Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study
Abstract
1. Introduction
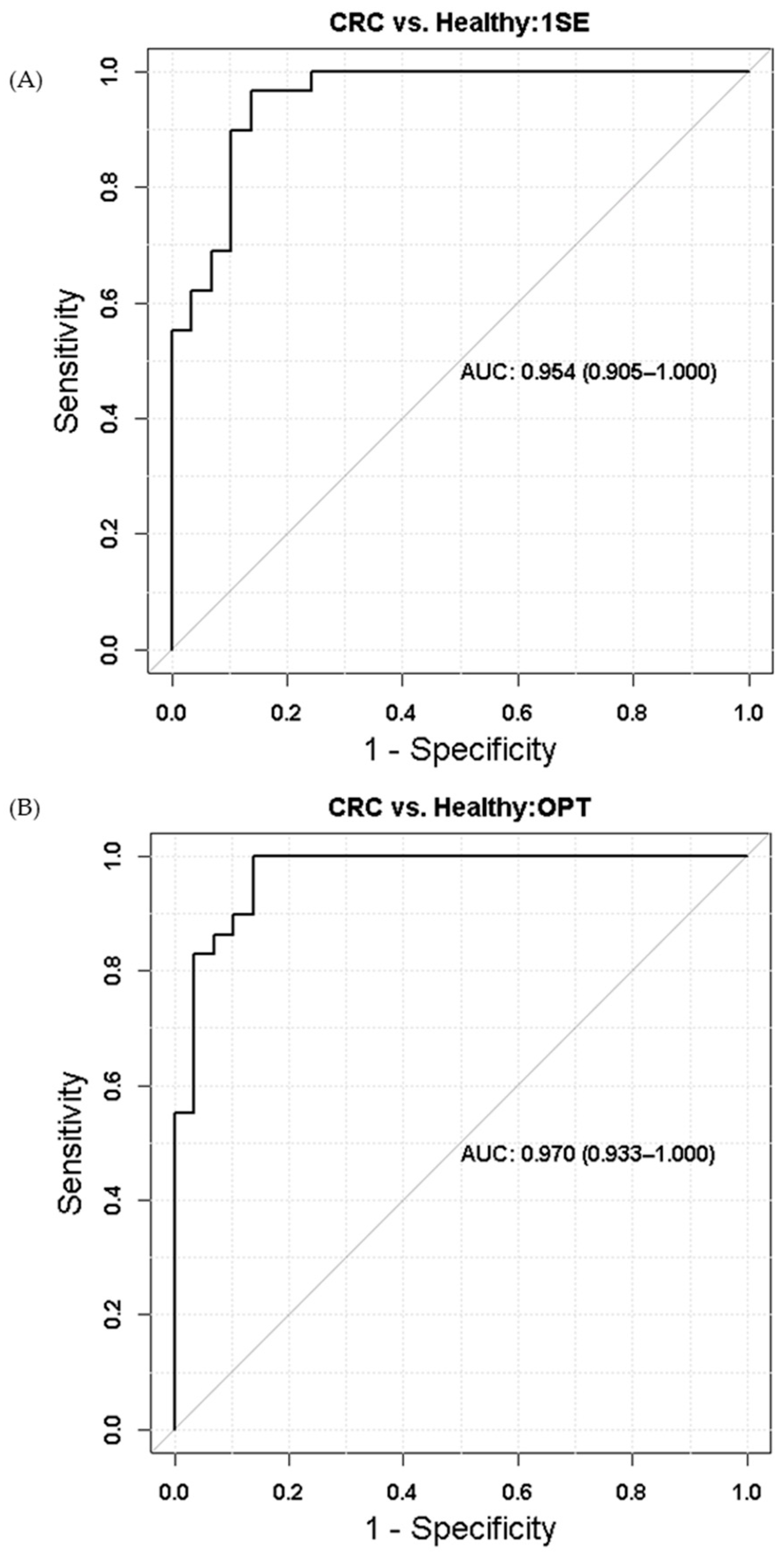
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Study Samples
4.3. Sample Preparation
4.4. GC × GC-MS Analysis
4.5. 2DLC-MS Analysis
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cui, L.; Lu, H.T.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.P.; Park, Y.; Ziegler, T.R. Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu. Rev. Nutr. 2012, 32, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Mishur, R.J.; Rea, S.L. Applications of mass spectrometry to metabolomics and metabonomics: Detection of biomarkers of aging and of age-related diseases. Mass Spectrom. Rev. 2012, 31, 70–95. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C. Systems biology—Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef]
- Yu, B.; Zanetti, K.A.; Temprosa, M.; Albanes, D.; Appel, N.; Barrera, C.B.; Ben-Shlomo, Y.; Boerwinkle, E.; Casas, J.P.; Clish, C.; et al. The consortium of metabolomics studies (COMETS): Metabolomics in 47 prospective cohort studies. Am. J. Epidemiol. 2019, 188, 991–1012. [Google Scholar] [CrossRef]
- Suhre, K.; Meisinger, C.; Doring, A.; Altmaier, E.; Belcredi, P.; Gieger, C.; Chang, D.; Milburn, M.V.; Gall, W.E.; Weinberger, K.M.; et al. Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS ONE 2010, 5, e13953. [Google Scholar] [CrossRef]
- Prodhan, M.A.I.; Shi, B.Y.; Song, M.; He, L.Q.; Yuan, F.; Yin, X.M.; Bohman, P.; McClain, C.J.; Zhang, X. Integrating comprehensive two-dimensional gas chromatography mass spectrometry and parallel two-dimensional liquid chromatography mass spectrometry for untargeted metabolomics. Analyst 2019, 144, 4331–4341. [Google Scholar] [CrossRef]
- Liesenfeld, D.B.; Habermann, N.; Owen, R.W.; Scalbert, A.; Ulrich, C.M. Review of mass spectrometry–based metabolomics in cancer research. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2182–2201. [Google Scholar] [CrossRef]
- Navarro-Reig, M.; Jaumot, J.; Baglai, A.; Vivó-Truyols, G.; Schoenmakers, P.J.; Tauler, R. Untargeted comprehensive two-dimensional liquid chromatography coupled with high-resolution mass spectrometry analysis of rice metabolome using multivariate curve resolution. Anal. Chem. 2017, 89, 7675–7683. [Google Scholar] [CrossRef]
- Shi, X.; Wahlang, B.; Wei, X.L.; Yin, X.M.; Falkner, K.C.; Prough, R.A.; Kim, S.H.; Mueller, E.G.; McClain, C.J.; Cave, M.; et al. Metabolomic analysis of the effects of polychlorinated biphenyls in nonalcoholic fatty liver disease. J. Proteome Res. 2012, 11, 3805–3815. [Google Scholar] [CrossRef]
