Distinguishing between Isobaric Ions Using Microdroplet Hydrogen–Deuterium Exchange Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
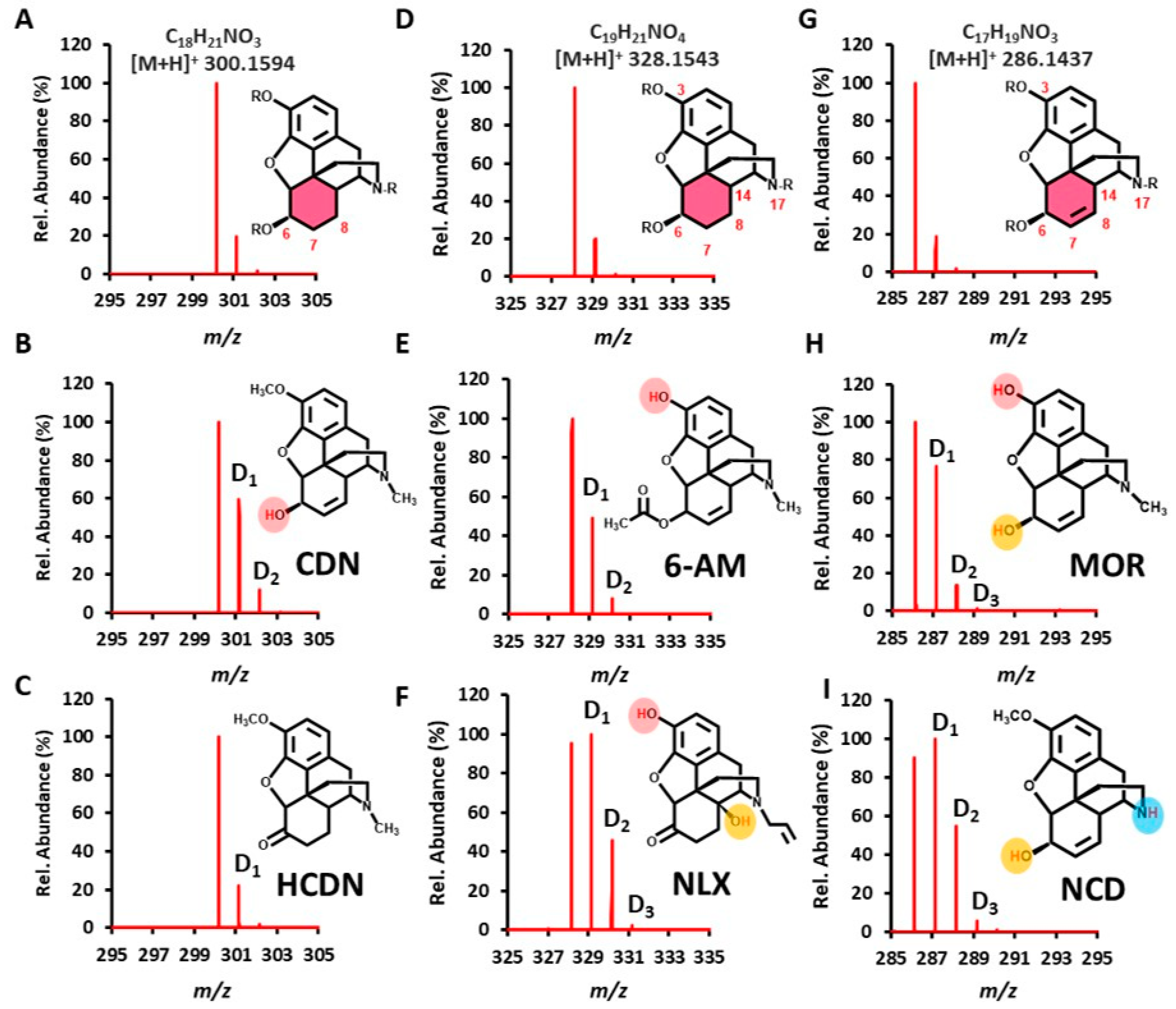
2.1. Rapid Recognition of Opioid Narcotics
2.2. Distinguishing Isobaric Ions That Are Challenging to Tell Apart by MS/MS
2.3. Correcting Falsely Matched Adduct Ions
2.4. Comparison with Gas-Phase HDX
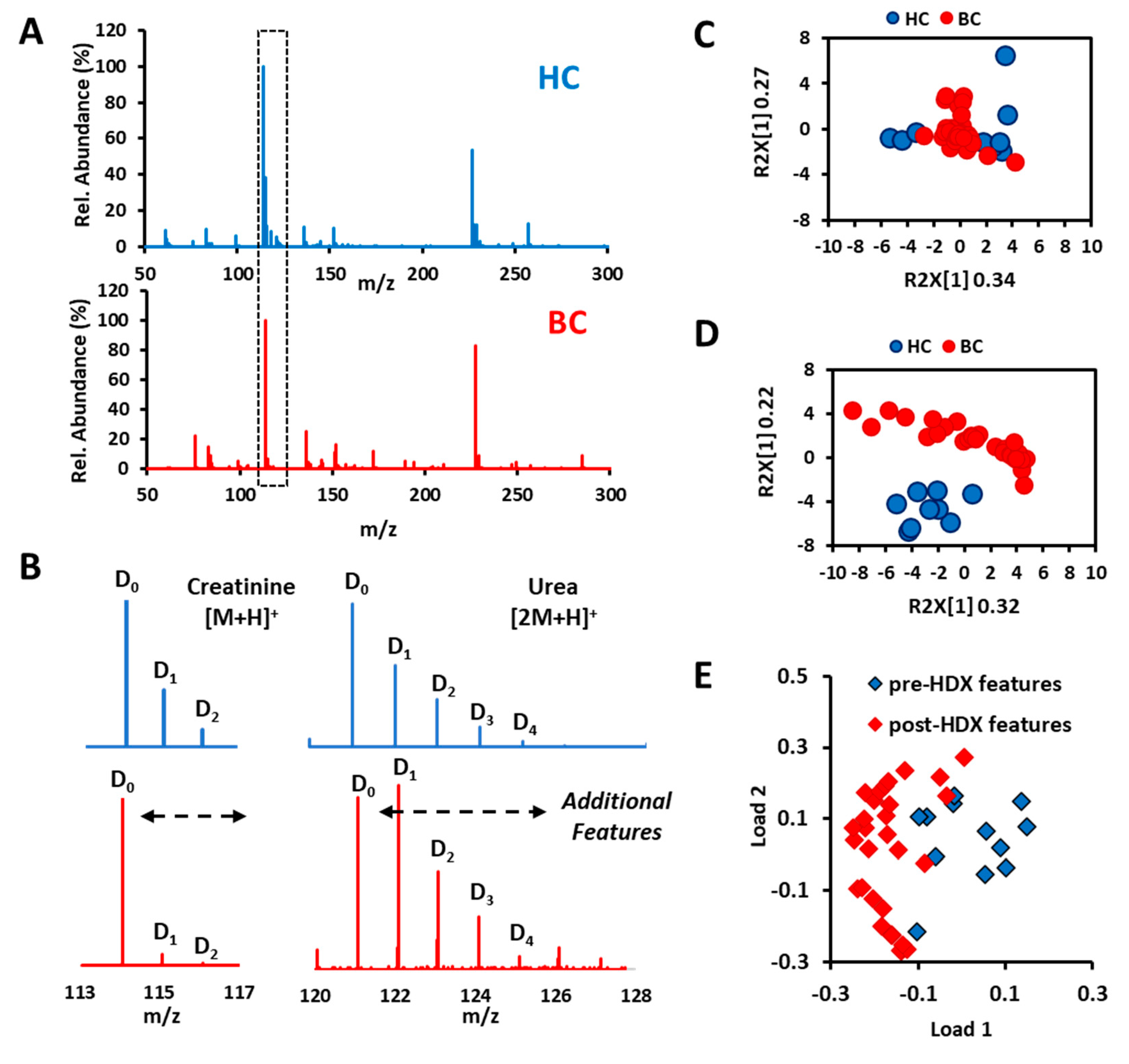
2.5. Creating New Dimension and Features for Metabolic Profiling
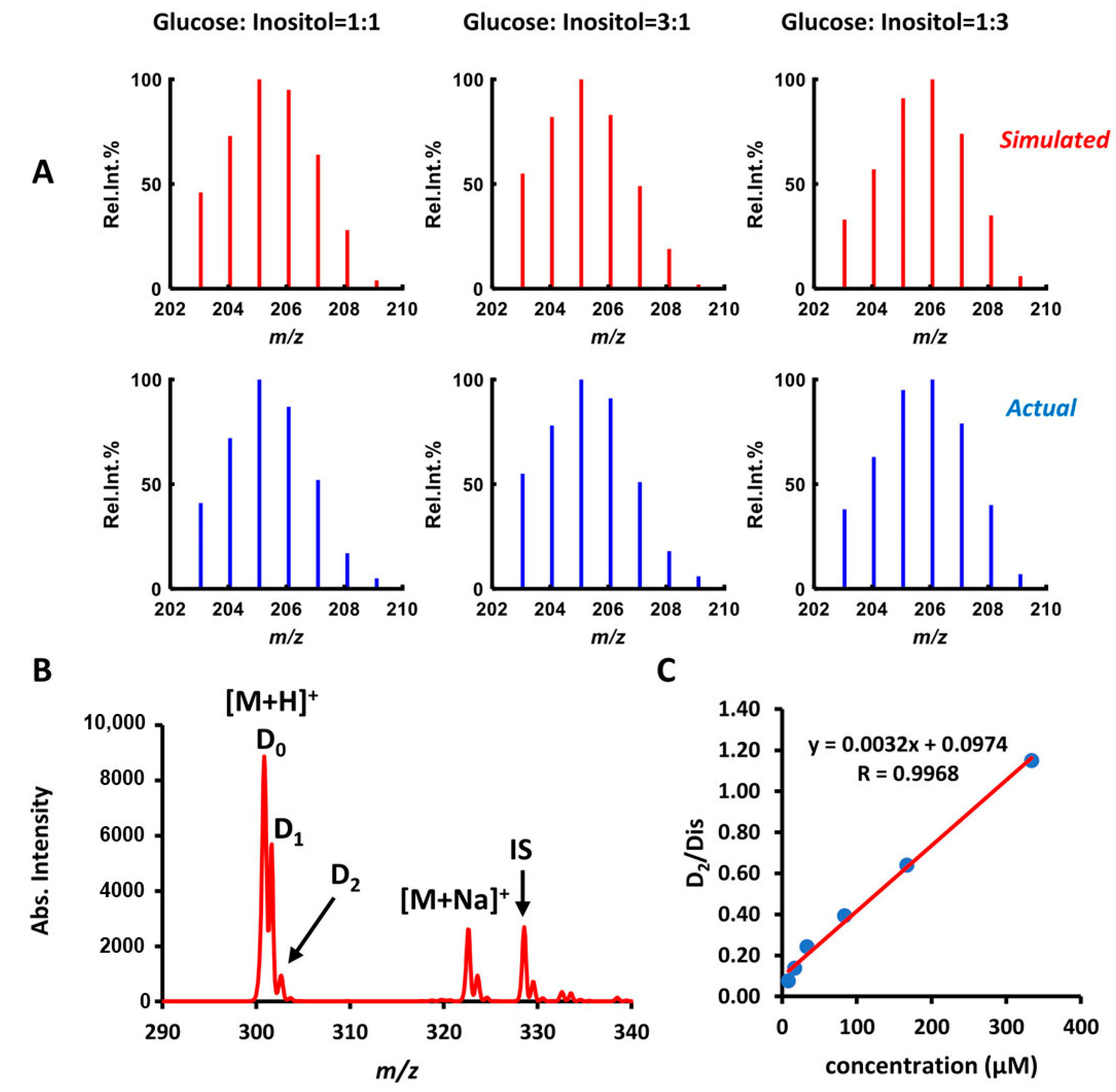
2.6. Relative and Absolute Quantitation
3. Methods
3.1. Reagents and Materials
3.2. Solution Preparation and Bio-Sample Collection
3.3. Droplet Spray Ionization
3.4. Data Acquisition and Processing
3.5. Isobaric Ion Discrimination and Assignment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feider, C.L.; Krieger, A.C.; DeHoog, R.J.; Eberlin, L.S. Ambient Ionization Mass Spectrometry: Recent Developments and Applications. Anal. Chem. 2019, 91, 4266–4290. [Google Scholar] [CrossRef]
- Takats, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass Spectrometry Sampling Under Ambient Conditions with Desorption Electrospray Ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Bhattarai, M.; Zhao, P.; Alnsour, T.; Held, M.; Faik, A.; Chen, H. Fast and Sensitive Detection of Oligosaccharides Using Desalting Paper Spray Mass Spectrometry (DPS-MS). J. Am. Soc. Mass Spectrom. 2020, 31, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, H.; Maas, J.D.; Chappell, W.J.; Manicke, N.E.; Cooks, R.G.; Ouyang, Z. Paper spray ionization devices for direct, biomedical analysis using mass spectrometry. Int. J. Mass Spectrom. 2012, 312, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Wang, H.; Liu, J.; Wei, P.; Cooks, R.G.; Ouyang, Z. Quantitative paper spray mass spectrometry analysis of drugs of abuse. Analyst 2013, 138, 4443–4447. