Comparative Metabolomic Analysis of Dendrobium officinale under Different Cultivation Substrates
Abstract
1. Introduction
2. Results
2.1. Data Evaluation
2.2. Widely Targeted Metabolomics Profiling Results
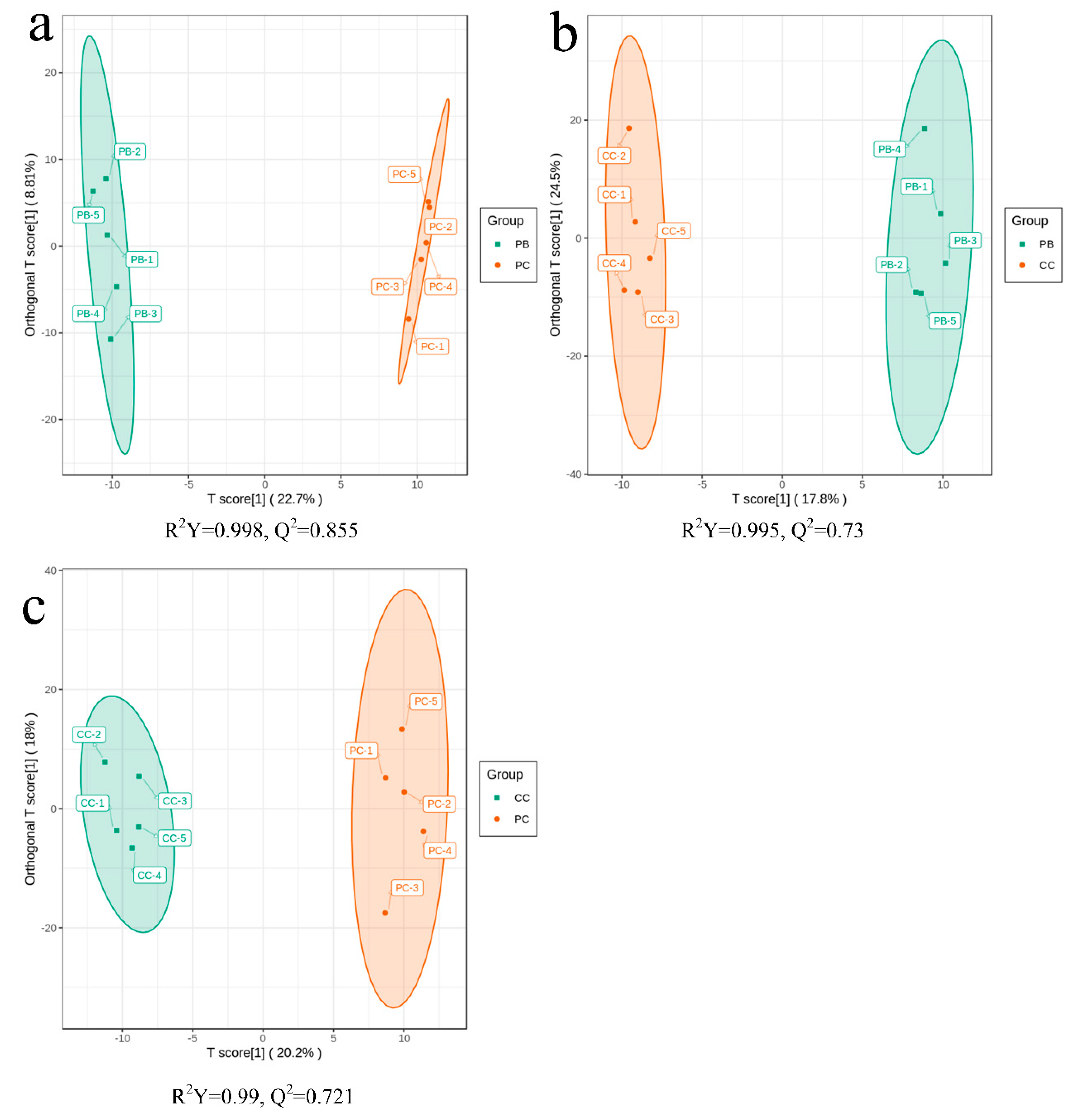
2.3. Principal Component Analysis
2.4. Orthogonal Partial Least Squares Discriminant Analysis
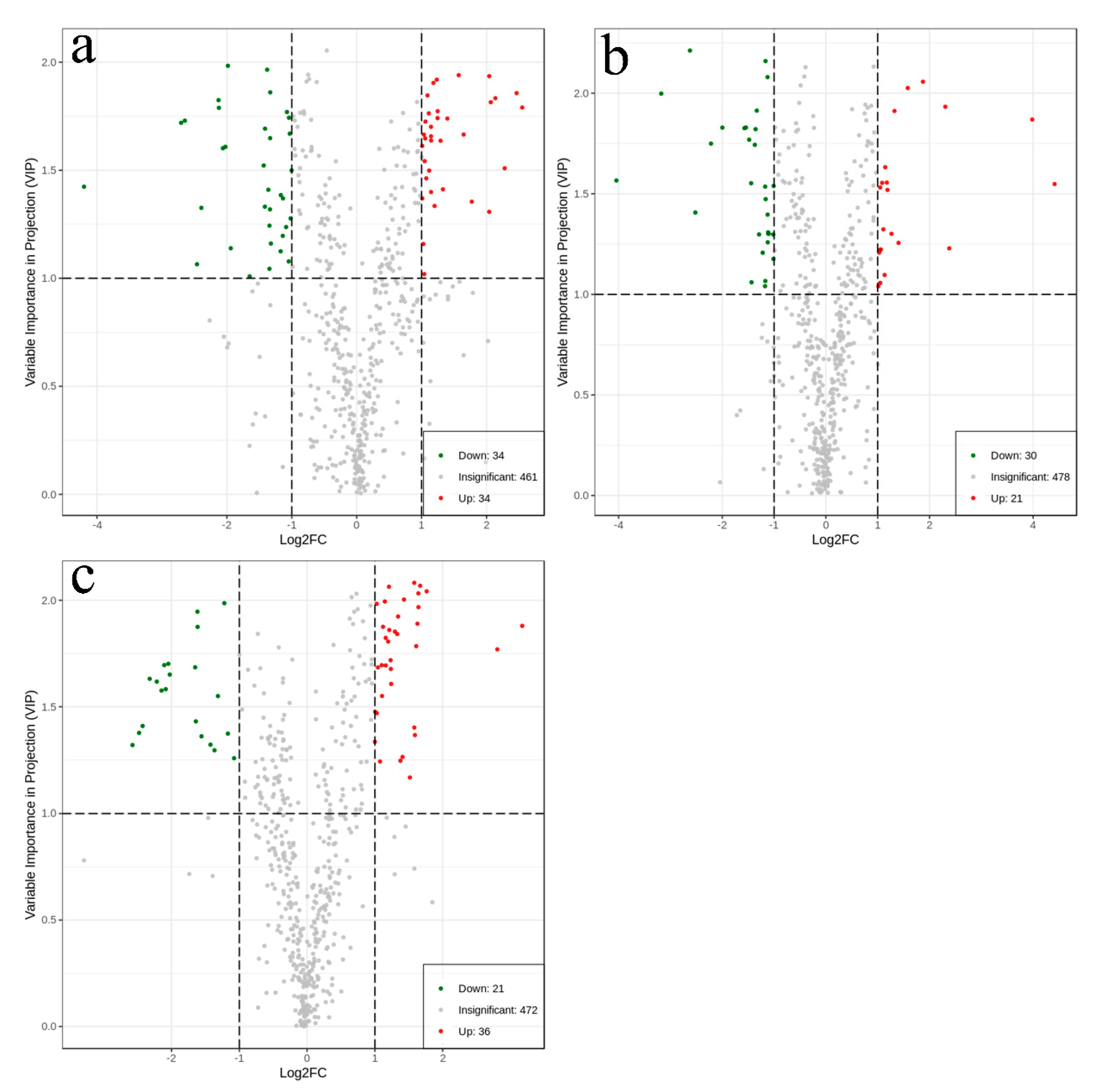
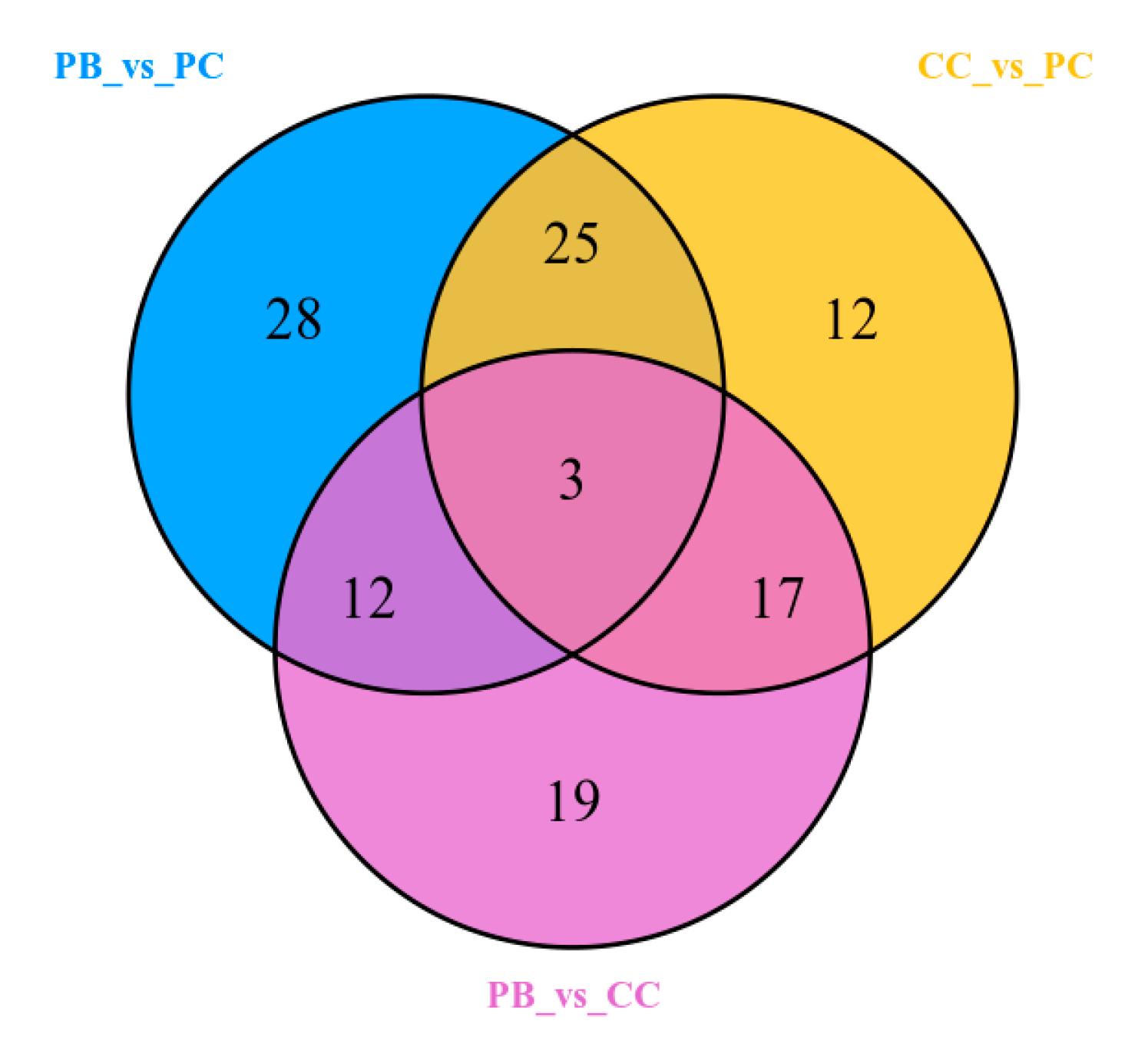
2.5. Identification of Differential Metabolites
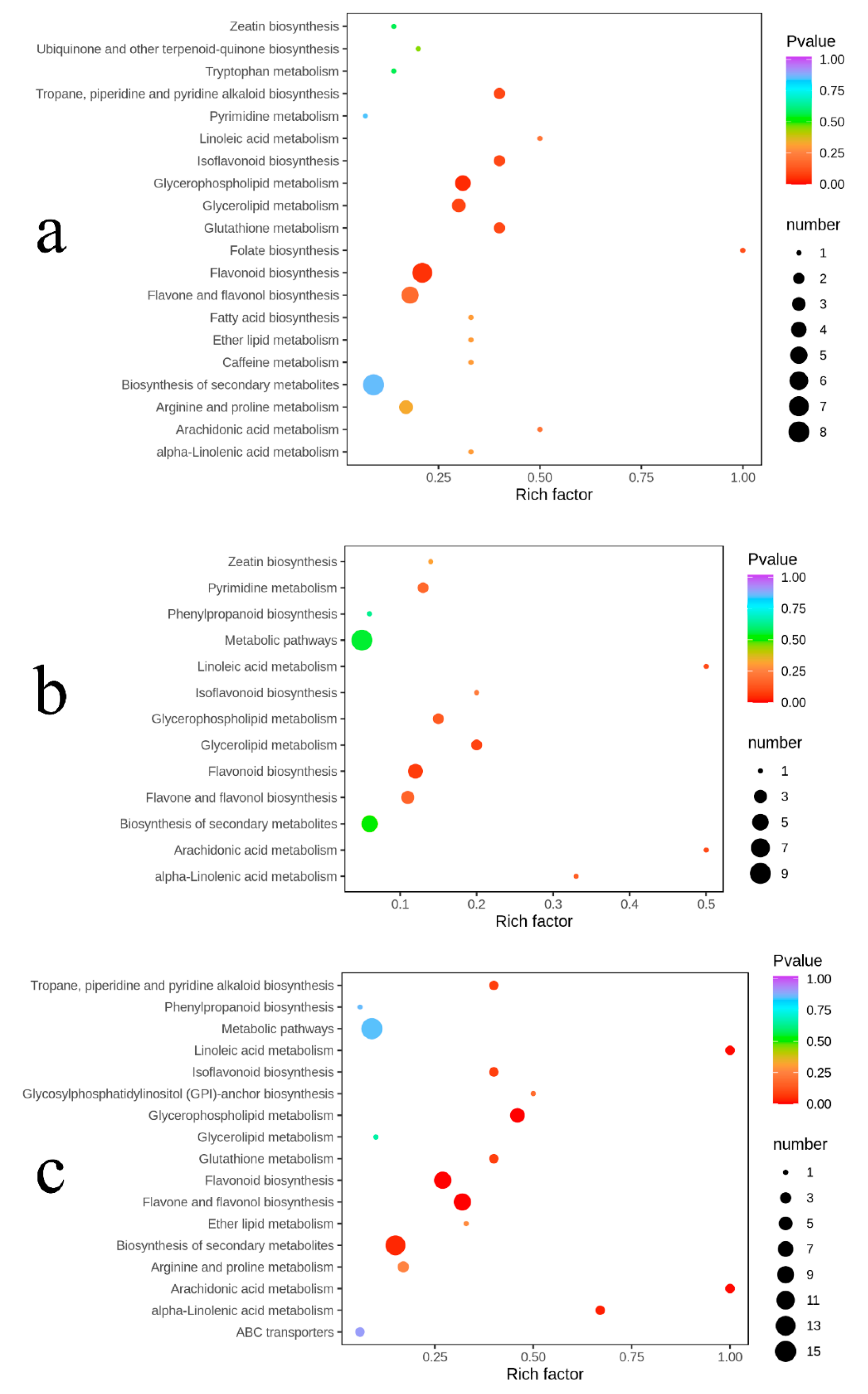
2.6. Pathway Enrichment Analysis of Differential Metabolites
3. Discussion
4. Materials and Methods
4.1. Plant Samples and Chemicals
4.2. Sample Preparation and Extraction
4.3. UPLC-MS/MS Analysis
4.4. Qualitative and Quantitative Analysis of Metabolites
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Wing Sze, S.C.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; He, R.; Yang, S.; Chen, Z.; Jiang, M.; Lu, J.; Wang, H. Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of Dendrobium species. Gene 2015, 567, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Lau, D.T.; Shaw, P.C.; Wang, J.; But, P.P. Authentication of medicinal Dendrobium species by the internal transcribed spacer of ribosomal DNA. Planta Med. 2001, 67, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Moretti, M.; Cossignani, L.; Messina, F.; Dominici, L.; Villarini, M.; Curini, M.; Marcotullio, M.C. Antigenotoxic