- He, L.Q.; Li, F.Y.; Yin, X.M.; Bohman, P.; Kim, S.; McClain, C.J.; Feng, W.K.; Zhang, X. Profiling of Polar Metabolites in mouse feces using four analytical platforms to study the effects Of cathelicidin-related antimicrobial peptide in alcoholic liver disease. J. Proteome Res. 2019, 18, 2875–2884. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Harder, J.; Ma, J.; Yin, X.; Zhang, X.; Kosiewicz, M.M. Using multiple analytical platforms to investigate the androgen depletion effects on fecal metabolites in a mouse model of systemic lupus erythematosus. J. Proteome Res. 2020, 19, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Pirok, B.W.J.; Stoll, D.R.; Schoenmakers, P.J. Recent developments in two-dimensional liquid chromatography: Fundamental improvements for practical applications. Anal. Chem. 2019, 91, 240–263. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Bellastella, G.; Scappaticcio, L.; Esposito, K.; Giugliano, D.; Maiorino, M.I. Metabolic syndrome and cancer: “The common soil hypothesis”. Diabetes Res. Clin. Pract. 2018, 143, 389–397. [Google Scholar] [CrossRef]
- Micucci, C.; Valli, D.; Matacchione, G.; Catalano, A. Current perspectives between metabolic syndrome and cancer. Oncotarget 2016, 7, 38959–38972. [Google Scholar] [CrossRef]
- Luo, Z.J.; Saha, A.K.; Xiang, X.Q.; Ruderman, N.B. AMPK, the metabolic syndrome and cancer. Trends Pharmacol. Sci. 2005, 26, 69–76. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review 1975–2016; National Cancer Institute: Bethesda, MD, USA, 2019. [Google Scholar]
- Nicholson, B.D.; Shinkins, B.; Pathiraja, I.; Roberts, N.W.; James, T.J.; Mallett, S.; Perera, R.; Primrose, J.N.; Mant, D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015, 2015, CD011134. [Google Scholar] [CrossRef]
- Klavins, K.; Drexler, H.; Hann, S.; Koellensperger, G. Quantitative metabolite profiling utilizing parallel column analysis for simultaneous reversed-phase and hydrophilic interaction liquid chromatography separations combined with tandem mass spectrometry. Anal. Chem. 2014, 86, 4145–4150. [Google Scholar] [CrossRef]
- Kim, S.; Yin, X.; Prodhan, M.A.I.; Zhang, X.; Zhong, Z.; Kato, I. Global plasma profiling for colorectal cancer-associated volatile organic compounds: A proof-of-principle study. J. Chromatogr. Sci. 2019, 57, 385–396. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, C.; Gui, Q.; Chen, Y.; Yang, Y. Ultraperformance liquid chromatography coupled with quadrupole timeofflight mass spectrometrybased metabolic profiling of human serum prior to and following radical resection of colorectal carcinoma. Mol. Med. Rep. 2015, 12, 6879–6886. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hashim, N.A.A.; Ab-Rahim, S.; Suddin, L.S.; Saman, M.S.A.; Mazlan, M. Global serum metabolomics profiling of colorectal cancer. Mol. Clin. Oncol. 2019, 11, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Sanchez-Espiridion, B.; Lin, M.B.; White, L.; Mishra, L.; Raju, G.S.; Kopetz, S.; Eng, C.; Hildebrandt, M.A.T.; Chang, D.W.; et al. Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer 2017, 123, 4066–4074. [Google Scholar] [CrossRef] [PubMed]
- Zamani, Z.; Arjmand, M.; Vahabi, F.; Eshaq Hosseini, S.M.; Fazeli, S.M.; Iravani, A.; Bayat, P.; Oghalayee, A.; Mehrabanfar, M.; Haj Hosseini, R.; et al. A metabolic study on colon cancer using 1h nuclear magnetic resonance spectroscopy. Biochem. Res. Int. 2014, 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Xiang, Y.B.; Rothman, N.; Yu, D.; Li, H.L.; Yang, G.; Cai, H.; Ma, X.; Lan, Q.; Gao, Y.T.; et al. Prospective study of blood metabolites associated with colorectal cancer risk. Int. J. Cancer 2018, 143, 527–534. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Cui, B.B.; Zhang, F.; Yang, Y.; Shen, X.T.; Li, Z.; Zhao, W.W.; Zhang, Y.Y.; Deng, K.; Rong, Z.W.; et al. Development of a correlative strategy to discover colorectal tumor tissue derived metabolite biomarkers in plasma using untargeted metabolomics. Anal. Chem. 2019, 91, 2401–2408. [Google Scholar] [CrossRef]
- Zaimenko, I.; Jaeger, C.; Brenner, H.; Chang-Claude, J.; Hoffmeister, M.; Grotzinger, C.; Detjen, K.; Burock, S.; Schmitt, C.A.; Stein, U.; et al. Non-invasive metastasis prognosis from plasma metabolites in stage II colorectal cancer patients: The DACHS study. Int. J. Cancer 2019, 145, 221–231. [Google Scholar] [CrossRef]
- Geijsen, A.J.M.R.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef]
- Deng, L.L.; Gu, H.W.; Zhu, J.J.; Gowda, G.A.N.; Djukovic, D.; Chiorean, E.G.; Raftery, D. Combining NMR and LC/MS using backward variable elimination: Metabolomics analysis of colorectal cancer, polyps and healthy controls. Anal. Chem. 2016, 88, 7975–7983. [Google Scholar] [CrossRef]
- Tan, B.B.; Qiu, Y.P.; Zou, X.; Chen, T.L.; Xie, G.X.; Cheng, Y.; Dong, T.T.; Zhao, L.J.; Feng, B.; Hu, X.F.; et al. Metabonomics Identifies Serum Metabolite Markers of Colorectal Cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef]
