (MMPs): Chemical-biological functions and (Q)SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, M.; Qin, L.; Li, X.; He, G.; Liu, X.; Xu, W. Deficiency of DICER reduces the invasion ability of trophoblasts and impairs the pro-angiogenic effect of trophoblast-derived microvesicles. J. Cell. Mol. Med. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetten, M.R. Some aspects of the metabolism of hydroxyproline, studied with the aid of isotopic nitrogen. J. Biol. Chem. 1949, 181, 31–37. [Google Scholar] [CrossRef]
- Valle, D.; Goodman, S.I.; Harris, S.C.; Phang, J.M. Genetic evidence for a common enzyme catalyzing the second step in the degradation of proline and hydroxyproline. J. Clin. Investig. 1979, 64, 1365–1370. [Google Scholar] [CrossRef]
- Galili, S.; Ran, H.; Dor, E.; Hershenhorn, J.; Harel, A.; Amir-Segev, O.; Bellalou, A.; Badani, H.; Smirnov, E.; Achdari, G. The history of chickpea cultivation and breeding in Israel. Israel J. Plant Sci. 2018, 65, 186–194. [Google Scholar] [CrossRef]
- Parsons, R.; Sunley, R.J. Nitrogen nutrition and the role of root–shoot nitrogen signalling particularly in symbiotic systems. J. Exp. Bot. 2001, 52, 435–443. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Araújo, S.; Gill, S.S. The Plant Family Fabaceae: Biology and Physiological Responses to Environmental Stresses; Springer: Singapore, 2020. [Google Scholar]
- Sulieman, S.; Tran, L.-S.P. Asparagine: An amide of particular distinction in the regulation of symbiotic nitrogen fixation of legumes. Crit. Rev. Biotechnol. 2013, 33, 309–327. [Google Scholar] [CrossRef]
- Miskevich, D.; Chaban, A.; Dronina, M.; Abramovich, I.; Gottlieb, E.; Shams, I. Comprehensive Analysis of 13C6 Glucose Fate in the Hypoxia-Tolerant Blind Mole Rat Skin Fibroblasts. Metabolites 2021, 11, 734. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Velez, J.; Hail, N., Jr.; Konopleva, M.; Zeng, Z.; Kojima, K.; Samudio, I.; Andreeff, M. Mitochondrial uncoupling and the reprograming of intermediary metabolism in leukemia cells. Front. Oncol. 2013, 3, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miskevich, D.; Chaban, A.; Dronina, M.; Abramovich, I.; Gottlieb, E.; Shams, I. Glutamine Homeostasis and Its Role in the Adaptive Strategies of the Blind Mole Rat, Spalax. Metabolites 2021, 11, 755. https://doi.org/10.3390/metabo11110755
Miskevich D, Chaban A, Dronina M, Abramovich I, Gottlieb E, Shams I. Glutamine Homeostasis and Its Role in the Adaptive Strategies of the Blind Mole Rat, Spalax. Metabolites. 2021; 11(11):755. https://doi.org/10.3390/metabo11110755
Chicago/Turabian StyleMiskevich, Dmitry, Anastasia Chaban, Maria Dronina, Ifat Abramovich, Eyal Gottlieb, and Imad Shams. 2021. "Glutamine Homeostasis and Its Role in the Adaptive Strategies of the Blind Mole Rat, Spalax" Metabolites 11, no. 11: 755. https://doi.org/10.3390/metabo11110755