effect, composition and antioxidant activity of Dendrobium speciosum. Food Chem. 2013, 140, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Li, Y.; Tao, S.; Wei, G.; Huang, Y.; Chen, D.; Wu, C. Purification, Characterization and Biological Activity of Polysaccharides from Dendrobium officinale. Molecules 2016, 21, 701. [Google Scholar] [CrossRef]
- Liang, J.; Li, H.; Chen, J.; He, L.; Du, X.; Zhou, L.; Xiong, Q.; Lai, X.; Yang, Y.; Huang, S.; et al. Dendrobium officinale polysaccharides alleviate colon tumorigenesis via restoring intestinal barrier function and enhancing anti-tumor immune response. Pharmacol. Res. 2019, 148, 104417. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Nie, S.P.; Huang, X.J.; Hu, J.L.; Cui, S.W.; Xie, M.Y.; Phillips, G.O. Study on Dendrobium officinale O-Acetyl-glucomannan (Dendronan). 7. Improving Effects on Colonic Health of Mice. J. Agric. Food Chem. 2016, 64, 2485–2491. [Google Scholar] [CrossRef]
- Chen, H.; Nie, Q.; Hu, J.; Huang, X.; Zhang, K.; Nie, S. Glucomannans Alleviated the Progression of Diabetic Kidney Disease by Improving Kidney Metabolic Disturbance. Mol. Nutr. Food Res. 2019, 63, 1801008. [Google Scholar] [CrossRef]