- Qiu, Y.P.; Cai, G.X.; Su, M.M.; Chen, T.L.; Zheng, X.J.; Xu, Y.; Ni, Y.; Zhao, A.H.; Xu, L.X.; Cai, S.J.; et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 2009, 8, 4844–4850. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.J.; Moore, S.C.; Boca, S.; Huang, W.Y.; Xiong, X.Q.; Stolzenberg-Solomon, R.; Sinha, R.; Sampson, J.N. A prospective study of serum metabolites and colorectal cancer risk. Cancer 2014, 120, 3049–3057. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Yagi, N.; Mizushima, K.; Higashimura, Y.; Hirai, Y.; Okayama, T.; Yoshida, N.; Katada, K.; Kamada, K.; Handa, O.; et al. Serum metabolomics analysis for early detection of colorectal cancer. J. Gastroenterol. 2017, 52, 677–694. [Google Scholar] [CrossRef]
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yamada, Y.; Sudo, K.; Yamaji, T.; Saito, Y.; et al. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115–17126. [Google Scholar] [CrossRef]
- Ma, Y.L.; Liu, W.J.; Peng, J.Y.; Huang, L.; Zhang, P.; Zhao, X.P.; Cheng, Y.Y.; Qin, H.L. A pilot study of gas chromatograph/mass spectrometry-based serum metabolic profiling of colorectal cancer after operation. Mol. Biol. Rep. 2010, 37, 1403–1411. [Google Scholar] [CrossRef]
- Ritchie, S.A.; Ahiahonu, P.W.K.; Jayasinghe, D.; Heath, D.; Liu, J.; Lu, Y.S.; Jin, W.; Kavianpour, A.; Yamazaki, Y.; Khan, A.M.; et al. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: Implications for early screening and detection. BMC Med. 2010, 8, 13. [Google Scholar] [CrossRef]
- Phua, L.C.; Chue, X.P.; Koh, P.K.; Cheah, P.Y.; Ho, H.K.; Chan, E.C.Y. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol. Ther. 2014, 15, 389–397. [Google Scholar] [CrossRef]
- Lin, Y.; Ma, C.C.; Bezabeh, T.; Wang, Z.N.; Liang, J.H.; Huang, Y.; Zhao, J.Y.; Liu, X.M.; Ye, W.; Tang, W.; et al. 1H NMR-based metabolomics reveal overlapping discriminatory metabolites and metabolic pathway disturbances between colorectal tumor tissues and fecal samples. Int. J. Cancer 2019, 145, 1679–1689. [Google Scholar] [CrossRef]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma-deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72. [Google Scholar] [CrossRef]
- Chan, E.C.Y.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa and stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.P.; Cai, G.X.; Su, M.M.; Chen, T.L.; Liu, Y.M.; Xu, Y.; Ni, Y.; Zhao, A.H.; Cai, S.J.; Xu, L.X.; et al. Urinary metabonomic study on colorectal cancer. J. Proteome Res. 2010, 9, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xie, G.X.; Chen, T.L.; Qiu, Y.P.; Zou, X.; Zheng, M.H.; Tan, B.B.; Feng, B.; Dong, T.T.; He, P.A.; et al. Distinct urinary metabolic profile of human colorectal cancer. J. Proteome Res. 2012, 11, 1354–1363. [Google Scholar] [CrossRef]
- Di Lena, M.; Porcelli, F.; Altomare, D.F. Volatile organic compounds as new biomarkers for colorectal cancer: A review. Colorectal Dis. 2016, 18, 654–663. [Google Scholar] [CrossRef]