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, Y.; Zhan, L.; Meng, L.; Huang, X.; Wang, T.; Li, Y.; Nie, Z. Point-of-Care Test Paper for Exhaled Breath Aldehyde Analysis via Mass Spectrometry. Anal. Chem. 2021, 93, 9158–9165. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.-Y.; Wang, Q.; Gunawardena, H.P.; Held, M.; Faik, A.; Chen, H. Desalting paper spay mass spectrometry (DPS-MS) for rapid detection of glycans and glycoconjugates. Int. J. Mass Spectrom. 2021, 469, 116688. [Google Scholar] [CrossRef]
- Rossini, E.L.; Kulyk, D.S.; Ansu-Gyeabourh, E.; Sahraeian, T.; Pezza, H.R.; Badu-Tawiah, A.K. Direct Analysis of Doping Agents in Raw Urine Using Hydrophobic Paper Spray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 1212–1222. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-N.; Di, D.; Yuan, Z.-C.; Wu, L.; Hu, B. Schirmer Paper Noninvasive Microsampling for Direct Mass Spectrometry Analysis of Human Tears. Anal. Chem. 2020, 92, 6207–6212. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zheng, Y.; Wang, X.; Austin, D.E.; Zhang, Z. Sub-ppt Mass Spectrometric Detection of Therapeutic Drugs in Complex Biological Matrixes Using Polystyrene-Microsphere-Coated Paper Spray. Anal. Chem. 2017, 89, 7988–7995. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, C.; Hatch, M.; Garrett, T. Extracellular Vesicle Analysis by Paper Spray Ionization Mass Spectrometry. Metabolites 2021, 11, 308. [Google Scholar] [CrossRef]
- Song, X.; Yang, X.; Narayanan, R.; Shankar, V.; Ethiraj, S.; Wang, X.; Duan, N.; Ni, Y.-H.; Hu, Q.; Zare, R.N. Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proc. Natl. Acad. Sci. USA 2020, 117, 16167–16173. [Google Scholar] [CrossRef]
- Li, C.; Li, K.; Xu, X.; Qi, W.; Hu, X.; Jin, P. A pilot study for colorectal carcinoma screening by instant metabolomic profiles using conductive polymer spray ionization mass spectrometry. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2021, 1867, 166210. [Google Scholar] [CrossRef] [PubMed]