- Zheng, H.; Pan, L.; Xu, P.; Zhu, J.; Wang, R.; Zhu, W.; Hu, Y.; Gao, H. An NMR-Based Metabolomic Approach to Unravel the Preventive Effect of Water-Soluble Extract from Dendrobium officinale Kimura & Migo on Streptozotocin-Induced Diabetes in Mice. Molecules 2017, 22, 1543. [Google Scholar] [CrossRef]
- Tang, H.; Zhao, T.; Sheng, Y.; Zheng, T.; Fu, L.; Zhang, Y. Dendrobium officinale Kimura et Migo: A Review on Its Ethnopharmacology, Phytochemistry, Pharmacology, and Industrialization. Evid. Based Complementary Altern. Med. 2017, 2017, 7436259. [Google Scholar] [CrossRef]
- Si, J.P.; Yu, Q.X.; Song, X.S.; Shao, W.J. Artificial cultivation modes for Dendrobium officinale. China J. Chin. Mater. Med. 2013, 38, 481–484. [Google Scholar]
- Fan, O.; Lin, Z.; Dong, W.B.; Wang, C.Z. Research progress on the rapid propagation of Dendrobium officinale. J. Food Saf. Qual. 2015, 11, 4378–4383. [Google Scholar]
- Lekala, C.S.; Madani, K.S.H.; Phan, A.D.T.; Maboko, M.M.; Fotouo, H.; Soundy, P.; Sultanbawa, Y.; Sivakumar, D. Cultivar-specific responses in red sweet peppers grown under shade nets and controlled-temperature plastic tunnel environment on antioxidant constituents at harvest. Food Chem. 2019, 275, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.Y.; Liang, Y.M.; Liu, H.Z.; Zhu, D.M.; Hou, S.Z.; Wu, Y.Y.; Huang, S.; Lai, X.P. Effect of Dendrobium officinale on D-galactose-induced aging mice. Chin. J. Integr. Med. 2017, 