- Kyle, J.E.; Casey, C.P.; Stratton, K.G.; Zink, E.M.; Kim, Y.M.; Zheng, X.Y.; Monroe, M.E.; Weitz, K.K.; Bloodsworth, K.J.; Orton, D.J.; et al. Comparing identified and statistically significant lipids and polar metabolites in 15-year old serum and dried blood spot samples for longitudinal studies. Rapid Commun. Mass Spectrom. 2017, 31, 447–456. [Google Scholar] [CrossRef]
- Anne, L. (Ed.) Glutamine metabolism in cancer. In The Heterogeneity of Cancer Metabolism; Springer International Publishing: Cham, Swizerland, 2018; pp. 13–32. [Google Scholar]
- Amelio, I.; Cutruzzola, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Budczies, J.; Brockmoller, S.F.; Muller, B.M.; Barupal, D.K.; Richter-Ehrenstein, C.; Kleine-Tebbe, A.; Griffin, J.L.; Oresic, M.; Dietel, M.; Denkert, C.; et al. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: Alterations in glutamine and beta-alanine metabolism. J. Proteom. 2013, 94, 279–288. [Google Scholar] [CrossRef]
- Hutschenreuther, A.; Birkenmeier, G.; Bigl, M.; Krohn, K.; Birkemeyer, C. Glycerophosphoglycerol, beta-alanine and pantothenic acid as metabolic companions of glycolytic activity and cell migration in breast cancer cell lines. Metabolites 2013, 3, 1084–1101. [Google Scholar] [CrossRef]
- Dai, X.F.; Luo, Y.N.; Xu, Y.; Zhang, J.Y. Key indexes and the emerging tool for tumor microenvironment editing. Am. J. Cancer Res. 2019, 9, 1027–1042. [Google Scholar]
- Bhowmik, S.K.; Ramirez-Pena, E.; Arnold, J.M.; Putluri, V.; Sphyris, N.; Michailidis, G.; Putluri, N.; Ambs, S.; Sreekumar, A.; Mani, S.A. EMT-induced metabolite signature identifies poor clinical outcome. Oncotarget 2015, 6, 42651–42660. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, M.P.; Cai, Q.P.; Ziegler, T.R.; Liu, K.H.; Tran, P.H.; Voss, M.B.; Martin, G.S.; Jones, D.P.; Yu, T.W.; Alvarez, J.A. Plasma high-resolution metabolomics differentiates adults with normal weight obesity from lean individuals. Obesity 2019, 27, 1729–1737. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.K.; Yeh, T.S.; Wu, R.C.; Lai, Y.C.; Chiang, M.H.; Lu, K.Y.; Hung, C.Y.; Ho, H.Y.; Cheng, M.L.; Lin, G.G. Metabolomic alterations and chromosomal instability status in gastric cancer. World J. Gastroenterol. 2018, 24, 3760–3769. [Google Scholar] [CrossRef] [PubMed]
- Cano, K.E.; Li, L.; Bhatia, S.; Bhatia, R.; Forman, S.J.; Chen, Y. NMR-based metabolomic analysis of the molecular pathogenesis of therapy-related myelodysplasia/acute myeloid leukemia. J. Proteome Res. 2011, 10, 2873–2881. [Google Scholar] [CrossRef]