- Mendes, T.P.P.; Pereira, I.; De Lima, L.A.S.; Morais, C.; Neves, A.C.; Martin, F.L.; Lima, K.M.G.; Vaz, B.G. Paper Spray Ionization Mass Spectrometry as a Potential Tool for Early Diagnosis of Cervical Cancer. J. Am. Soc. Mass Spectrom. 2020, 31, 1665–1672. [Google Scholar] [CrossRef]
- Mahmud, I.; Pinto, F.G.; Rubio, V.Y.; Lee, B.; Pavlovich, C.P.; Perera, R.J.; Garrett, T.J. Rapid Diagnosis of Prostate Cancer Disease Progression Using Paper Spray Ionization Mass Spectrometry. Anal. Chem. 2021, 93, 7774–7780. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Chung, H.-H.; Dutkiewicz, E.P.; Chen, C.-L.; Hsieh, H.-Y.; Chen, B.-R.; Wang, M.-Y.; Hsu, C.-C. Predicting Breast Cancer by Paper Spray Ion Mobility Spectrometry Mass Spectrometry and Machine Learning. Anal. Chem. 2019, 92, 1653–1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burnum-Johnson, K.E.; Zheng, X.; Dodds, J.N.; Ash, J.; Fourches, D.; Nicora, C.D.; Wendler, J.P.; Metz, T.O.; Waters, K.M.; Jansson, J.K.; et al. Ion mobility spectrometry and the omics: Distinguishing isomers, molecular classes and contaminant ions in complex samples. TrAC Trends Anal. Chem. 2019, 116, 292–299. [Google Scholar] [CrossRef]
- Maleki, H.; Maurer, M.M.; Ronaghi, N.; Valentine, S.J. Ion Mobility, Hydrogen/Deuterium Exchange, and Isotope Scrambling: Tools to Aid Compound Identification in ‘Omics Mixtures. Anal. Chem. 2017, 89, 6399–6407. [Google Scholar] [CrossRef]
- Uppal, S.S.; Beasley, S.E.; Scian, M.; Guttman, M. Gas-Phase Hydrogen/Deuterium Exchange for Distinguishing Isomeric Carbohydrate Ions. Anal. Chem. 2017, 89, 4737–4742. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.; Bell, S.; Maleki, H.; Valentine, S.J. Synthetic Small Molecule Characterization and Isomer Discrimination Using Gas-Phase Hydrogen–Deuterium Exchange IMS-MS. Anal. Chem. 2019, 91, 6259–6265. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Huang, Y.; Yu, F.; Li, Z.H.; Ding, C.-F. Effect of Transition-Metal Ions on the Conformation of Encephalin Investigated by Hydrogen/Deuterium Exchange and Theoretical Calculations. J. Phys. Chem. B 2020, 124, 101–109. [Google Scholar] [CrossRef]
- Trabjerg, E.; Nazari, Z.E.; Rand, K.D. Conformational analysis of complex protein states by hydrogen/deuterium exchange mass spectrometry (HDX-MS): Challenges and emerging solutions. TrAC Trends Anal. Chem. 2018, 106, 125–138. [Google Scholar] [CrossRef]
- Majuta, S.N.; Li, C.; Jayasundara, K.; Karanji, A.K.; Attanayake, K.; Ranganathan, N.; Li, P.; Valentine, S.J. Rapid Solution-Phase Hydrogen/Deuterium Exchange for Metabolite Compound Identification. J. Am. Soc. Mass Spectrom. 2019, 30, 1102–1114. [Google Scholar] [CrossRef]
- Narayanan, R.; Song, X.; Chen, H.; Zare, R.N. Teflon Spray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 234–239. [Google Scholar] [CrossRef]
- Dulay, M.T.; Boeser, C.R.; Walker, K.L.; Feider, C.; Zare, R.N. Polymer Substrate with Surface Solvent Reservoir for Polymer-Spray Mass Spectrometric Analysis of Hydrophilic Drugs. Talanta Open 2021, 4, 100048. [Google Scholar] [CrossRef]
- Dulay, M.T.; Zare, R.N. Polymer-spray mass spectrometric detection and quantitation of hydrophilic compounds and some narcotics. Rapid Commun. Mass Spectrom. 2017, 31, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Chen, H.; Zare, R.N. Coulometry--assisted quantitation in spray ionization mass spectrometry. J. Mass Spectrom. 2021, 56, e4628. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Chen, H.; Zare, R.N. Conductive Polymer Spray Ionization Mass Spectrometry for Biofluid Analysis. Anal. Chem. 2018, 90, 12878–12885. [Google Scholar] [CrossRef]