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kroslakova, I.; Pedrussio, S.; Wolfram, E. Direct Coupling of HPTLC with MALDI-TOF MS for Qualitative Detection of Flavonoids on Phytochemical Fingerprints. Phytochem. Anal. 2016, 27, 222–228. [Google Scholar] [CrossRef]
- Meng, Q.; Fan, H.; Li, Y.; Zhang, L. Effect of drying methods on physico-chemical properties and antioxidant activity of Dendrobium officinale. J. Food Meas. Charact. 2017, 12, 1–10. [Google Scholar] [CrossRef]
- Zhang, X.L.; Si, J.P.; Wu, L.S.; Guo, Y.Y.; Yu, J.; Wang, L.H. Field experiment of F1 generation and superior families selection of Dendrobium officinale. China J. Chin. Mater. Med. 2013, 38, 3861–3865. [Google Scholar]
- Si, J.P.; He, B.W.; Yu, Q.X. Progress and countermeasures of Dendrobium officinale breeding. China J. Chin. Mater. Med. 2013, 38, 475–480. [Google Scholar]
- Ma, Y.; Hou, Z.; Liang, Z.; Liu, L. Quality Evaluation of the Rare Medicinal Plant Dendrobium officinale Based on Volatile Constituents, Methanol Extracts and Polysaccharides. Curr. Pharm. Anal. 2018, 14, 121–132. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Gao, B.; Qian, Z.; Liu, J.; Wu, S.; Si, J. Identification and quantitative analysis of phenolic glycosides with antioxidant activity in methanolic extract of Dendrobium catenatum flowers and selection of quality control herb-markers. Food Res. Int. 2019, 123, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Sawada, Y.; Akiyama, K.; Sakata, A.; Kuwahara, A.; Otsuki, H.; Sakurai, T.; Saito, K.; Hirai, M.Y. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 2009, 50, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X.; Zhang, H.; Dong, H.; et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014, 46, 714–721. [Google Scholar] [CrossRef]
- Qin, Z.; Liao, D.; Chen, Y.; Zhang, C.; An, R.; Zeng, Q.; Li, X. A Widely Metabolomic Analysis Revealed Metabolic Alterations of Epimedium Pubescens Leaves at Different Growth Stages. Molecules 2019, 25. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Z.; Zeng, Q.; Wang, S.; Luo, Y.; Huang, Y.; Xin, Y.; He, N. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 2020, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Tang, X.; Jia, Z.; Li, C.; Forests, J.Z.J. The Effects of Ecological Factors on the Main Medicinal Components of Dendrobium officinale under Different Cultivation Modes. Forests 2020, 11, 94. [Google Scholar] [CrossRef]
- Yu, M.M.; Wu, L.S.; Si, J.P.; Liu, J.J.; Yu, Z.X.; Wang, H.C.; Zhang, M.; Rong, S. Screening of meaningful endophytic fungi in Dendrobium officinale based on polysaccharides and flavonoids. China J. Chin. Mater. Med. 2016, 41, 2208–2212. [Google Scholar] [CrossRef]
- Cao, H.; Ji, Y.; Li, S.; Lu, L.; Tian, M.; Yang, W.; Li, H. Extensive Metabolic Profiles of Leaves and Stems from the Medicinal Plant Dendrobium officinale Kimura et Migo. Metabolites 2019, 9, 215. [Google Scholar] [CrossRef]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Kohl, M. Introduction to Statistical Data Analysis with R; Bookboon: London, UK, 2015; p. 228. [Google Scholar]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ying, H.; Pingcuo, G.; Wang, S.; Zhao, F.; Cui, Y.; Shi, J.; Zeng, H.; Zeng, X. Identification of Potential Metabolites Mediating Bird’s Selective Feeding on Prunus mira Flowers. BioMed Res. Int. 2019, 2019, 1395480. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]


| Group | Average Pearson’s Correlation Coefficient |
|---|---|
| PB vs. PB | 0.9403 |
| PC vs. PC | 0.9422 |
| CC vs. CC | 0.9519 |
| PB vs. PC | 0.8636 |
| PB vs. CC | 0.9078 |
| PC vs. CC | 0.9108 |
| Group | PB vs. PC | PB vs. CC | PC vs. CC | |||
|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |
| Flavonoids (53/135) | 11 | 21 | 4 | 17 | 12 | 11 |
| Phenolic acid (16/75) | 3 | 5 | 3 | 6 | 5 | 3 |
| Lipids (22/72) | 13 | 3 | 4 | 2 | 13 | 1 |
| Amino acids and derivatives (3/64) | 0 | 0 | 3 | 0 | 0 | 0 |
| Alkaloids (7/41) | 4 | 0 | 0 | 3 | 4 | 0 |
| Nucleotides and derivatives (3/37) | 1 | 1 | 2 | 0 | 0 | 0 |
| Organic acids (3/34) | 1 | 0 | 2 | 1 | 1 | 1 |
| Lignans and Coumarins (6/17) | 0 | 3 | 2 | 1 | 0 | 4 |