- Li, F.; Li, X.; Miao, Y.; Shan, C.; Yuan, F.; Ma, C.; Wang, Q.; Chen, J.; Chen, Y. Uhplc–ms-based metabolomics analysis on mice bearing neoplasm (h22) for hispidulin. J. Pharm. Biomed. Anal. 2016, 125, 310–318. [Google Scholar] [CrossRef]
- Chen, W.Y.; Wu, F.; You, Z.Y.; Zhang, Z.M.; Guo, Y.L.; Zhong, L.X. Analyzing the differentially expressed genes and pathway cross-talk in aggressive breast cancer. J. Obstet. Gynaecol. Res. 2015, 41, 132–140. [Google Scholar] [CrossRef]
- Kim, S.; You, S.; Hwang, D. Aminoacyl-tRNA synthetases and tumorigenesis: More than housekeeping. Nat. Rev. Cancer 2011, 11, 708–718. [Google Scholar] [CrossRef]
- Kim, D.; Kwon, N.H.; Kim, S. Association of Aminoacyl-tRNA Synthetases with Cancer. In Aminoacyl-Trna Synthetases in Biology and Medicine; Springer: Dordrecht, The Netherlands, 2013; pp. 207–245. [Google Scholar] [CrossRef]
- Kushner, J.P.; Boll, D.; Quagliana, J.; Dickman, S. Elevated methionine-trna synthetase activity in human colon cancer. Proc. Soc. Exp. Biol. Med. 1976, 153, 273–276. [Google Scholar] [CrossRef]
- Pavon-Eternod, M.; Gomes, S.; Rosner, M.R.; Pan, T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 2013, 19, 461–466. [Google Scholar] [CrossRef]
- Yao, P.; Fox, P.L. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 2013, 5, 332–343. [Google Scholar] [CrossRef]
- Wei, X.L.; Shi, X.; Koo, I.; Kim, S.; Schmidt, R.H.; Arteel, G.E.; Watson, W.H.; McClain, C.; Zhang, X. MetPP: A computational platform for comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry-based metabolomics. Bioinformatics 2013, 29, 1786–1792. [Google Scholar] [CrossRef] [PubMed]
- Lustgarten, M.S.; Price, L.L.; Phillips, E.M.; Fielding, R.A. Serum Glycine Is Associated with Regional Body Fat and Insulin Resistance in Functionally-Limited Older Adults. PLoS ONE 2013, 8, e84034. [Google Scholar] [CrossRef] [PubMed]
- Palsson-McDermott, E.M.; O’Neill, L.A.J. The Warburg effect then and now: From cancer to inflammatory diseases. Bioessays 2013, 35, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kauppinen, A.; Hiltunen, M.; Kaarniranta, K. Krebs cycle intermediates regulate DNA and histone methylation: Epigenetic impact on the aging process. Ageing Res. Rev. 2014, 16, 45–65. [Google Scholar] [CrossRef]
- Altomare, D.F.; Di Lena, M.; Porcelli, F.; Travaglio, E.; Longobardi, F.; Tutino, M.; Depalma, N.; Tedesco, G.; Sardaro, A.; Memeo, R.; et al. Effects of Curative Colorectal Cancer Surgery on Exhaled Volatile Organic Compounds and Potential Implications in Clinical Follow-up. Ann. Surg. 2015, 262, 862–867. [Google Scholar] [CrossRef]