- Song, X.; Zang, Q.; Zare, R.N. Hydrogen–Deuterium Exchange Desorption Electrospray Ionization Mass Spectrometry Visualizes an Acidic Tumor Microenvironment. Anal. Chem. 2021, 93, 10411–10417. [Google Scholar] [CrossRef]
- Hamuro, Y. Tutorial: Chemistry of Hydrogen/Deuterium Exchange Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2021, 32, 133–151. [Google Scholar] [CrossRef]
- Liyanage, O.T.; Brantley, M.R.; Calixte, E.I.; Solouki, T.; Shuford, K.L.; Gallagher, E.S. Characterization of Electrospray Ionization (ESI) Parameters on In-ESI Hydrogen/Deuterium Exchange of Carbohydrate-Metal Ion Adducts. J. Am. Soc. Mass Spectrom. 2019, 30, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Kostyukevich, Y.; Acter, T.; Zherebker, A.; Ahmed, A.; Kim, S.; Nikolaev, E. Hydrogen/deuterium exchange in mass spectrometry. Mass. Spec. Rev. 2018, 37, 811–853. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Li, J.; Mofidfar, M.; Zare, R.N. Distinguishing between Isobaric Ions Using Microdroplet Hydrogen–Deuterium Exchange Mass Spectrometry. Metabolites 2021, 11, 728. https://doi.org/10.3390/metabo11110728
Song X, Li J, Mofidfar M, Zare RN. Distinguishing between Isobaric Ions Using Microdroplet Hydrogen–Deuterium Exchange Mass Spectrometry. Metabolites. 2021; 11(11):728. https://doi.org/10.3390/metabo11110728
Chicago/Turabian StyleSong, Xiaowei, Jia Li, Mohammad Mofidfar, and Richard N. Zare. 2021. "Distinguishing between Isobaric Ions Using Microdroplet Hydrogen–Deuterium Exchange Mass Spectrometry" Metabolites 11, no. 11: 728. https://doi.org/10.3390/metabo11110728
APA StyleSong, X., Li, J., Mofidfar, M., & Zare, R. N. (2021). Distinguishing between Isobaric Ions Using Microdroplet Hydrogen–Deuterium Exchange Mass Spectrometry. Metabolites, 11(11), 728. https://doi.org/10.3390/metabo11110728