| Quinones (1/2) | 0 | 1 | 0 | 0 | 0 | 1 |
| Others (2/49) | 1 | 0 | 1 | 0 | 1 | 0 |
| Significantly differential metabolites | 34 | 34 | 21 | 30 | 36 | 21 |
| Total metabolites that differed significantly in content (116) | 68 | 51 | 57 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, S.-M.; Yu, H.-D.; Zhang, W.; Zhong, Q.; Chen, W.; Chen, W.; Yun, Y.-H.; Chen, H. Comparative Metabolomic Analysis of Dendrobium officinale under Different Cultivation Substrates. Metabolites 2020, 10, 325. https://doi.org/10.3390/metabo10080325
Zuo S-M, Yu H-D, Zhang W, Zhong Q, Chen W, Chen W, Yun Y-H, Chen H. Comparative Metabolomic Analysis of Dendrobium officinale under Different Cultivation Substrates. Metabolites. 2020; 10(8):325. https://doi.org/10.3390/metabo10080325
Chicago/Turabian StyleZuo, Si-Min, Hai-Dong Yu, Weimin Zhang, Qiuping Zhong, Wenxue Chen, Weijun Chen, Yong-Huan Yun, and Haiming Chen. 2020. "Comparative Metabolomic Analysis of Dendrobium officinale under Different Cultivation Substrates" Metabolites 10, no. 8: 325. https://doi.org/10.3390/metabo10080325
APA StyleZuo, S.-M., Yu, H.-D., Zhang, W., Zhong, Q., Chen, W., Chen, W., Yun, Y.-H., & Chen, H. (2020). Comparative Metabolomic Analysis of Dendrobium officinale under Different Cultivation Substrates. Metabolites, 10(8), 325. https://doi.org/10.3390/metabo10080325