- Kato, I.; Land, S.; Majumdar, A.P.; Barnholtz-Sloan, J.; Severson, R.K. Functional polymorphisms to modulate luminal lipid exposure and risk of colorectal cancer. Cancer Epidemiol. 2010, 34, 291–297. [Google Scholar] [CrossRef]
- Koo, I.; Shi, X.; Kim, S.; Zhang, X. iMatch2: Compound identification using retention index for analysis of gas chromatography-mass spectrometry data. J. Chromatogr. A 2014, 1337, 202–210. [Google Scholar] [CrossRef]
- Wei, X.L.; Shi, X.; Kim, S.; Patrick, J.S.; Binkley, J.; Kong, M.Y.; McClain, C.; Zhang, X. Data Dependent Peak Model Based Spectrum Deconvolution for Analysis of High Resolution LC-MS Data. Anal. Chem. 2014, 86, 2156–2165. [Google Scholar] [CrossRef]
- Wei, X.L.; Sun, W.L.; Shi, X.; Koo, I.; Wang, B.; Zhang, J.; Yin, X.M.; Tang, Y.N.; Bogdanov, B.; Kim, S.; et al. MetSign: A Computational Platform for High-Resolution Mass Spectrometry-Based Metabolomics. Anal. Chem. 2011, 83, 7668–7675. [Google Scholar] [CrossRef]
- Wei, X.L.; Shi, X.; Kim, S.; Zhang, L.; Patrick, J.S.; Binkey, J.; McClain, C.; Zhang, X. Data Preprocessing Method for Liquid Chromatography-Mass Spectrometry Based Metabolomics. Anal. Chem. 2012, 84, 7963–7971. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
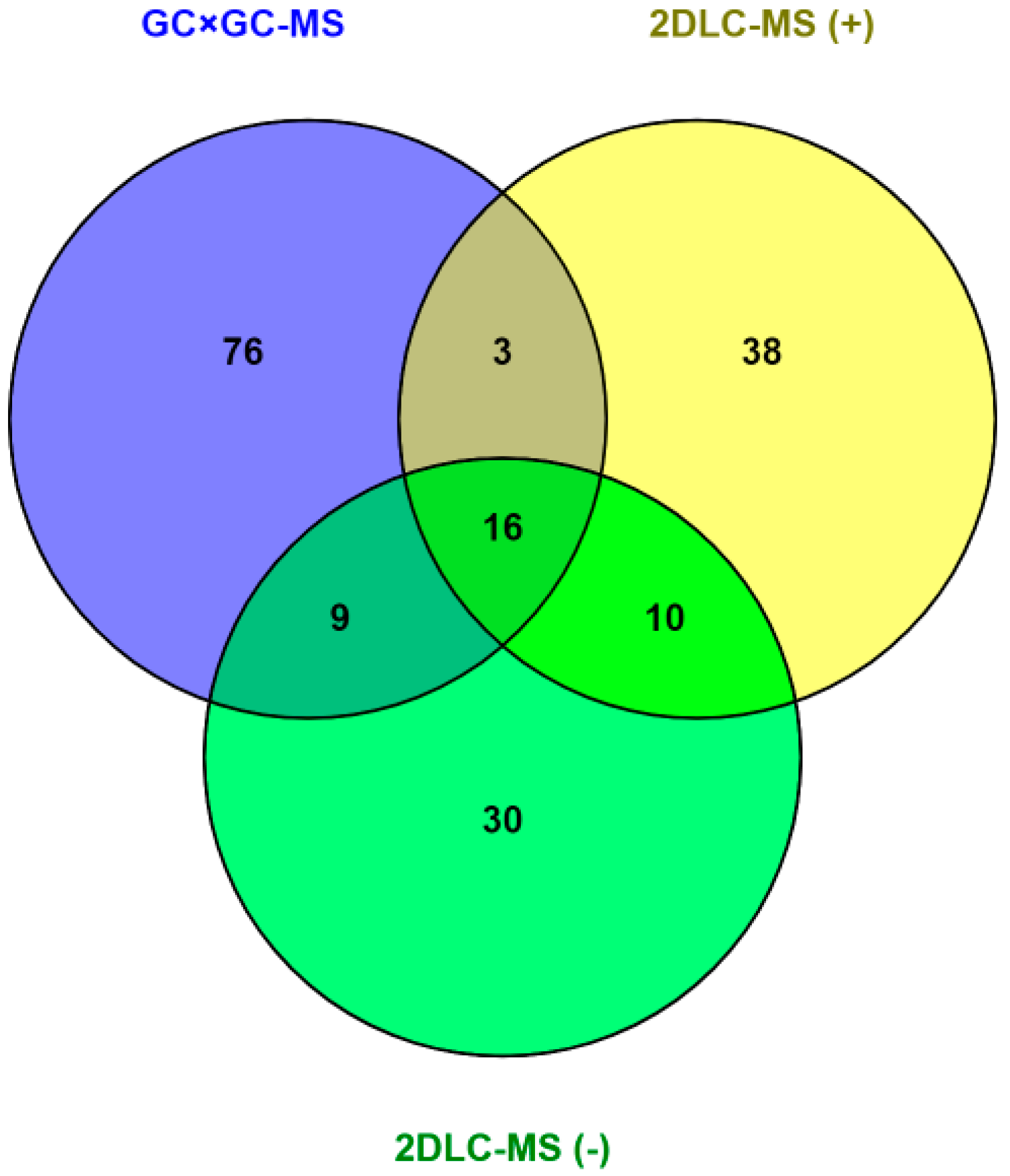
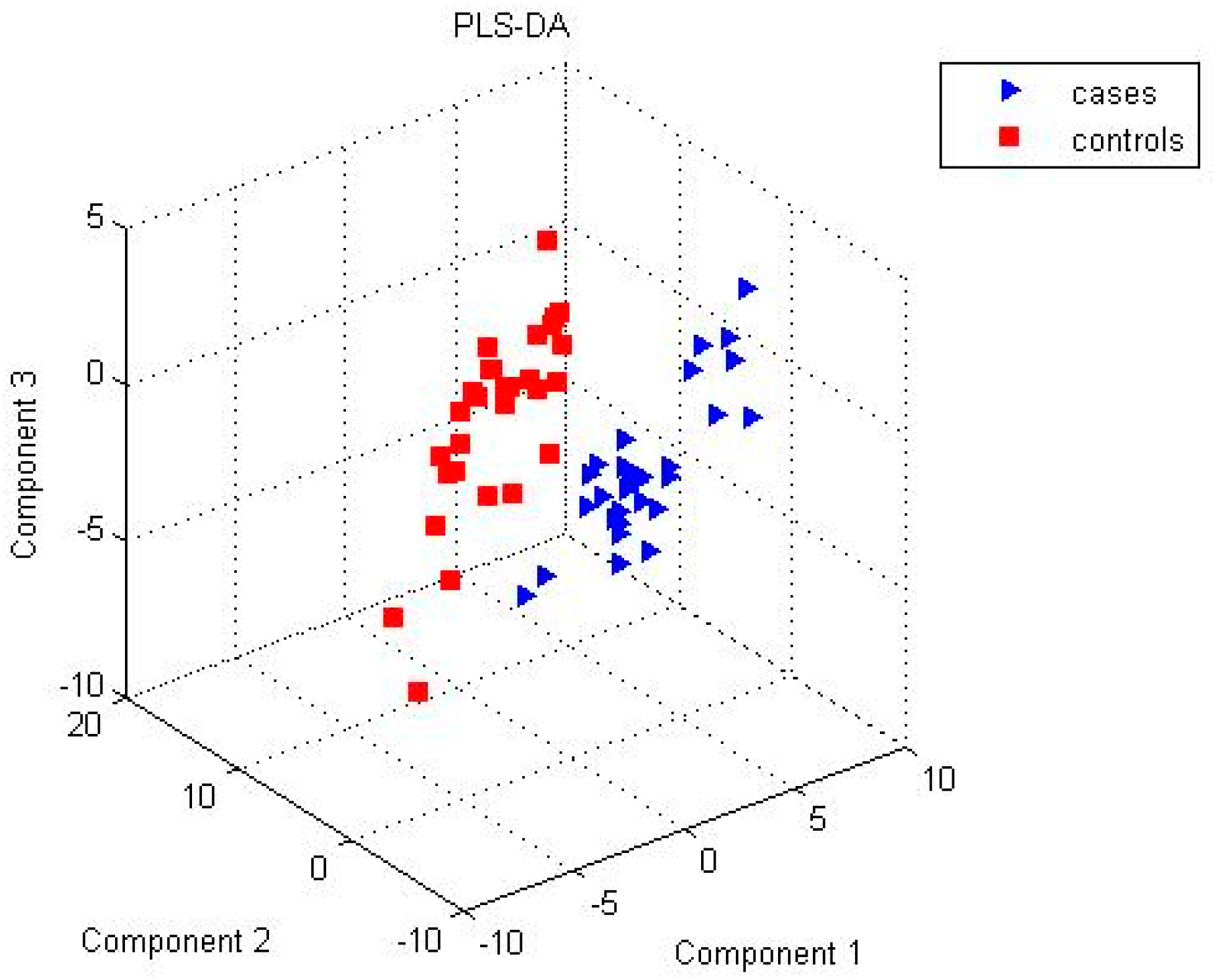
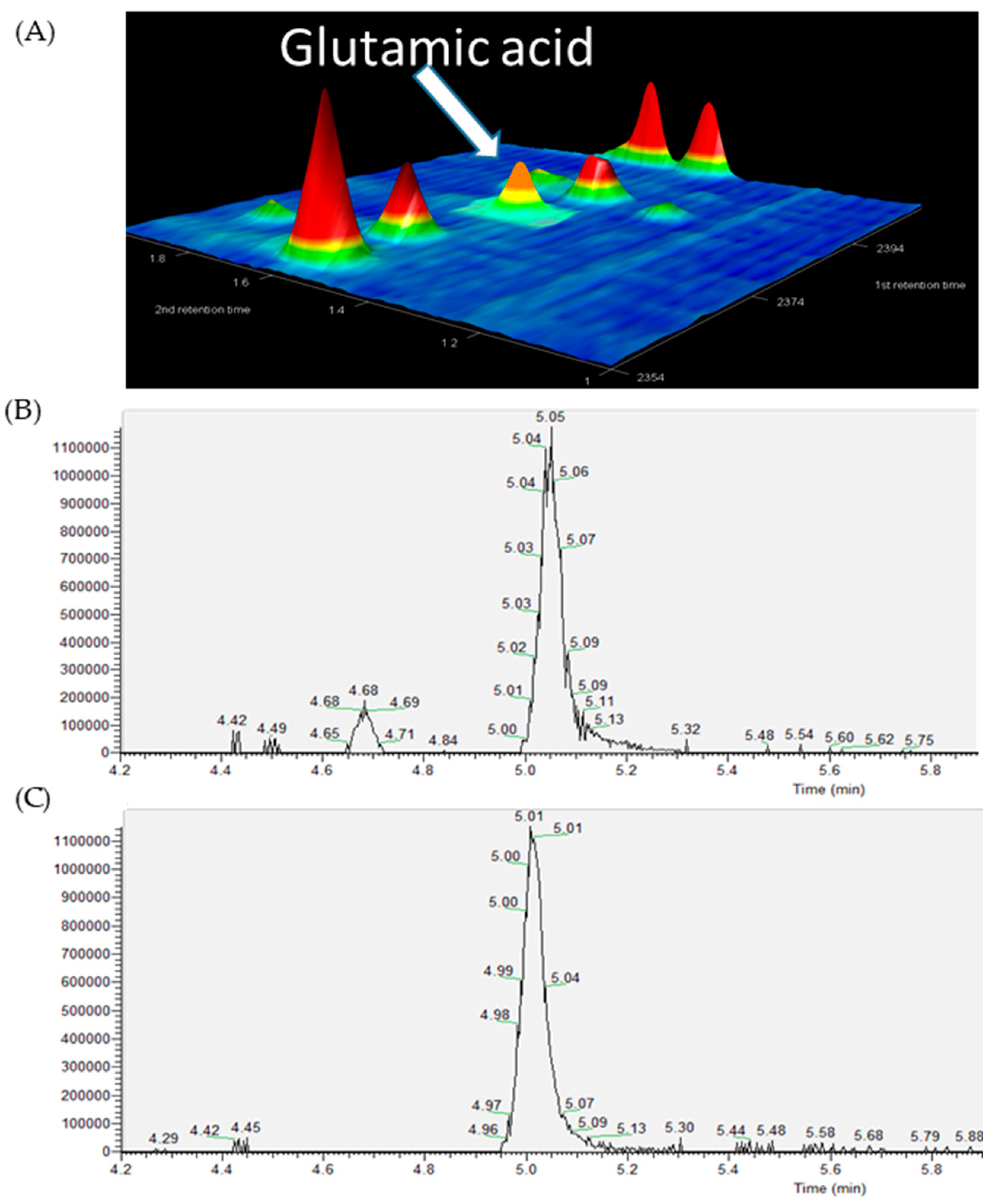
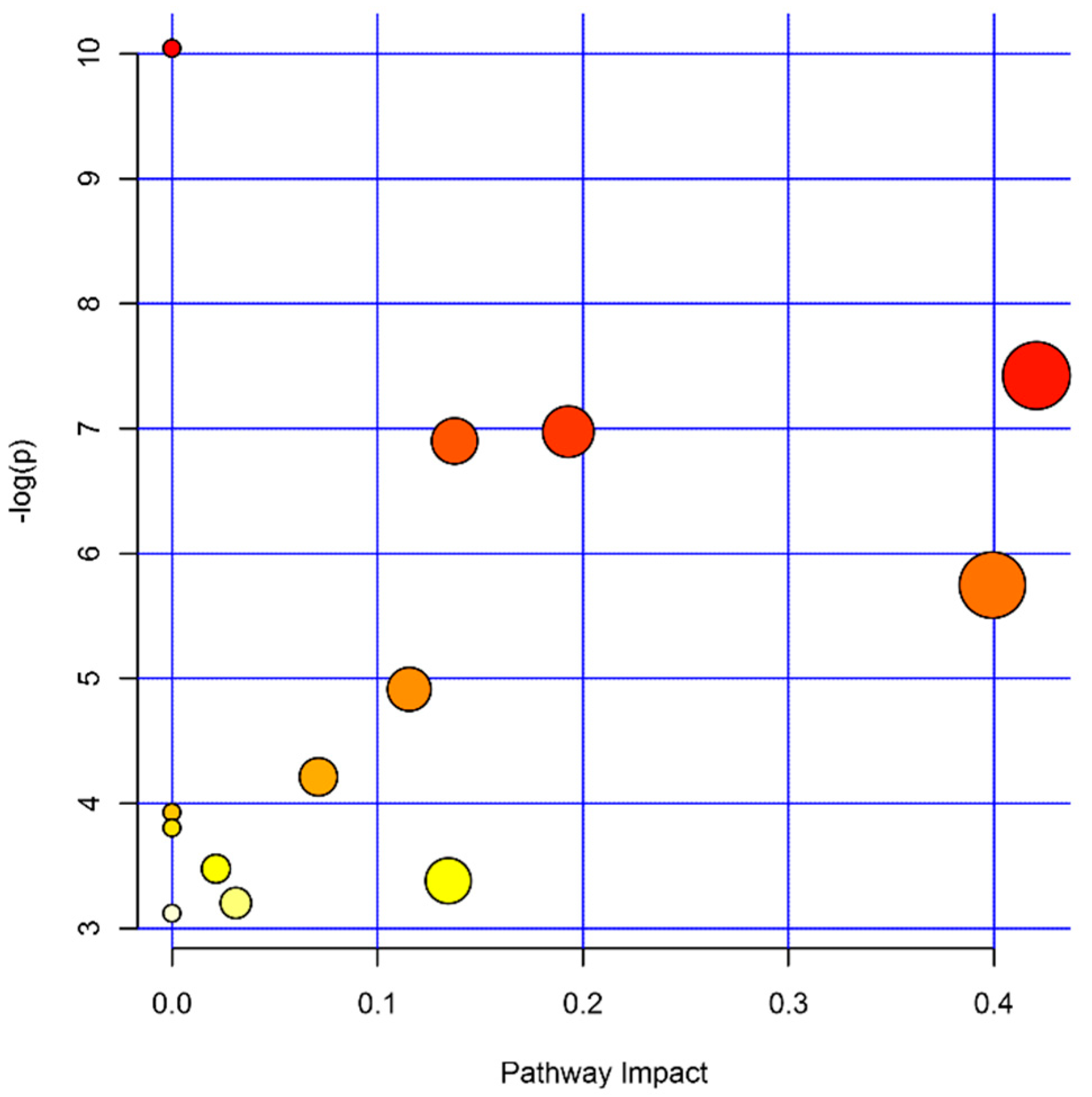
| Name | p-Value | Fold Change (Cases/Controls) | Platform | %FO * | |
|---|---|---|---|---|---|
| Controls n = 29 | Cases n = 29 | ||||
| 3-Hydroxybutyric acid | 9.98 × 10−3 | 2.12 | 2DLC-MS (−) | 96.55 | 96.55 |
| 4-Methyl-2-oxovaleric acid | 4.62 × 10−3 | 0.71 | 2DLC-MS (−) | 100.00 | 100.00 |
| Adonitol | 1.07 × 10−2 | 1.27 | 2DLC-MS (−) | 62.07 | 62.07 |
| Alanine | 4.98 × 10−3 | 0.52 | 2DLC-MS (−) | 100.00 | 100.00 |
| Arginine | 3.83 × 10−2 | 0.78 | 2DLC-MS (+) | 82.76 | 82.76 |
| Aspartic Acid | 5.09 × 10−3 | 0.74 | 2DLC-MS (+) | 79.31 | 79.31 |
| beta-Alanine | 4.15 × 10−2 | 1.76 | GC × GC-MS | 100.00 | 100.00 |
| Choline | 8.56 × 10−3 | 1.19 | 2DLC-MS (+) | 68.97 | 72.41 |
| Citric acid | 2.13 × 10−2 | 1.51 | 2DLC-MS (−) | 100.00 | 100.00 |
| Ethanolamine | 3.17 × 10−2 | 1.31 | 2DLC-MS (+) | 100.00 | 100.00 |
| Glutamic acid | 2.58 × 10−3 | 2.00 | GC × GC-MS | 68.97 | 72.41 |
| Glutaric acid | 4.86 × 10−2 | 1.16 | 2DLC-MS (−) | 96.55 | 96.55 |
| Glycine | 1.44 × 10−3 | 1.34 | 2DLC-MS (+) | 93.10 | 89.66 |
| Hydracrylic acid | 4.32 × 10−2 | 1.28 | GC × GC-MS | 100.00 | 100.00 |
| Lactic acid | 3.11 × 10−2 | 1.13 | GC × GC-MS | 100.00 | 100.00 |
| Lysine | 1.86 × 10−2 | 1.30 | GC × GC-MS | 79.31 | 82.76 |
| Lysophosphatidic acid | 4.57 × 10−2 | 1.23 | 2DLC-MS (+) | 100.00 | 100.00 |
| Malate | 2.09 × 10−2 | 1.37 | 2DLC-MS (−) | 100.00 | 100.00 |
| Malate | 4.91 × 10−2 | 1.51 | GC × GC-MS | 72.41 | 72.41 |
| Neopentyl glycol | 3.24 × 10−2 | 1.16 | GC × GC-MS | 100.00 | 100.00 |
| Oleamide | 3.40 × 10−2 | 1.11 | 2DLC-MS (+) | 72.41 | 72.41 |
| Pinolenic Acid | 2.26 × 10−2 | 0.56 | 2DLC-MS (+) | 72.41 | 75.86 |
| Piperine | 4.29 × 10−2 | 0.63 | 2DLC-MS (+) | 100.00 | 100.00 |
| Propylene glycol | 4.03 × 10−3 | 0.50 | GC × GC-MS | 96.55 | 100.00 |
| Pyroglutamic acid | 1.21 × 10−2 | 1.55 | 2DLC-MS (−) | 72.41 | 72.41 |
| Testosterone sulfate | 2.83 × 10−2 | 0.54 | 2DLC-MS (−) | 100.00 | 100.00 |
| Tricine | 1.81 × 10−2 | 1.60 | GC × GC-MS | 86.21 | 82.76 |
| Uridine | 2.48 × 10−2 | 1.25 | 2DLC-MS (−) | 96.55 | 96.55 |
| Name | Total | Hits | p-Value | Impact |
|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 48 | 6 | 4.35 × 10−5 | 0 |
| Alanine, aspartate, and glutamate metabolism | 28 | 4 | 5.97 × 10−4 | 0.42068 |
| Arginine biosynthesis | 14 | 3 | 9.35 × 10−4 | 0.19289 |
| Glyoxylate and dicarboxylate metabolism | 32 | 4 | 1.01× 10−3 | 0.13757 |
| beta-Alanine metabolism | 21 | 3 | 3.19 × 10−3 | 0.39925 |
| Glutathione metabolism | 28 | 3 | 7.34 × 10−3 | 0.11548 |
| Glycerophospholipid metabolism | 36 | 3 | 1.48 × 10−2 | 0.07130 |
| Butanoate metabolism | 15 | 2 | 1.97 × 10−2 | 0 |
| Histidine metabolism | 16 | 2 | 2.23 × 10−2 | 0 |
| Pantothenate and CoA biosynthesis | 19 | 2 | 3.09 × 10−2 | 0.02143 |
| Citrate cycle (TCA cycle) | 20 | 2 | 3.40 × 10−2 | 0.13450 |
| Pyruvate metabolism | 22 | 2 | 4.06 × 10−2 | 0.03110 |
| Propanoate metabolism | 23 | 2 | 4.41 × 10−2 | 0.00000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, F.; Kim, S.; Yin, X.; Zhang, X.; Kato, I. Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study. Metabolites 2020, 10, 343. https://doi.org/10.3390/metabo10090343
Yuan F, Kim S, Yin X, Zhang X, Kato I. Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study. Metabolites. 2020; 10(9):343. https://doi.org/10.3390/metabo10090343
Chicago/Turabian StyleYuan, Fang, Seongho Kim, Xinmin Yin, Xiang Zhang, and Ikuko Kato. 2020. "Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study" Metabolites 10, no. 9: 343. https://doi.org/10.3390/metabo10090343
APA StyleYuan, F., Kim, S., Yin, X., Zhang, X., & Kato, I. (2020). Integrating Two-Dimensional Gas and Liquid Chromatography-Mass Spectrometry for Untargeted Colorectal Cancer Metabolomics: A Proof-of-Principle Study. Metabolites, 10(9), 343. https://doi.org/10.3390/metabo10090343





